Abstract
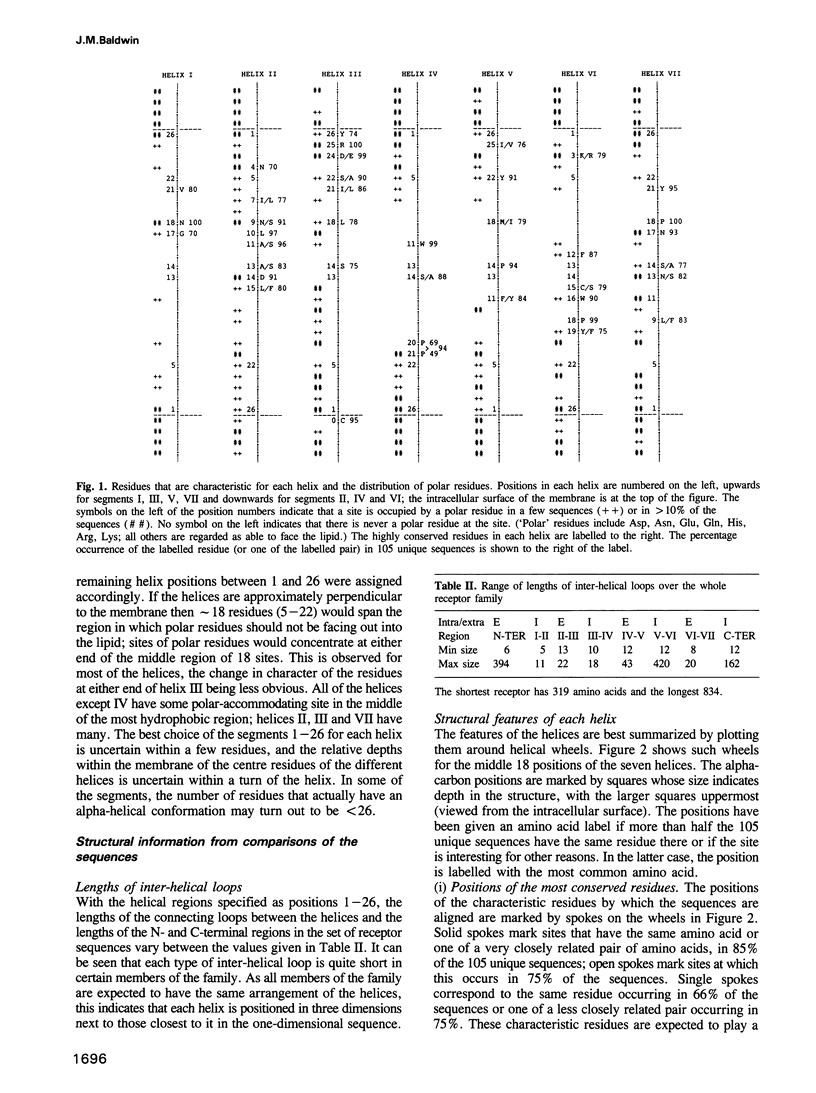
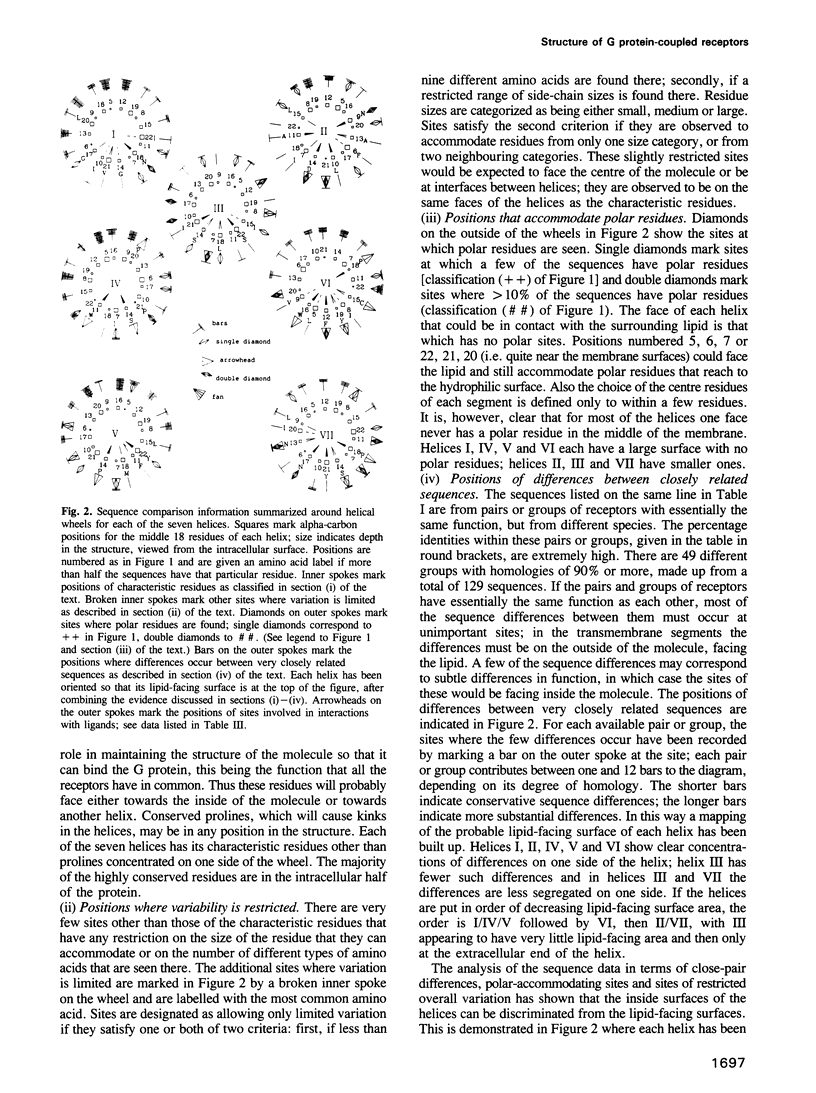
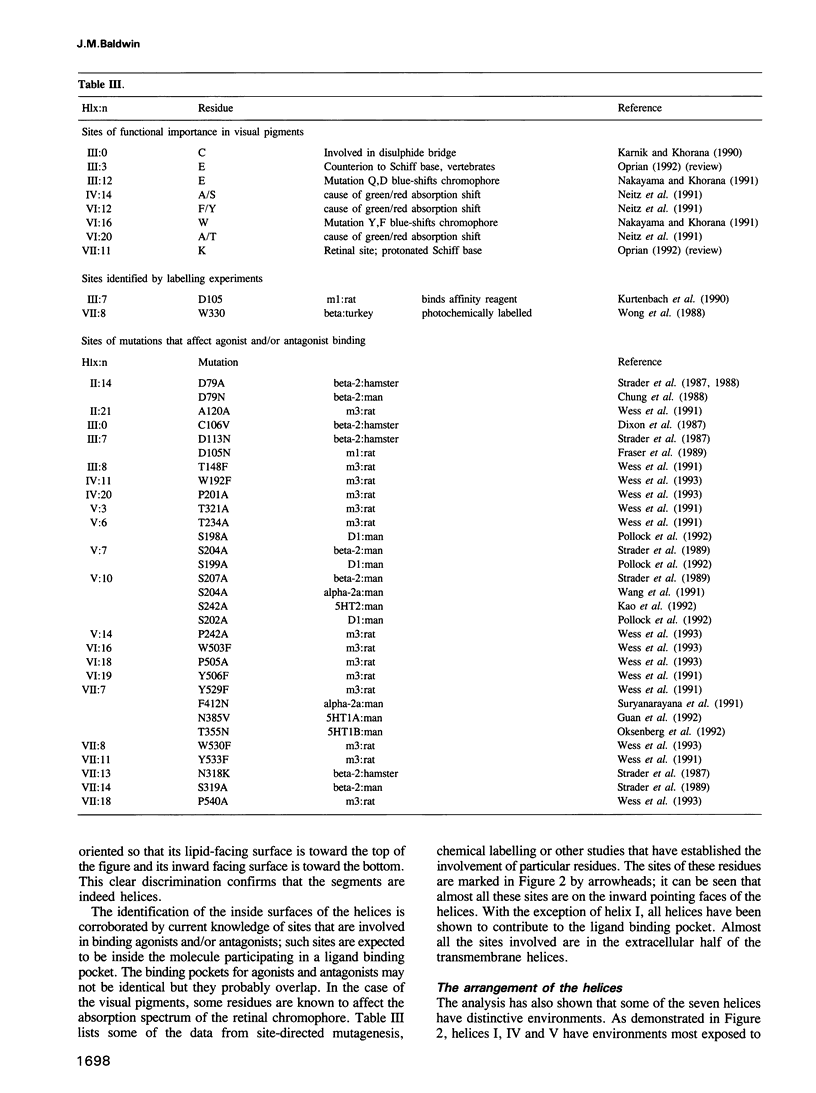
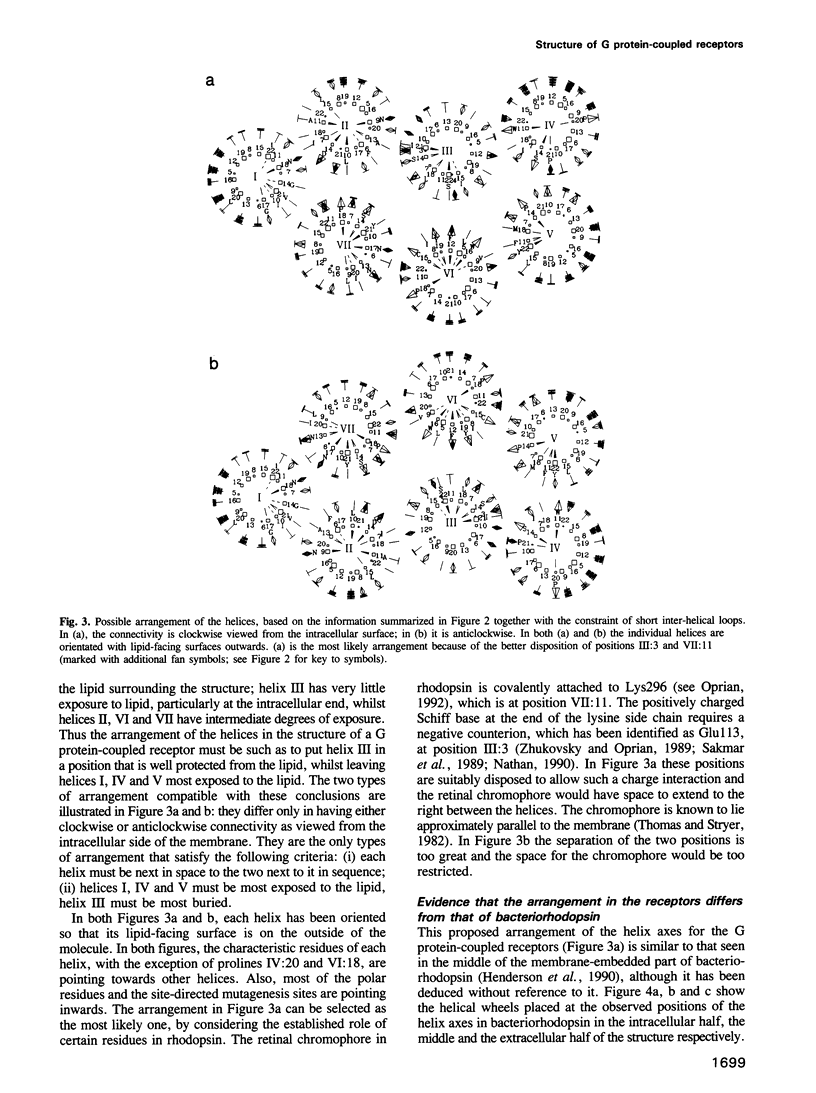
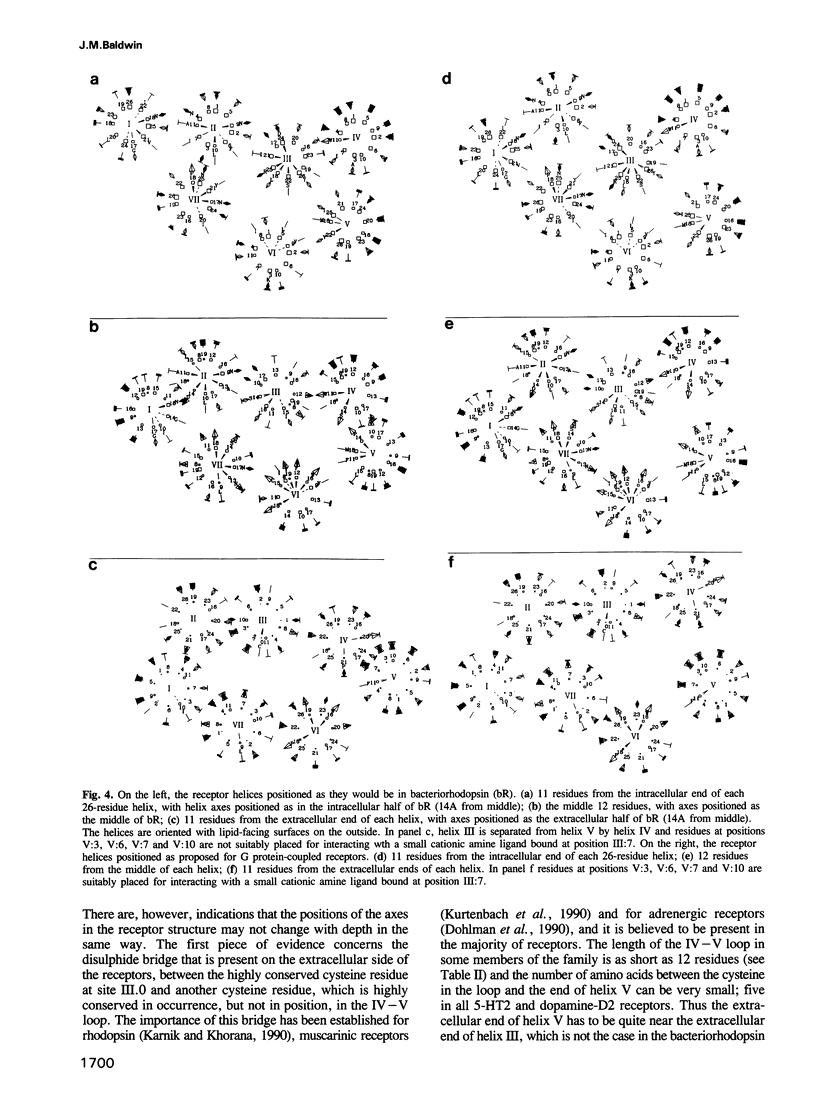
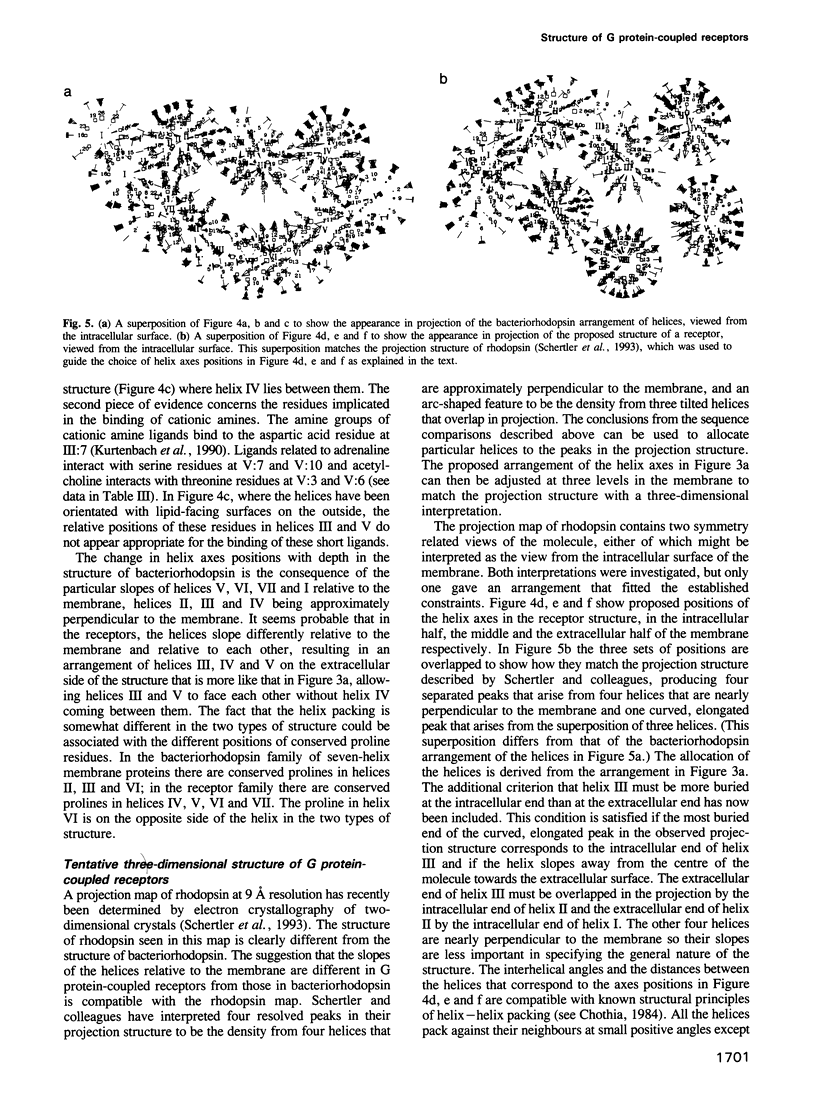
G protein-coupled receptors form a large family of integral membrane proteins whose amino acid sequences have seven hydrophobic segments containing distinctive sequence patterns. Rhodopsin, a member of the family, is known to have transmembrane alpha-helices. The probable arrangement of the seven helices, in all receptors, was deduced from structural information extracted from a detailed analysis of the sequences. Constraints established include: (1) each helix must be positioned next to its neighbours in the sequence; (2) helices I, IV and V must be most exposed to the lipid surrounding the receptor and helix III least exposed. (1) is established from the lengths of the shortest loops. (2) is determined by considering: (i) sites of the most conserved residues; (ii) other sites where variability is restricted; (iii) sites that accommodate polar residues; (iv) sites of differences in sequence between pairs or within groups of closely related receptors. Most sites in the last category should be in unimportant positions and are most useful in determining the position and extent of lipid-facing surface in each helix. The structural constraints for the receptors are used to allocate particular helices to the peaks in the recently published projection map of rhodopsin and to propose a tentative three-dimensional arrangement of the helices in G protein-coupled receptors.
