Abstract
Apoptosis, programmed cell death, is a process involved in the development and maintenance of cell homeostasis in multicellular organisms. It is typically accompanied by the activation of a class of cysteine proteases called caspases. Apoptotic caspases are classified into the initiator caspases and the executioner caspases, according to the stage of their action in apoptotic processes. Although caspase-3, a typical executioner caspase, has been studied for its mechanism and substrates, little is known of caspase-6, one of the executioner caspases. To understand the biological functions of caspase-6, we performed proteomics analyses, to seek for novel caspase-6 substrates, using recombinant caspase-6 and HepG2 extract. Consequently, 34 different candidate proteins were identified, through 2-dimensional electrophoresis/MALDI-TOF analyses. Of these identified proteins, 8 proteins were validated with in vitro and in vivo cleavage assay. Herein, we report that HAUSP, Kinesin5B, GEP100, SDCCAG3 and PARD3 are novel substrates for caspase-6 during apoptosis. [BMB Reports 2013; 46(12): 588-593]
Keywords: Apoptosis, Caspase-6, Degradomics, Proteomic screening, Substrate
INTRODUCTION
Apoptosis is a programmed cell death in multicellular organisms, employed for the development and maintenance of homeostasis, and removal of damaged cells (1). It has been demonstrated that caspases play a pivotal role in apoptosis (2). Caspases cleave their substrates after aspartic acid at P1 sites, by using cysteine residue in the catalytic site (3). P4-P1 residue in the target proteins determines the substrate specificity of caspases (4,5). In apoptosis, caspases are classified into initiator caspases and executioner caspases (6). Once the extrinsic death signal is given by the ligand-induced activation of death receptors, initiator caspase-8 and caspase-10 are activated by dimerization (7,8). The death signal is delivered by an internal factor, such as DNA damage, and the release of cytochrome c from the mitochondria induces the formation of the apoptosome. Initiator caspase-9 is recruited to the apoptosome complex, and activated by dimerization (9). In turn, this activated initiator caspases activate the executioner caspases. The activated executioner caspases cleave their specific target proteins (10). Executioner caspases include caspase-3, -7 and caspase-6. Among them, it has been well known that caspase-3 cleaves a large set of substrates, and plays a major role in apoptosis (11,12). Caspase-7, sharing the high sequence homology with caspase-3, recognizes the same cleavage site of caspase-3 (4,13), suggesting that caspase-3 and -7 have redundant roles (11). However, caspase-6 has somewhat unique roles as an executioner in apoptosis. Caspase-6 prefers leucine or valine at the P4 position, whereas caspase-3 and -7 prefer aspartate at the P4 position in target substrates (14,15). Therefore, caspase-6 has unique substrates that are not cleaved by either caspase-3 or caspase-7, such as lamin A/C (16), special AT-rich binding protein-1 (SATB1) (17), the neurodegenerative disease proteins Huntingtin (18), amyloid precursor protein (19), Presenilin 1, 2 (20), and DJ-1 (21). The most distinctive feature of caspase-6 is that it is activated by caspase-3 and -7 (22), and inversely activates caspase-3 and -7 (23). Also, caspase-6 acts as an initiator caspase, by processing initiator caspase-8 and -10 during apoptosis (22,24). Although the role of caspase-6 has been proposed in a number of reports, its substrates and its function are still unclear during apoptosis.
Thus, to better understand the biological roles of caspase-6 during apoptosis, we carried out proteomic analyses to seek for novel sets of caspase-6 substrates. In our present study, we report novel putative 34 substrates for caspase-6, and show that 5 of them, HAUSP, Kinesin5B, GEP100, SDCCAG3, and PARD3, are directly cleaved by caspase-6 during apoptosis.
