Abstract
Although BMP6 is highly capable of inducing osteogenic differentiation of mesenchymal progenitor cells (MPCs), the molecular mechanism involved remains to be fully elucidated. Using dominant negative (dn) mutant form of type I and type II TGFβ receptors, we demonstrated that three dn-type I receptors (dnALK2, dnALK3, dnALK6), and three dn-type II receptors (dnBMPRII, dnActRII, dnActRIIB), effectively diminished BMP6-induced osteogenic differentiation of MPCs. These findings suggested that ALK2, ALK3, ALK6, BMPRII, ActRII and ActRIIB are essential for BMP6-induced osteogenic differentiation of MPCs. However, MPCs in this study do not express ActRIIB. Moreover, RNA interference of ALK2, ALK3, ALK6, BMPRII and ActRII inhibited BMP6-induced osteogenic differentiation in MPCs. Our results strongly suggested that BMP6-induced osteogenic differentiation of MPCs is mediated by its functional TGFβ receptors including ALK2, ALK3, ALK6, BMPRII, and ActRII. [BMB Reports 2013; 46(2): 107-112]
Keywords: BMP6, Bone morphogenetic proteins, Mesenchymal progenitor cells, Osteogenic differentiation, Transforming growth factor β receptor
INTRODUCTION
Mesenchymal progenitor cells (MPCs) are pluripotent stem cell capable of differentiating into osteoblast, chondrocyte, myocyte or adipocyte (1). Bone morphogenetic proteins (BMPs), belonging to the transforming growth factors beta (TGFβ) superfamily, are known as pivotal regulators of embryogenesis, multiple growth and differentiation processes (2). To date, more than twenty BMPs isoforms have been identified in mammals. Several forms of recombinant BMPs, most notably BMP2 and BMP7, have been shown to promote osteogenesis and are now used as adjunctive therapy in the clinical setting (3).
BMP6 is a member of the BMPs proteins. Although expression of BMP6 was detected in various cell types, BMP6 protein is predominantly expressed in mature chondrocytes during endochondral ossification (4), strongly supporting the important roles of BMP6 in osteogenesis and bone development. Our previously study has shown that BMP6 was highly capable of inducing osteogenic differentiation of MPCs both in vitro and in vivo (5). However, the signaling mechanism through which BMP6 regulates osteogenic differentiation of MPCs is largely unknown and warrants extensive studies.
BMPs fulfill their signaling activity by interacting with receptor complexes composed of type I and type II TGFβ receptors. This interaction brings both type I and type II receptors into close proximity, permitting the constitutively active type II receptor to cross-phosphorylate type I receptor, and subsequently triggers downstream signal cascade (2). To date, seven type I (ALK1toALK7) and four type II TGFβ receptors (TGFβRII, BMPRII, ActRII, ActRIIB) have been identified (6,7). Several studies have demonstrated that TGFβ receptors are essential for osteogenic activity of BMPs (2,8). Unfortunately, the functional TGFβ receptor specific for BMP6-induced osteogenic differentiation of MPCs remains to be elucidated.
In the current study, we sought to determine the functional TGFβ receptors required for BMP6-induced osteogenic differentiation of MPCs. Using a panel of dominant negative (dn) TGFβ receptors (dnR), the functional TGFβ receptors essential in BMP6-induced osteogenic differentiation of MPCs was comprehensively analyzed. It was found that dnALK2, dnALK3, dnALK6, dnBMPRII, dnActRII, dnActRIIB not only effectively inhibited BMP6-induced osteogenic differentiation, but also decreased BMP6-activated Smads signaling of MPCs. These results implied that the corresponding wild receptors, ALK2, ALK3, ALK6, BMPRII, ActRII and ActRIIB, may play a functional role in BMP6-induced osteogenic differentiation. However, MPCs been used in this study do not express ActRIIB. Then, when ALK2, ALK3, ALK6, BMPRII and ActRII were silenced by RNAi in MPCs, BMP6-induced osteogenic differentiation and Smads signaling activation were found to be accordingly inhibited. Together, our results intensively indicated ALK2, ALK3, ALK6, BMPRII and ActRII are the functional TGFβ receptors required for BMP6-induced osteogenic differentiation of MPCs.