Full text
PDF




Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Applebury M. L., Hargrave P. A. Molecular biology of the visual pigments. Vision Res. 1986;26(12):1881–1895. doi: 10.1016/0042-6989(86)90115-x. [DOI] [PubMed] [Google Scholar]
- Applebury M. L. Molecular determinants of visual pigment function. Curr Opin Neurobiol. 1991 Aug;1(2):263–269. doi: 10.1016/0959-4388(91)90088-o. [DOI] [PubMed] [Google Scholar]
- Archer S. N., Lythgoe J. N., Hall L. Rod opsin cDNA sequence from the sand goby (Pomatoschistus minutus) compared with those of other vertebrates. Proc Biol Sci. 1992 Apr 22;248(1321):19–25. doi: 10.1098/rspb.1992.0037. [DOI] [PubMed] [Google Scholar]
- Birnbaumer L., Abramowitz J., Brown A. M. Receptor-effector coupling by G proteins. Biochim Biophys Acta. 1990 May 7;1031(2):163–224. doi: 10.1016/0304-4157(90)90007-y. [DOI] [PubMed] [Google Scholar]
- Buck L., Axel R. A novel multigene family may encode odorant receptors: a molecular basis for odor recognition. Cell. 1991 Apr 5;65(1):175–187. doi: 10.1016/0092-8674(91)90418-x. [DOI] [PubMed] [Google Scholar]
- Chabre M. Trigger and amplification mechanisms in visual phototransduction. Annu Rev Biophys Biophys Chem. 1985;14:331–360. doi: 10.1146/annurev.bb.14.060185.001555. [DOI] [PubMed] [Google Scholar]
- Chothia C. Principles that determine the structure of proteins. Annu Rev Biochem. 1984;53:537–572. doi: 10.1146/annurev.bi.53.070184.002541. [DOI] [PubMed] [Google Scholar]
- Chung F. Z., Wang C. D., Potter P. C., Venter J. C., Fraser C. M. Site-directed mutagenesis and continuous expression of human beta-adrenergic receptors. Identification of a conserved aspartate residue involved in agonist binding and receptor activation. J Biol Chem. 1988 Mar 25;263(9):4052–4055. [PubMed] [Google Scholar]