RESULTS AND DISCUSSION
Screening of putative substrates for caspase-6 by proteomic approaches
To seek for novel caspase-6 substrates, we performed two-dimensional gel electrophoresis based proteomics analyses (Fig. 1A). HepG2, a human hepatoma cell line was used, since HepG2 cells have not been studied to screen for caspase substrates (14). To increase the possibility of finding more candidates in 2DE analyses, HepG2 cell extract was fractionated into cytosol, membrane and nucleus fractions, and each subcellular fraction was incubated with either recombinant active caspase-6 or catalytic mutant substituted 163-cysteine into serine in active site (inactive caspase-6). Then, incubated proteins were separated by isoelectric focusing and SDS-PAGE (Fig. 1A). The experiments were repeated three times, independently, to establish reproducibility. By comparing two-dimensional gel images of active caspase-6 and inactive caspase-6 mutant, appeared or disappeared protein spots were selected. Consequently, we found different 39 spots in cytosolic (Fig. 1B), 33 spots in membrane (Fig. 1C), and 62 spots in nucleus fraction (Fig. 1D). These protein spots were identified by MALDI-TOF- MS analysis. To obtain reliable results, proteins that have >95% confidence level in the Mascot program (Matrix Science) were selected. To determine whether the identified candidates were novel or not, we carried out a database search at The CASBAH (http://bioinf.gen.tcd.ie/casbah) and the CutDB, a proteolytic event database (http://cutdb.burnham.org), and a literature search at Pubmed. Finally, we obtained a list of novel candidates of caspase-6 substrates (Table 1). It is worth noting that the representative caspase-6 substrates, such as lamin A/C, vimentin, actin, tubulin, and ezrin, were identified in our screening system indicating that our screening method was functioning properly (Table 1).
Fig. 1. (A) The workflow for screening to identify novel caspase-6 substrates by proteomics approaches. (B-D) 2-DE gel images showing the differentially displayed spots. (B) Cytosol fractions, (C) membrane fractions, or (D) nucleus fractions, treated with either active caspase-6 or inactive casspase-6, were analyzed using 2-DE. The experiments were repeated for three times, independently. The representative gel images are shown.
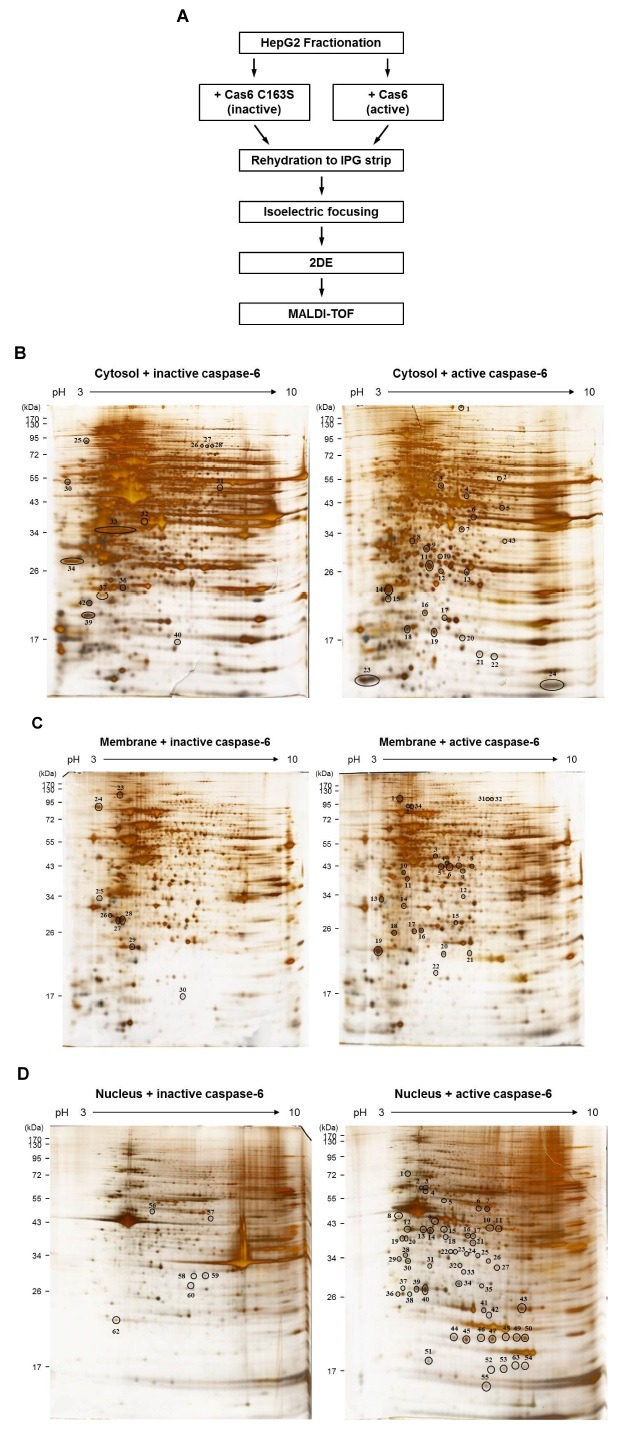
Table 1.