RESULTS
Endogenous expression of the seven type I and four type II TGFβ receptors in MPCs
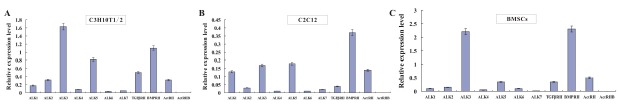
Here, we sought to determine the obligate TGFβ receptors required for BMP6-mediated osteogenic signaling in MPCs. First of all, we examined the endogenous expression of all seven type I and four type II TGFβ receptors in C3H10T1/2, C2C12 and primary bone marrow stem cells (BMSCs). As shown in Fig. 1, although the expression levels are different, all seven endogenous wild type I receptors can totally be detected. For the type II receptors, however, C3H10T1/2, C2C12 and BMSCs expressed TGFβRII, BMPRII and ActRII, but not ActRIIB.
Fig. 1. Endogenous expressions of wild TGFβ receptors in MPCs. (A). C3H10T1/2 were cultured for 24 hours. Endogenous expression of wild TGFβ was detected by qPCR. Data were means ± SD of three independent experiments. (B). C2C12 were cultured for 24 hours. Endogenous expression of wild TGFβ was detected by qPCR. Data were means ± SD of three independent experiments. (C). BMSCs were cultured for 24 hours. Endogenous expression of wild TGFβ was detected by qPCR. Data were means ± SD of three independent experiments.
Dominant negative mutant form of ALK2, ALK3, ALK6, BMPRII, ActRII and ActRIIB attenuated BMP6-induced early osteogenic differentiation of MPCs
We have made a panel of adenoviruses expressing seven type I dnR and four type II dnR (9,10). Here, we thought to use these dnR to identify the TGFβ receptors essential for BMP6-induced osteogenic differentiation of MPCs. C3H10T1/2 cells were infected by adenoviruses expressing dnR, and the expression of exogenous dnR was confirmed by RT-PCR using appropriate primers to distinguish expression of the dnR from endogenous wild receptors, as demonstrated in Fig. 2A. These PCR products should be specifically derived from the adenoviral vector-mediated dnR expression, as the 3'-end primer was derived from the SV40 polyA cassette of shutter vector.
Fig. 2. dnALK2, dnALK3, dnALK6, dnBMPRII, dnActRII and dnActRIIB inhibited BMP6-induced ALP activity of MPCs. (A) C3H10T1/2 were infected with a comparable titer of AdR-dnR or Ad-RFP. At 36 hours post infection, total RNA was isolated, and then exogenous expression of dnR was validated by RT-PCR. ‘RFP’: red fluorescent protein; ‘+’: PCR products from +RT reactions of the original cDNA synthesis; ‘−’: PCR products from −RT reactions of the original cDNA synthesis. (B) C3H10T1/2, C2C12 and BMSCs were co-infected with Ad-BMP6 and AdR-dnR. ALP activity was assessed at 7 days (C3H10T1/2) or 5 days (C2C12 and BMSCs) post infection. Data were means ± SD of three independent experiments. **P < 0.01 vs RFP, *P < 0.05 vs RFP. (C) C3H10T1/2 were co-infected with a fixed titer of Ad-BMP6 and varying titers of AdR-dnALK2 or AdR-dnALK3 or dnALK6. ALP activity was assessed at 7 days post infection. Data were means ± SD of three independent experiments. (D) C3H10T1/2 were co-infected with a fixed titer of Ad-BMP6 and varying titers of AdR-dnBMPRII or AdR-dnActRII or AdR-dnActRIIB. ALP activity was assessed at 7 days post infection. Data were means ± SD of three independent experiments.
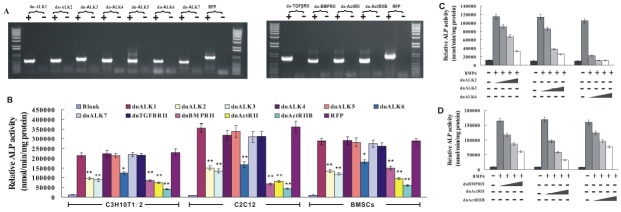
We next tested the effects of exogenous dnR on BMP6-induced early osteogenic differentiation. As demonstrated in Fig. 2B, dnALK2, dnALK3, dnALK6, dnBMPRII, dnActRII and dnActRIIB remarkably inhibited BMP6-induced activity of ALP, a well established marker of early osteogenic differentiation (9,10). Then, C3H10T1/2 cells were co-stimulated with a fixed titer of BMP6 and varying titers of dnALK2, dnALK3, dnALK6, dnBMPRII, dnActRII and dnActRIIB respectively. As shown in Fig. 2C and 2D, an increase in dnALK2, dnALK3, dnALK6, dnBMPRII, dnActRII and dnActRIIB led to a significantly higher reduction in BMP6-induced ALP activity of C3H10T1/2 cells. These data confirmed that dnALK2, dnALK3, dnALK6, dnBMPRII, dnActRII and dnActRIIB inhibited BMP6-induced ALP activity in a dose-dependent manner. Taken together, the above findings strongly demonstrated that dnALK2, dnALK3, dnALK6, dnBMPRII, dnActRII and dnActRIIB inhibited BMP6- induced early stage of osteogenic differentiation of MPCs.