- Cowan S. W., Schirmer T., Rummel G., Steiert M., Ghosh R., Pauptit R. A., Jansonius J. N., Rosenbusch J. P. Crystal structures explain functional properties of two E. coli porins. Nature. 1992 Aug 27;358(6389):727–733. doi: 10.1038/358727a0. [DOI] [PubMed] [Google Scholar]
- Deisenhofer J., Michel H. The Photosynthetic Reaction Center from the Purple Bacterium Rhodopseudomonas viridis. Science. 1989 Sep 29;245(4925):1463–1473. doi: 10.1126/science.245.4925.1463. [DOI] [PubMed] [Google Scholar]
- Dixon R. A., Sigal I. S., Candelore M. R., Register R. B., Scattergood W., Rands E., Strader C. D. Structural features required for ligand binding to the beta-adrenergic receptor. EMBO J. 1987 Nov;6(11):3269–3275. doi: 10.1002/j.1460-2075.1987.tb02645.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dohlman H. G., Caron M. G., DeBlasi A., Frielle T., Lefkowitz R. J. Role of extracellular disulfide-bonded cysteines in the ligand binding function of the beta 2-adrenergic receptor. Biochemistry. 1990 Mar 6;29(9):2335–2342. doi: 10.1021/bi00461a018. [DOI] [PubMed] [Google Scholar]
- Dohlman H. G., Thorner J., Caron M. G., Lefkowitz R. J. Model systems for the study of seven-transmembrane-segment receptors. Annu Rev Biochem. 1991;60:653–688. doi: 10.1146/annurev.bi.60.070191.003253. [DOI] [PubMed] [Google Scholar]
- Donnelly D., Johnson M. S., Blundell T. L., Saunders J. An analysis of the periodicity of conserved residues in sequence alignments of G-protein coupled receptors. Implications for the three-dimensional structure. FEBS Lett. 1989 Jul 17;251(1-2):109–116. doi: 10.1016/0014-5793(89)81438-3. [DOI] [PubMed] [Google Scholar]