Identified proteins from 2DE-Proteomics and MALDI-TOF-MS analysis
| Fraction | Spot no. | Identified protein description | Symbol | Accession no. | Mascot scorea | Matched peptide | Sequence coverage (%) | Molecular weight (Da) | pI |
|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||
| Cytosol | 2 | Dual specificity protein kinase CLK3 | CLK3 | gi|194097436 | 67 | 13 | 28 | 74,267 | 9.94 |
| 5 | Tryptophanyl-Trna Synthetase | WARS | gi|50513261 | 66 | 13 | 27 | 43,586 | 7.12 | |
| 9 | Chain A, Human Ubiquitous Kinesin Motor Domain | Kinesin5B | gi|157830287 | 66 | 9 | 40 | 36,792 | 5.88 | |
| 11 | Actin, cytoplasmic 1 | Beta-actin | gi|4501885 | 748 | 24 | 29 | 42,052 | 5.29 | |
| 12 | Alpha-actin | Alpha-actin | gi|178027 | 118 | 2 | 7 | 42,480 | 5.23 | |
| 21 | GTP-binding protein Rit1 isoform 2 | Rit1 | gi|5902050 | 67 | 10 | 55 | 25,357 | 9.2 | |
| 24 | Serologically defined colon cancer antigen 3 | SDCCAG3 | gi|55961008 | 66 | 9 | 69 | 18,921 | 5.74 | |
| Membrane | 2 | IQ motif and Sec7 domain 1 | GEP100 | gi|119584552 | 66 | 15 | 24 | 121,073 | 9.2 |
| 8 | Herpesvirus associated ubiquitin-specific protease | HAUSP | gi|1545952 | 70 | 20 | 20 | 129,274 | 5.33 | |
| 10 | Tubulin, gamma complex associated protein 2 variant | TUBGCP2 | gi|62898904 | 67 | 16 | 27 | 103,011 | 6.38 | |
| Nucleus | 3 | Alpha-tubulin | Alpha-tubulin | gi|340021 | 90 | 14 | 35 | 50,804 | 4.94 |
| 5 | Antigen identified by monoclonal antibody Ki-67 | Ki-67 | gi|119569563 | 76 | 32 | 14 | 360,616 | 9.48 | |
| 6 | Translin-associated factor X interacting protein 1 | TSNAX | gi|119603582 | 67 | 14 | 25 | 72,374 | 5.11 | |
| 9 | Dynein, axonemal, heavy polypeptide 1 | DNAH1 | gi|119585621 | 70 | 32 | 10 | 497,514 | 5.62 | |
| 10 | Gephyrin | Gephyrin | gi|16605466 | 67 | 14 | 33 | 80,410 | 5.25 | |
| 11 | Olfactomedin-like protein 1 precursor | OLFL1 | gi|284172520 | 66 | 11 | 41 | 46,378 | 8.29 | |
| 12 | Alcohol dehydrogenase 4 | ADH4 | gi|825623 | 67 | 9 | 41 | 41,094 | 8.25 | |
| 13 | Actin, beta, partial | Beta-actin | gi|14250401 | 70 | 13 | 52 | 41,321 | 5.56 | |
| 15 | Peroxisomal bifunctional enzyme isoform 1 | EHHADH | gi|68989263 | 66 | 12 | 25 | 80,072 | 9.24 | |
| 16 | PR domain containing 5 | PRDM5 | gi|119625672 | 66 | 12 | 36 | 50,017 | 9.49 | |
| 20 | Regulatory factor X, 5 (influences HLA class II expression) | RFX5 | gi|122889236 | 67 | 13 | 31 | 63,473 | 9.41 | |
| 21 | KIAA0524SARM protein | - | gi|7711002 | 67 | 12 | 26 | 76,260 | 8.58 | |