Dominant negative mutant form of ALK2, ALK3, ALK6, BMPRII, ActRII and ActRIIB inhibited BMP6-induced late osteogenic differentiation and Smads signaling activation of MPCs
Next, we sought to further determine the effects of dnALK2, dnALK3, dnALK6, dnBMPRII, dnActRII and dnActRIIB on BMP6-induced late osteogenic marker, such as matrix mineralization and osteopontin (OPN) expression. As shown in Fig. 3A, it was found that dnALK2, dnALK3, dnALK6, dnBMPRII, dnActRII and dnActRIIB effectively inhibited BMP6-induced matrix mineralization of MPCs, resulting in less calcium deposition. Likewise, BMP6-induced OPN expression was decreased in the presence of dnALK2, dnALK3, dnALK6, dnBMPRII, dnActRII and dnActRIIB, respectively (Fig. 3B). These Results suggested that dnALK2, dnALK3, dnALK6, dnBMPRII, dnActRII and dnActRIIB inhibited BMP6-induced late stage of osteogenic differentiation of MPCs.
Fig. 3. dnALK2, dnALK3, dnALK6, dnBMPRII, dnActRII and dnActRIIB inhibited BMP6-induced late stage of osteogenic differentiation and Smads signaling activation of MPCs. (A) C3H10T1/2, C2C12 and BMSCs were co-infected with Ad-BMP6 and AdR-dnR. Matrix mineralization was assessed at 21 days post infection by Alizarin Red S stain. Magnification, 100×. (B) C3H10T1/2 were co-infected with Ad-BMP6 and AdR-dnR. OPN expression was assessed at 12 days post infection by immunocytochemical stain. Magnification, 100×. (C) C3H10T1/2 were co-infected with Ad-BMP6 and AdR-dnR. Smad1/5/8 transcription activity was assessed at indicated time point post infection by luciferase reporter assay. **P < 0.01 vs RFP. (D) C3H10T1/2 were co-infected with Ad-BMP6 and AdR-dnR. Phosphorylation of Smad1/5/8 was detected at 24 hours post infection by Western blot.
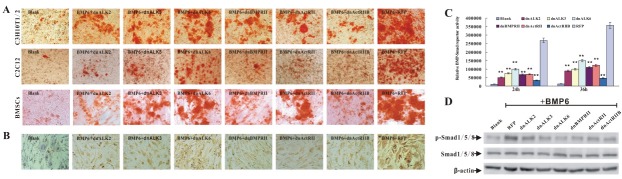
Smad1, 5, and 8 (Smad1/5/8) are classic mediators for BMPs osteoinductive signaling (2). Therefore, we asked whether BMP6-activated Smads signaling was inhibited by dnALK2, dnALK3, dnALK6, dnBMPRII, dnActRII and dnActRIIB, respectively. Using the BMPs responsive Smads reporter, p12xSBE- luc (11), we found that dnALK2, dnALK3, dnALK6, dnBMPRII, dnActRII and dnActRIIB were able to neutralize BMP6-induced transcription activity of Smad1/5/8 (Fig. 3C). Moreover, BMP6-induced phosphorylation of Smad1/5/8 was always effectively attenuated by dnALK2, dnALK3, dnALK6, dnBMPRII, dnActRII and dnActRIIB (Fig. 3D). Taken together, these above data intensively implied that dnALK2, dnALK3, dnALK6, dnBMPRII, dnActRII and dnActRIIB inhibited BMP6-induced osteogenic differentiation and Smads signaling. Therefore, we postulated that the corresponding wild type I receptor ALK2, dnALK3 and ALK6, the wild type II receptor BMPRII, ActRII and ActRIIB may be the functional TGFβ receptors essential for BMP6-induced osteogenic differentiation of MPCs.