- Fraser C. M., Wang C. D., Robinson D. A., Gocayne J. D., Venter J. C. Site-directed mutagenesis of m1 muscarinic acetylcholine receptors: conserved aspartic acids play important roles in receptor function. Mol Pharmacol. 1989 Dec;36(6):840–847. [PubMed] [Google Scholar]
- Guan X. M., Peroutka S. J., Kobilka B. K. Identification of a single amino acid residue responsible for the binding of a class of beta-adrenergic receptor antagonists to 5-hydroxytryptamine1A receptors. Mol Pharmacol. 1992 Apr;41(4):695–698. [PubMed] [Google Scholar]
- Henderson R., Baldwin J. M., Ceska T. A., Zemlin F., Beckmann E., Downing K. H. Model for the structure of bacteriorhodopsin based on high-resolution electron cryo-microscopy. J Mol Biol. 1990 Jun 20;213(4):899–929. doi: 10.1016/S0022-2836(05)80271-2. [DOI] [PubMed] [Google Scholar]
- Hibert M. F., Trumpp-Kallmeyer S., Bruinvels A., Hoflack J. Three-dimensional models of neurotransmitter G-binding protein-coupled receptors. Mol Pharmacol. 1991 Jul;40(1):8–15. [PubMed] [Google Scholar]
- Hulme E. C., Birdsall N. J., Buckley N. J. Muscarinic receptor subtypes. Annu Rev Pharmacol Toxicol. 1990;30:633–673. doi: 10.1146/annurev.pa.30.040190.003221. [DOI] [PubMed] [Google Scholar]
- Ishihara T., Shigemoto R., Mori K., Takahashi K., Nagata S. Functional expression and tissue distribution of a novel receptor for vasoactive intestinal polypeptide. Neuron. 1992 Apr;8(4):811–819. doi: 10.1016/0896-6273(92)90101-i. [DOI] [PubMed] [Google Scholar]