| 22 | Partitioning-defective 3-like protein splice variant c | PARD3 | gi|18874468 | 66 | 16 | 19 | 125,435 | 8.57 | |
| 24 | Dynein, axonemal, heavy polypeptide 1 | DNAH1 | gi|119585622 | 67 | 21 | 12 | 280,703 | 5.74 | |
| 25 | Kinesin-like protein KIF16B | KIF16B | gi|41327691 | 74 | 23 | 19 | 152,488 | 5.86 | |
| 25 | Ezrin | Ezrin | gi|46249758 | 64 | 17 | 25 | 69,313 | 5.94 | |
| 26 | Cytoplasmic linker associated protein 1 | CLASP1 | gi|119615659 | 66 | 15 | 23 | 102,275 | 9.53 | |
| 27 | Adenosine deaminase | ADA | gi|1197210 | 67 | 11 | 42 | 35,335 | 5.6 | |
| 28 | Herpesvirus associated ubiquitin-specific protease | HAUSP | gi|1545952 | 112 | 26 | 24 | 129,274 | 5.33 | |
| 29 | Alpha tubulin | Alpha-tubulin | gi|109093209 | 72 | 13 | 40 | 46,767 | 5.01 | |
| 30 | Bullous pemphigoid antigen 1 | BPAG1 | gi|27923959 | 69 | 34 | 12 | 374,544 | 6.38 | |
| 31 | Vimentin | VIM | gi|62414289 | 70 | 16 | 27 | 53,676 | 5.06 | |
| 33 | Inter-alpha-trypsin inhibitor family heavy chain-related protein | IHRP | gi|1483187 | 97 | 21 | 28 | 103,549 | 6.51 | |
| 34 | Lamin A/C | LMNA | gi|21619981 | 193 | 27 | 40 | 53,222 | 6.03 | |
| 35 | Lamin A/C | LMNA | gi|21619981 | 107 | 19 | 30 | 53,222 | 6.03 | |
| 37 | GT mitochondrial solute carrier protein homologue; putative, partial | - | gi|386960 | 66 | 9 | 37 | 38,645 | 9.9 | |
| 38 | MIF4G domain-containing protein isoform 2 | MIF4GD | gi|23510352 | 66 | 7 | 33 | 29,941 | 5.29 | |
| 39 | Actin, cytoplasmic 2-like | ACTG1 | gi|354468985 | 111 | 17 | 62 | 28,478 | 5.2 | |
| 40 | Actin, beta | Beta-actin | gi|14250401 | 126 | 19 | 44 | 41,321 | 5.56 | |
| 43 | Glutaryl-Coenzyme A dehydrogenase | GCDH | gi|119604731 | 67 | 11 | 38 | 47,923 | 8.73 | |
| 47 | Malate dehydrogenase 1B | MDH1 | gi|89886456 | 71 | 13 | 22 | 59,013 | 5.85 | |
| 49 | ATP synthase, H+ transporting, mitochondrial F1 complex, alpha subunit 1, cardiac muscle | ATP5A | gi|15030240 | 66 | 13 | 28 | 59,886 | 9.07 | |
| 52 | Protein FAM73A | FAM73A | gi|38348384 | 67 | 13 | 33 | 71,930 | 5.42 | |
| 56 | Rab GDP dissociation inhibitor beta isoform 1 | GDI2 | gi|6598323 | 67 | 10 | 44 | 51,087 | 6.11 | |
| 57 | WD repeat domain 18 | WDR18 | gi|12804481 | 67 | 11 | 28 | 47,960 | 6.22 | |
| 58 | Coiled-coil domain-containing protein 148 | CCDC148 | gi|76779830 | 66 | 8 | 42 | 33,401 | 7.63 | |
| 63 | ATP/GTP binding protein-like 3 | AGBL3 | gi|22658305 | 67 | 14 | 31 | 73,571 | 7.22 | |
aproteins scores are significant at 95% confidence level.