Gene silencing of ALK2, ALK3, ALK6, BMPRII, and ActRII reduced BMP6-induced osteogenic differentiation and Smads signaling activation
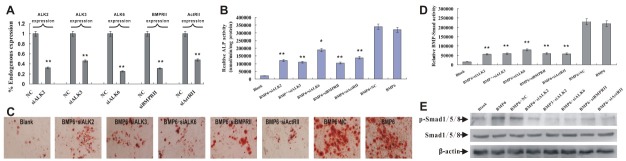
Although dominant negative mutant technology provides useful method to analyze the functions of wild receptors, it also presents some limitations. Besides binding to concerned ligand, the over-expressed dnR may interact with other known/unknown proteins, leading to some side and/or overlap effects. For this reason, we thought to further ascertain our above finding by using RNAi. As shown in Fig. 1, ActRIIB was not expressed in MPCs. Therefore, we employed adenoviruses expressing small interference RNA (siRNA) targeted ALK2, ALK3, ALK6, BMPRII and ActRII to infect C3H10T1/2 cells. Inhibitory efficiency of these siRNAs on expressions of ALK2, ALK3, ALK6, BMPRII and ActRII was assessed by qPCR (Fig. 4A). As shown in Fig. 4B and 4C, gene silencing of ALK2, ALK3, ALK6, BMPRII, and ActRII diminished BMP6-induced ALP activity and matrix mineralization. Furthermore, knockdown of ALK2, ALK3, ALK6, BMPRII, and ActRII effectively reduced BMP6-activated Smads signaling, leading to reduction of BMP6-Smad transcriptional activity and less phosphorylation of Smad1/5/8. These findings implied us that ALK2, ALK3, ALK6, BMPRII and ActRII may play a private role in regulating BMP6-induced osteogenic differentiation and Smads signal activation.
Fig. 4. Silencing of ALK2, ALK3, ALK6, BMPRII, and ActRII attenuated BMP6-induced osteogenic differentiation and Smads signaling of C3H10T1/2 cells. (A) Effective knockdown of ALK2, ALK3, ALK6, BMPRII and ActRII was confirmed by qPCR. **P < 0.01 vs NC (negative control). (B) After achieving effective knockdown of ALK2, ALK3, ALK6, BMPRII and ActRII, ALP activity was assessed at 7 days post BMP6-sitmulation. Data were means ± SD of three experiments. **P < 0.01 vs BMP6 + NC, *P < 0.05 vs BMP6 + NC. (C) After achieving effective knockdown of ALK2, ALK3, ALK6, BMPRII and ActRII, BMP6-induced matrix mineralization was detected at 21 days post BMP6-stimulation. Magnification, 100×. (D) After achieving effective knockdown of ALK2, ALK3, ALK6, BMPRII and ActRII, luciferase activity was assessed at 24 hour post BMP6-stimulation. Data were means ± SD of three experiments. **P < 0.01 vs BMP6 + NC. (E) After achieving effective knockdown of ALK2, ALK3, ALK6, BMPRII, and ActRII, phosphorylation of Smad1/5/8 was detected at 24 hours post infection by western blot.
In summary, using dominant negative mutant assay and RNAi technology, we have demonstrated that ALK2, dnALK3, ALK6, BMPRII and ActRII are probably the functional receptors for BMP6-induced osteogenic differentiation of MPCs. It should be noted, however, ActRIIB is not expressed endogenously and ectopic expression of dominant negative form of ActRIIB suppresses BMP6-induced osteogenic differentiation, characterized by low calcium deposition, decreased ALP and Smad activity and reduced OPN expression. These results implied that ActRIIB may also interact with BMP6. The potential role of ActRIIB in BMP6-induced osteogenic differentiation can not be rule out rashly without careful validation.
DISCUSSION
BMP6 was originally isolated from murine embryonic cDNA library (12). The human and bovine BMP6 were subsequently isolated from bone (13). BMP6 was capable of regulating glucose metabolism, maintaining the serum iron level, and sustaining the integrity of joint (14). Previous study has established BMP6 as an important regulator of osteogenic differentiation. Unfortunately, BMP6 osteoinductive signaling remains uncharacterized currently.