- Johnson R. L., Grant K. B., Zankel T. C., Boehm M. F., Merbs S. L., Nathans J., Nakanishi K. Cloning and expression of goldfish opsin sequences. Biochemistry. 1993 Jan 12;32(1):208–214. doi: 10.1021/bi00052a027. [DOI] [PubMed] [Google Scholar]
- Kao H. T., Adham N., Olsen M. A., Weinshank R. L., Branchek T. A., Hartig P. R. Site-directed mutagenesis of a single residue changes the binding properties of the serotonin 5-HT2 receptor from a human to a rat pharmacology. FEBS Lett. 1992 Aug 3;307(3):324–328. doi: 10.1016/0014-5793(92)80705-l. [DOI] [PubMed] [Google Scholar]
- Karnik S. S., Khorana H. G. Assembly of functional rhodopsin requires a disulfide bond between cysteine residues 110 and 187. J Biol Chem. 1990 Oct 15;265(29):17520–17524. [PubMed] [Google Scholar]
- Kobilka B. Adrenergic receptors as models for G protein-coupled receptors. Annu Rev Neurosci. 1992;15:87–114. doi: 10.1146/annurev.ne.15.030192.000511. [DOI] [PubMed] [Google Scholar]
- Kurtenbach E., Curtis C. A., Pedder E. K., Aitken A., Harris A. C., Hulme E. C. Muscarinic acetylcholine receptors. Peptide sequencing identifies residues involved in antagonist binding and disulfide bond formation. J Biol Chem. 1990 Aug 15;265(23):13702–13708. [PubMed] [Google Scholar]
- Nakayama T. A., Khorana H. G. Mapping of the amino acids in membrane-embedded helices that interact with the retinal chromophore in bovine rhodopsin. J Biol Chem. 1991 Mar 5;266(7):4269–4275. [PubMed] [Google Scholar]
- Nathans J. Determinants of visual pigment absorbance: identification of the retinylidene Schiff's base counterion in bovine rhodopsin. Biochemistry. 1990 Oct 16;29(41):9746–9752. doi: 10.1021/bi00493a034. [DOI] [PubMed] [Google Scholar]