HAUSP, Kinesin5B, GEP100, SDCCAG3 and PARD3 are directly cleaved by caspase-6 in vitro
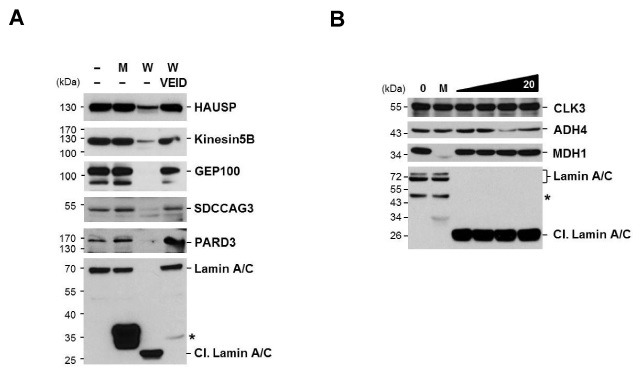
In order to confirm whether the candidate proteins listed in Table 1 would be cleaved by caspase-6, we performed the in vitro cleavage assay. HepG2 cell extracts were incubated with either active caspase-6, or catalytic mutant caspase-6. The cleavage was detected by western blotting, using the specific antibodies. Eight candidate proteins (HAUSP, Kinesin 5B, GEP100, SDCCAG3, PARD3, CLK3, ADH4, and MDH1), of which commercial antibodies are available, were tested. Lamin A/C, a well known caspase-6 substrate, was used as a positive control. As a result, the level of HAUSP, Kinesin 5B, GEP100, SDCCAG3 and PARD3 reduced only when active caspase-6 was treated. This indicates that these proteins were cleaved directly by caspase-6, according to Lamin A/C cleavage (Fig. 2A). However, these cleavages were blocked in the presence of caspase-6 inhibitor, Z-VEID-FMK (Fig. 2A). Therefore, these results indicate that HAUSP, Kinesin5B, GEP100, SDCCAG3 and PARD3 are directly cleaved by caspase-6. On the other hand, CLK3, ADH4 and MDH1 were not cleaved by caspase-6, even though twice the amounts of caspase-6 were used (Fig. 2B), implying that CLK3, ADH4 and MDH1 are not direct substrates for caspase-6.
Fig. 2. Cleavage of caspase-6 substrate candidates was confirmed by in vitro cleavage assay. (A) HepG2 extract was incubated with active caspase-6 (10 U) or inactive capsase-6 (10 U) in the presence or absence of 30 μM Z-VEID-FMK. The reaction mixture was analyzed by immunoblotting with the specific antibodies against (A) HAUSP, Kinesin5B, GEP100, SDCCAG3, PARD3 and Lamin A/C. (B) HepG2 extract was incubated with increasing amount of active caspase-6 (5, 10, 15, 20 U) or inactive capsase-6 (20 U). The reaction mixture was analyzed by immunoblotting with the specific antibodies against CLK3, ADH4, MDH1, and Lamin A/C (Cl, cleaved Lamin A/C). Lamin A/C cleavage was used as a positive control. The asterisk indicates a nonspecific band. The representative blot of at least two independent experiments is shown. 1 U is the amount of enzyme required to cleave 1nmole of caspase substrate completely in 1 hr.
Candidate proteins were proteolytically processed by caspases during apoptosis
Then we performed the in vivo cleavage assay to verify whether the candidates would be cleaved in vivo during apoptosis. HeLa cells were treated with staurosporine (STS), which induces apoptosis by inhibiting cellular protein kinase (25). Apoptosis was confirmed by observing of PARP cleavage (Fig. 3A and 3B), which is a well-established apoptosis hallmark, and is also a caspase-3 and caspase-7 substrate (26,27). To examine the specificity of caspases, three different caspase inhibitors (Z-VEID-FMK; caspase-6 inhibitor, Z-DEVD-FMK; caspase-3/-7 inhibitor and Z-VAD-FMK; pan-caspase inhibitor) were treated together with STS. In STS stimulated HeLa cells, all candidate proteins (HAUSP, Kinesin5B, GEP100, SDCCAG3, and PARD3) were processed (Fig. 3A). In the presence of Z-VEID-FMK and STS, the cleavage of Kinesin5B and GEP100 was blocked, and recovered to the level of DMSO treated (Fig. 3A). Meanwhile, the cleavage of HAUSP, SDCCAG3, and PARD3 was inhibited slightly. Upon Z-DEVD-FMK and STS treatment, the cleavages of Kinesin5B, GEP100, and SDCCAG3 were inhibited completely, but HAUSP and PARD3 were still cleaved, but less than STS alone (Fig. 3A). Upon STS and Z-VAD-FMK treatment, the cleavage of all candidate proteins was inhibited completely (Fig. 3A). Accordingly, these data suggest that HAUSP, Kinesin5B, GEP100, SDCCAG3, and PARD3 are cleaved somewhat, not only by caspase-6, but also by other caspases, and their cleavages seem not to be mutually exclusive in vivo.