BMPs signaling are triggered when a dimeric ligand binds to the type I and type II TGFβ receptors, which have serine/threonine kinase activities in their cytoplasmic domains and play a key role in BMPs-induced osteogenic signal transduction (2,15). After binding to type I and type II TGFβ receptor, osteogenic BMPs could activate its receptor and then invoke Smad-dependent and/or Smad-independent pathways (2,9,10,16,17). Receptor-activated Smad-dependent and/or Smad-independent pathways subsequently regulated various proteins, and transmit specific osteoinductive signals (2,17). It has been well characterized that BMP2 utilized ALK2, ALK3, ALK6, BMPRII and ActRII to mediate osteoinductive signaling, whereas BMP7 employed ALK3, ALK6, BMPRII and ActRII (6,18). Moreover, our recently studies have shown that BMP9, one of the least characterized BMPs, utilized ALK1, ALK2, BMPRII and ActRII to induced osteogenic signaling of MPCs (9,10). However, the functional TGFβ receptors essential for BMP6-induced osteogenic differentiation of MPCs remains to be elucidated.
TGFβ receptors contain three domains: extracellular ligand binding domain, transmembrane domain and intracellular serine/threonine kinase domain. The kinase domain of TGFβ receptor is essential to signal transduction (19). Dominant negative TGFβ receptors (dnR) were generated by deletions of this kinase domain. dnR inhibits endogenous receptors function, most likely by interfering with endogenous receptors complex formation and signal transduction (20,21). Thus, dnR can compete with endogenous receptors for binding of ligands, thereby acting as an inhibitor of BMPs signaling pathway.
Seven type I TGFβ receptors have been identified in mammals, and are divided into three groups: the ALK1 group (ALK1andALK2), the ALK3 group (ALK3andALK6), and the TGFβRI group (ALK4, ALK5, and ALK7) (2,6). Generally, the receptors of the ALK1 group and ALK3 group activate Smad1/5/8, transduce similar intracellular signals and are consider as BMPs type I receptors (2). However, these BMPs type I receptors are also shared by certain other members of the TGFβ superfamily. For example, TGFβ binds to ALK1 as well as ALK5 in endothelial cells (22). MIS binds to ALK2, ALK3 and ALK6 in the presence of its specific type II receptor, MISR-II (23). In this study, we found that the type I TGFβ receptors essential for BMP6- induced osteogenic differentiation of MPCs are ALK2, ALK3 and ALK6.
Three receptors, BMPRII, ActRII and ActRIIB, serve as type II TGFβ receptors for BMPs in mammals, and are widely expressed in various tissues. These type II receptors appear to bind most BMPs ligands and affect the binding preferences of BMPs to type I receptors (2,6). BMPRII, ActRII and ActRIIB have a similar domain organization, however, BMPRII has an extraordinary long C-terminal tail region which can interact with several proteins and might have a regulatory role in the BMP pathway (24). BMPRII has been confirmed as a specific receptor for BMP2 and BMP4 (25). ActRII and ActRIIB are receptors for activin, however, they work also as functional receptor for BMPs, notable BMP7 (26). Our result showed that BMPRII and ActRII are functional type II TGFβ receptors for BMP6-induced osteogenic differentiation of MPCs. However, the potential role of ActRIIB in BMP6-induced osteogenic differentiation can not be rule out rashly.
In conclusion, we have conducted a comprehensive analysis of the functional role of receptors in BMP6-induced osteogenic signaling of MPCs. Using dominant negative mutants assay and RNAi, we reported that ALK2, ALK3, ALK6, BMPRII and ActRII may be the functional TGFβ receptors essential for BMP6- induced osteogenic differentiation of MPCs. Future studies should be devoted to the elucidation of detailed mechanism behind BMP6 and its receptors interaction in the context of osteogenic differentiation.
MATERIALS AND METHODS
Cell culture and chemicals
HEK293, C3H10T1/2 and C2C12 were obtained from ATCC and maintained in complete DMEM (Dulbeccos modified Eagle medium) supplemented with 10% fetal bovine serum and 100 units/ml streptomycin/penicillin at 37℃ in a humidified atmosphere of 5% CO2.
Isolation of primary bone marrow stem cells (BMSCs)
A single-step primary BMSCs purification method using adhesion to cell culture plastic was employed as described in the reference (27). Cells were utilized for experimentation between passages 3-9.