- Neitz M., Neitz J., Jacobs G. H. Spectral tuning of pigments underlying red-green color vision. Science. 1991 May 17;252(5008):971–974. doi: 10.1126/science.1903559. [DOI] [PubMed] [Google Scholar]
- Oksenberg D., Marsters S. A., O'Dowd B. F., Jin H., Havlik S., Peroutka S. J., Ashkenazi A. A single amino-acid difference confers major pharmacological variation between human and rodent 5-HT1B receptors. Nature. 1992 Nov 12;360(6400):161–163. doi: 10.1038/360161a0. [DOI] [PubMed] [Google Scholar]
- Oprian D. D. The ligand-binding domain of rhodopsin and other G protein-linked receptors. J Bioenerg Biomembr. 1992 Apr;24(2):211–217. doi: 10.1007/BF00762679. [DOI] [PubMed] [Google Scholar]
- Pollock N. J., Manelli A. M., Hutchins C. W., Steffey M. E., MacKenzie R. G., Frail D. E. Serine mutations in transmembrane V of the dopamine D1 receptor affect ligand interactions and receptor activation. J Biol Chem. 1992 Sep 5;267(25):17780–17786. [PubMed] [Google Scholar]
- Sakmar T. P., Franke R. R., Khorana H. G. Glutamic acid-113 serves as the retinylidene Schiff base counterion in bovine rhodopsin. Proc Natl Acad Sci U S A. 1989 Nov;86(21):8309–8313. doi: 10.1073/pnas.86.21.8309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savarese T. M., Fraser C. M. In vitro mutagenesis and the search for structure-function relationships among G protein-coupled receptors. Biochem J. 1992 Apr 1;283(Pt 1):1–19. doi: 10.1042/bj2830001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simchen G., Hugerat Y. What determines whether chromosomes segregate reductionally or equationally in meiosis? Bioessays. 1993 Jan;15(1):1–8. doi: 10.1002/bies.950150102. [DOI] [PubMed] [Google Scholar]