Fig. 3. Cleavage of caspase-6 substrate candidates was confirmed by in vivo cleavage assay. HeLa cells were treated with 1 μM STS for 24 hr in the presence or absence of the 100 μM caspase inhibitors (Z-VEID-FMK; caspase-6 inhibitor, Z-DEVD-FMK; caspase-3/-7 inhibitor, Z-VAD-FMK; pan-caspase inhibitor) pretreated to cells for 3 hr. After 24 hr, cells were harvested and lysed. Cell lysates were analyzed by immunoblotting with the specific antibodies against (A) HAUSP, Kinesin5B, GEP100, SDCCAG3, PARD3, Lamin A/C (Cl, cleaved Lamin A/C), and PARP (Cl, cleaved PARP); and (B) CLK3, ADH4, MDH1, Lamin A/C, and PARP. A coomassie blue stain of PVDF membrane was the loading control. The representative blot of at least two independent experiments is shown.
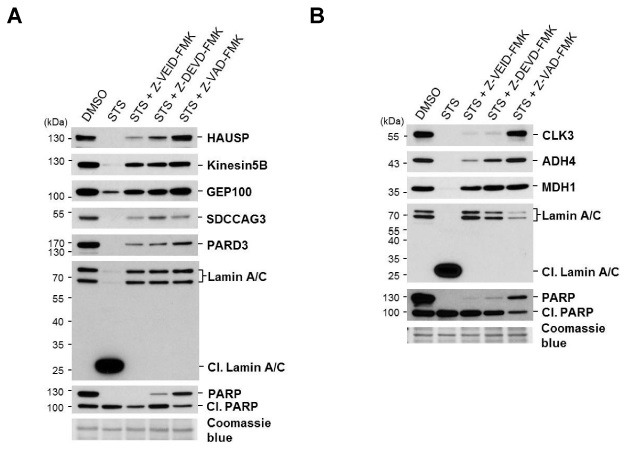
We also checked the rest of the candidates that were not cleaved directly by caspase-6, in the same way (Fig. 3B). CLK3 and ADH4 were proteolytically processed upon STS treatment, but these cleavages were slightly blocked by Z-VEID-FMK or Z-DEVD-FMK, and completely blocked by Z-VAD-FMK. Meanwhile the cleavage of MDH1 was almost completely inhibited by either Z-VEID-FMK or Z-DEVD-FMK. Taken together, CLK3, ADH4, and MDH1 are processed during apoptosis, but they are not direct substrates for caspase-6.
Among the identified caspase-6 substrates, HAUSP is a deubiquitinating enzyme that removes ubiquitin from p53 and MDM2 (28). HAUSP plays a role in cell survival, proliferation and apoptosis, through regulating the p53 level (28). It has been reported that HAUSP is processed in a caspase-dependent manner during thymocyte apoptosis (29). In our study, we demonstrate that HAUSP is directly processed by caspase-6 in human cells, the cervical cancer cell HeLa (Fig. 2A and 3A) and hepatocarcinoma cell HepG2 (data not shown), indicating that HAUSP is caspase-6 specific substrate, and is not cell type specific. Another identified caspase-6 substrate, Kinesin 5B is microtubule associated motor protein transporting vesicles along the microtubule (30). It has been reported that kinectin, kinesin receptor protein, is cleaved by caspase, and the transport system becomes destroyed during apoptosis (31). Based on our results, caspase-6 indeed cleaves kinesin 5B transporting cellular cargoes, as well as cytoskeletal protein, such as tubulin and actin, suggesting that caspase-6 might be involved in the disruption of intracellular vesicle transport process, by inactivating kinesin 5B.
In conclusion, we identified 34 putative caspase-6 substrates, through 2-dimesional electrophoresis/MALDI-TOF analyses. Furthermore, we confirmed that HAUSP, Kinesin 5B, GEP100, SDCCAG3 and PARD3 are directly cleaved by caspase-6 during apoptosis. These newly identified substrates seem to be involved in membrane trafficking, cell polarization, and p53 stability, suggesting that our results may provide a clue to understanding the roles of caspase-6 in apoptosis.
MATERIALS AND METHODS
Cell culture
HeLa and HepG2 were maintained in Dulbecco’s Modified Eagle’s Medium (DMEM, Gibco, USA), supplemented with 10% fetal bovine serum (FBS, Gibco, USA) in a humidified atmosphere of 5% CO2 in air, at 37℃. HepG2 cells were fractionated using Qproteome Cell Compartment Kit (QIAGEN, USA), according to the manufacture’s instruction.