Construction of recombinant adenoviruses
Recombinant adenovirus expressing BMP6 (Ad-BMP6) were generated previously, as demonstrated (5). Recombinant adenoviruses harboring dominant negative TGFβ receptors (AdR-dnR) were generated as previously described (9,10). Recombinant adenoviruses expressing siRNA targeted ALK2 (AdR-siALK2), ALK3 (AdR-siALK3), ALK6 (AdR-siALK6), BMPRII (AdR-siBMPRII), ActRII (AdR-siActRII) were kindly provided by Dr. Tong-chuan He of University of Chicago Medical Center. Adenoviruses expressing only RFP (Ad-RFP) were used as negative controls.
Determination of ALP activity
ALP activity was assessed by a modified Great Escape SEAP chemiluminescence assay (BD Clontech, Mountain View, CA), as described previously (10). Each assay condition was performed in triplicate. ALP activity was normalized by total cellular protein concentrations among the samples.
Alizarin Red S stain
Mitrix mineralization was detected by Alizarin Red S stain, as described previously (10). Briefly, cells were cultured in the presence of ascorbic acid (50 μg/ml) and β-glycerophosphate (10 mmol/L). At 21 days after cultured, cells were fixed with 0.05% (v/v) glutaraldehyde at room temperature for 10 min. After being washed with distilled water, fixed cells were incubated with 0.4% Alizarin Red S for 5 min, followed by extensive washing with distilled water. The staining of matrix mineralization was recorded under bright field microscopy.
Western blotting
Briefly, cells were collected and lysed in RAPI buffer. Cleared total cell lysate was denatured by boiling and loaded onto a 10% gradient SDS-PAGE. After electrophoresis separation, proteins were transferred to an Immobilon-P membrane. Membrane was blocked with Super-Block Blocking Buffer, and probed with the primary antibody, followed by incubation with a secondary antibody conjugated with horseradish peroxidase. The proteins of interest were detected by using SuperSignal West Pico Chemiluminescent Substrate kit. Primary antibodies were obtained from Santa Cruz, as follows: anti-phosphor-Smad1/5/8, anti-Smad1/5/8, and anti-β-actin.
Immunocytochemical stain
Cultured cells were co-infected with Ad-BMP6 and AdR-dnR. At the indicated time points, cells were fixed with 4% formalin and washed with PBS. The fixed cells were permeabilized with 0.25% Triton X-100 and blocked with 10% goat serum, followed by incubation with an anti-OPN antibody (Santa Cruz Biotechnology) overnight. After washing, cells were incubated with biotin-labeled secondary antibody for 30 min, followed by incubating cells with streptavidin-HRP conjugate for 20 min at room temperature. The presence of the expected protein was visualized by DAB staining and examined under a microscope.
Luciferase reporter assay
Exponentially growing cells were seeded in 25 cm2 cell culture flasks and transfected with 2 mg per flask of BMP receptor Smad-responsive luciferase reporter (12), p12xSBE-luc using LipofectAmine. At 16 hours after transfection, cells were replated to 24 well plates and co-infected with Ad-BMP6 and AdR-dnR. Cells were lysed and cell lysates were collected for luciferase assays using luciferase assay kit (Promega).
Statistical analysis
Data are expressed as means ± SD. Statistical analysis was performed using SAS (version 8.1; SAS Institute, Cary, NC), P <0.05 was taken as the level of significance.
Acknowledgments
This work was supported by grants from the National Natural Science Foundation of China (No.31071304, 30800658).