- Strader C. D., Candelore M. R., Hill W. S., Sigal I. S., Dixon R. A. Identification of two serine residues involved in agonist activation of the beta-adrenergic receptor. J Biol Chem. 1989 Aug 15;264(23):13572–13578. [PubMed] [Google Scholar]
- Strader C. D., Sigal I. S., Candelore M. R., Rands E., Hill W. S., Dixon R. A. Conserved aspartic acid residues 79 and 113 of the beta-adrenergic receptor have different roles in receptor function. J Biol Chem. 1988 Jul 25;263(21):10267–10271. [PubMed] [Google Scholar]
- Strader C. D., Sigal I. S., Register R. B., Candelore M. R., Rands E., Dixon R. A. Identification of residues required for ligand binding to the beta-adrenergic receptor. Proc Natl Acad Sci U S A. 1987 Jul;84(13):4384–4388. doi: 10.1073/pnas.84.13.4384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suryanarayana S., Daunt D. A., Von Zastrow M., Kobilka B. K. A point mutation in the seventh hydrophobic domain of the alpha 2 adrenergic receptor increases its affinity for a family of beta receptor antagonists. J Biol Chem. 1991 Aug 15;266(23):15488–15492. [PubMed] [Google Scholar]
- Thomas D. D., Stryer L. Transverse location of the retinal chromophore of rhodopsin in rod outer segment disc membranes. J Mol Biol. 1982 Jan 5;154(1):145–157. doi: 10.1016/0022-2836(82)90422-3. [DOI] [PubMed] [Google Scholar]
- Wang C. D., Buck M. A., Fraser C. M. Site-directed mutagenesis of alpha 2A-adrenergic receptors: identification of amino acids involved in ligand binding and receptor activation by agonists. Mol Pharmacol. 1991 Aug;40(2):168–179. [PubMed] [Google Scholar]
- Wang H., Lipfert L., Malbon C. C., Bahouth S. Site-directed anti-peptide antibodies define the topography of the beta-adrenergic receptor. J Biol Chem. 1989 Aug 25;264(24):14424–14431. [PubMed] [Google Scholar]
- Weiss M. S., Abele U., Weckesser J., Welte W., Schiltz E., Schulz G. E. Molecular architecture and electrostatic properties of a bacterial porin. Science. 1991 Dec 13;254(5038):1627–1630. doi: 10.1126/science.1721242. [DOI] [PubMed] [Google Scholar]
- Wess J., Gdula D., Brann M. R. Site-directed mutagenesis of the m3 muscarinic receptor: identification of a series of threonine and tyrosine residues involved in agonist but not antagonist binding. EMBO J. 1991 Dec;10(12):3729–3734. doi: 10.1002/j.1460-2075.1991.tb04941.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wess J., Nanavati S., Vogel Z., Maggio R. Functional role of proline and tryptophan residues highly conserved among G protein-coupled receptors studied by mutational analysis of the m3 muscarinic receptor. EMBO J. 1993 Jan;12(1):331–338. doi: 10.1002/j.1460-2075.1993.tb05661.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong S. K., Slaughter C., Ruoho A. E., Ross E. M. The catecholamine binding site of the beta-adrenergic receptor is formed by juxtaposed membrane-spanning domains. J Biol Chem. 1988 Jun 15;263(17):7925–7928. [PubMed] [Google Scholar]
- Zhukovsky E. A., Oprian D. D. Effect of carboxylic acid side chains on the absorption maximum of visual pigments. Science. 1989 Nov 17;246(4932):928–930. doi: 10.1126/science.2573154. [DOI] [PubMed] [Google Scholar]