References
- 1.Pittenger M. F., Mackay A. M., Beck S. C., Jaiswal R. K., Douglas R., Mosca J. D., Moorman M. A., Simonetti D. W., Craig S., Marshak D. R. Multilineage potential of adult human mesenchymal stem cells. Science. (1999);284:143–147. doi: 10.1126/science.284.5411.143. [DOI] [PubMed] [Google Scholar]
- 2.Chen D., Zhao M., Mundy G. R. Bone morphogenetic proteins. Growth Factors. (2004);22:233–241. doi: 10.1080/08977190412331279890. [DOI] [PubMed] [Google Scholar]
- 3.Bostrom M. P., Camacho N. P. Potential role of bone morphogenetic proteins in fracture healing. Clin. Orthop. Relat. Res. (1998);355:S274–282. doi: 10.1097/00003086-199810001-00028. [DOI] [PubMed] [Google Scholar]
- 4.Gitelman S. E., Kobrin M. S., Ye J.-Q., Lopez A. R., Lee A., Derynck R. Recombinant Vgr-1/BMP-6-expressing tumors induce fibrosis and enchondral bone formation in vivo. J. Cell Biol. (1994);126:1595–1609. doi: 10.1083/jcb.126.6.1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Luu H. H., Song W. X., Luo X., Manning D., Luo J., Deng Z. L., Sharff K. A., Montag A. G., Haydon R. C., He T. C. Distinct roles of bone morphogenetic proteins in osteogenic differentiation of mesenchymal stem cells. J. Orthop. Res. (2007);25:665–677. doi: 10.1002/jor.20359. [DOI] [PubMed] [Google Scholar]
- 6.Miyazono K., Maeda S., Imamura T. BMP receptor signaling: transcriptional targets, regulation of signals, and signaling cross-talk. Cytokine Growth Factor Rev. (2005);16:251–263. doi: 10.1016/j.cytogfr.2005.01.009. [DOI] [PubMed] [Google Scholar]
- 7.Sieber C., Kopf J., Hiepen C., Knaus P. Recent advances in BMP receptor signaling. Cytokine Growth Factor Rev. (2009);20:343–355. doi: 10.1016/j.cytogfr.2009.10.007. [DOI] [PubMed] [Google Scholar]
- 8.Ten D. P., Korchynskyi O., Valdimarsdottir G., Goumans M. J. Controlling cell fate by bone morphogenetic protein receptors. Mol. Cell Endocrinol. (2003);211:105–113. doi: 10.1016/j.mce.2003.09.016. [DOI] [PubMed] [Google Scholar]
- 9.Luo J., Tang M., Huang J., He B. C., Gao J. L., Chen L., Zuo G. W., Zhang W., Luo Q., Shi Q., Zhang B. Q., Bi Y., Luo X., Jiang W., Su Y., Shen J., Kim S. H., Huang E., Gao Y., Zhou J. Z., Yang K., Luu H. H., Pan X., Haydon R. C., Deng Z. L., He T. C. TGFbeta/BMP type I receptors ALK1 and ALK2 are essential for BMP9-induced osteogenic signaling in mesenchymal stem cells. J. Biol. Chem. (2010);285:29588–29598. doi: 10.1074/jbc.M110.130518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wu N., Zhao Y., Yin Y., Zhang Y., Luo J. Identification and analysis of type II TGF-{beta} receptors in BMP-9-induced osteogenic differentiation of C3H10T1/2 mesenchymal stem cells. Acta. Biochim. Biophys. Sin. (Shanghai). (2010);42:699–708. doi: 10.1093/abbs/gmq075. [DOI] [PubMed] [Google Scholar]
- 11.Zhao M., Qiao M., Harris S. E., Chen D., Oyajobi B. O., Mundy G. R. The zinc finger transcription factor Gli2 mediates bone morphogenetic protein 2 expression in osteoblasts in response to hedgehog signaling. Mol. Cell Biol. (2006);26:6197–6208. doi: 10.1128/MCB.02214-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lyons K. M., Pelton R. W., Hogan B. L. M. Patterns of expression of murine Vgr-1 and BMP-2 RNA suggest that transforming growth factor-β-like genes coordinately regulate aspects of embryonic development. Genes Dev. (1989);3:1657–1668. doi: 10.1101/gad.3.11.1657. [DOI] [PubMed] [Google Scholar]
- 13.Celeste A. J., Iannazzi J. A., Taylor R. C., Hewick R. M., Rosen V., Wang E. A., Wozney J. M. Identification of transforming growth factor-β family members present in bone-inductive protein purified from bovine bone. Proc. Nat. Acad. Sci. U.S.A. (1990);86:9843–9847. doi: 10.1073/pnas.87.24.9843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Slobodan V., Lovorka G. BMP-6 and mesenchymal stem cell differentiation. Cytokine Growth Factor Rev. (2009);20:441–448. doi: 10.1016/j.cytogfr.2009.10.020. [DOI] [PubMed] [Google Scholar]
- 15.Ten Dijke P., Korchynskyi O., Valdimarsdottir G., Goumans M. J. Controlling cell fate by bone morphogenetic protein receptors. Mol. Cell Endocrinol. (2003);211:105–113. doi: 10.1016/j.mce.2003.09.016. [DOI] [PubMed] [Google Scholar]
- 16.Wu C. J., Lu H. K. Smad signal pathway in BMP-2-induced osteogenesis-a mini review. J. Dent. Sci. (2008);3:13–21. [Google Scholar]
- 17.Xu D. J., Zhao Y. Z., Wan J., He J. W., Weng Y. G., Luo J. Y. Smads, p38 and ERK1/2 are involved in BMP9-induced osteogenic differentiation of C3H10T1/2 mesenchymal stem cells. BMB Rep. (2012);45:247–252. doi: 10.5483/BMBRep.2012.45.4.247. [DOI] [PubMed] [Google Scholar]
- 18.Lavery K., Swain P., Falb D., Alaoui-Ismaili M. H. BMP-2/4 and BMP-6/7 differentially utilize cell surface receptors to induce osteoblastic differentiation of human bone marrow-derived mesenchymal stem cells. J. Biol. Chem. (2008);283:20948–20958. doi: 10.1074/jbc.M800850200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cao X., Chen D. The BMP signaling and in vivo bone formation. Gene. (2005);357:1–8. doi: 10.1016/j.gene.2005.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yamamoto H., Ueno H., Ooshima A., Takeshita A. Adenovirus-mediated transfer of a truncated transforming growth factor-beta (TGF-beta) type II receptor completely and specifically abolishes diverse signaling by TGF-beta in vascular wall cells in primary culture. J. Biol. Chem. (1996);271:16253–16259. doi: 10.1074/jbc.271.27.16253. [DOI] [PubMed] [Google Scholar]
- 21.Choi M. E., Ballermann B. J. Inhibition of capillary morphogenesis and associated apoptosis by dominant negative mutant transforming growth factor-beta receptors. J. Biol. Chem. (1995);270:21144–21150. doi: 10.1074/jbc.270.36.21144. [DOI] [PubMed] [Google Scholar]
- 22.Goumans M. J., Valdimarsdottir G., Itoh S., Lebrin F., Larsson J., Mummery C., Karlsson S., ten Dijke P. Activin receptor-like kinase (ALK)1 is an antagonistic mediator of lateral TGFb/ALK5 signaling. Mol. Cell. (2003);12:817–828. doi: 10.1016/S1097-2765(03)00386-1. [DOI] [PubMed] [Google Scholar]
- 23.Corinne B., Soazik P J., Jean-Yves P., Nathalie J., Nathalie di C. Role of type I receptors for anti- Mullerian hormone in the SMAT-1 sertoli cell line. Oncogen. (2005);24:4984–4992. doi: 10.1038/sj.onc.1208686. [DOI] [PubMed] [Google Scholar]
- 24.Chan M. C., Nguyen P. H., Davis B. N., Ohoka N., Hayashi H., Du K., Lagna G., Akiko H. A novel regulatory mechanism of the bone morphogenetic protein (BMP) signaling pathway involving the carboxyl-terminal tail domain of BMP type II receptor. Mol. Cell Biol. (2007);27:5776–5789. doi: 10.1128/MCB.00218-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mazerbourg S., Sangkuhl K., Luo C. W., Sudo S., Klein C., Hsueh A. J. Identification of receptors and signaling pathways for orphan bone morphogenetic protein/growth differentiation factor ligands based on genomic analyses. J. Biol. Chem. (2005);280:32122–32132. doi: 10.1074/jbc.M504629200. [DOI] [PubMed] [Google Scholar]
- 26.Ebisawa T., Tada K., Kitajima I., Tojo K., Sampath T. K., Kawabata M., Miyazono K., Imamura T. Characterization of bone morphogenetic protein-6 signaling pathways in osteoblast differentiation. J. Cell Sci. (1999);112:3519–3527. doi: 10.1242/jcs.112.20.3519. [DOI] [PubMed] [Google Scholar]
- 27.Peister A., Mellad J. A., Larson B. L., Hall B. M., Gibson L. F., Prockop D. J. Adult stem cells from bone marrow (MSCs) isolated from different strains of inbred mice vary in surface epitopes, rates of proliferation, and differentiation potential. Blood. (2004);103:1662–1668. doi: 10.1182/blood-2003-09-3070. [DOI] [PubMed] [Google Scholar]