Antibodies and reagents
Anti-PARP (#9542), anti-Lamin A/C (#2032), and anti-HAUSP (#3277) were purchased from Cell Signaling (USA). Anti-ADH4 (ab83819), anti-CLK3 (ab54683), anti-Kinesin5B (ab42492), anti-PARD3 (ab40769), and anti-SDCCAG3 (ab97666) were from Abcam (USA). Anti-GEP100 (G4798) was from Sigma. Anti- MDHC (sc-166879) was from Santa Cruz (USA). Staurosporine (#1048-1) was from BioVision (CA). Z-VEID-FMK (FMK006), Z-DEVD-FMK (FMK004), and Z-VAD-FMK (FMK001) were purchased from R&D System (USA).
Cloning and mutagenesis of caspase-6
Δ23 caspase-6, prodomain deleted mutant, was subcloned to pET21a(+), using BamH I and Xho I sites. This construct was mutated using the QuikChange Site-Directed Mutagenesis Kit (Stratagene, CA), according to the manufacture’s instructions, with the following oligonucleotides: Caspase-6 Forward C163S 5’-TTTATCATTCAGGCAAGTCGGGGAAACCAGCAC-3’, Caspase-6 Reverse C163S 5’-GTGCTGGTTTCCCCGACTTGCCTGAATGATAAA-3’. The constructs were confirmed by sequencing at the Solgent (Daejeon, Korea).
Expression and purification of recombinant caspase-6
Recombinant Δ23 caspase-6 and Δ23 caspase-6 C163S with C-terminal His-tag were expressed and purified from E. coli BL21 codon plus (DE3) RIL (stratagene, CA), using affinity chromatography. In brief, cells were lysed in the lysis buffer (50 mM NaH2PO4, 500 mM NaCl, pH 8.0) with 0.5% sarcosine and EDTA free protease inhibitor (Roche, Germany). The lysate was clarified by centrifuging at 13,000 rpm, 4℃ for 15min and the supernatant was incubated with Ni-NTA agarose beads (QIAGEN, USA). After extensive washing, bound proteins were eluted with the lysis buffer containing 250 mM imidazole. Protein concentration was determined by the Bradford (Bio-Rad, USA) protein assay.
2-DE analysis and MALDI-TOF-MS
The protein samples were dissolved in 9 M urea, 2 M thiourea, 4% CHAPS, 50 mM DTT, 0.003% bromophenol blue, and 2% IPG buffer pH3-10 (GE healthcare, USA). The dissolved samples were applied onto immobilized 18 cm, pH 3-10 gradient strips (GE healthcare, USA). Strips were rehydrated, and isoelectric focusing was performed in a stepwise fashion (0:01 h at 300 V; 1:30 h at 3,500 V; 13:00 hr at 3,500 V). After the focused strips were equilibrated with DTT and iodoacetamide equilibration solution for 15 min respectively, the second-dimensional gel electrophoresis was performed on 10% SDS-PAGE. The gels were silver-stained, using PlusOne Silver Staining Kit, Protein (GE healthcare, USA), according to the manufacturer’s instructions. For mass spectrometry analysis, silver stained proteins were excised from the gels. The spots were destained and digested with trypsin, and the peptides from the spot were analyzed by MALDI-TOF, at Yonsei Proteome Research Center (Seoul, Korea). The database search was performed at The CASBAH and CutDB. The CASBAH (The CAspase Substrate DataBAse Homepage) is an online database containing all of the reported mammalian caspase substrates (32). CutDB is an online proteolytic event database that provides the list of protease and corresponding substrates and cleavage sites (33).
In vitro cleavage assay
HepG2 cells were lysed using NP40 lysis buffer (137 mM NaCl, 20 mM Tris-HCl pH 8.0, 10% Glycerol, 1% NP40, 2 mM EDTA) containing protease inhibitor (Roche, Germany). Cell lysates were incubated with either recombinant Δ23 caspase-6 protein or Δ23 caspase-6 C163S protein, at 37℃ for 24 hr. Reactions were stopped by adding Laemmli buffer, and were boiled at 100℃ for 5 min.
Acknowledgments
We thank S. H. Kim and K. H. Bae for critical reading of manuscript. This work was supported by KRIBB, and research grants from the National Research Foundation of Korea (NRF-2011-0028172).
References
- 1.Ellis R. E., Yuan J. Y., Horvitz H. R. Mechanisms and functions of cell death. Annu. Rev. Cell Biol. (1991);7:663–698. doi: 10.1146/annurev.cb.07.110191.003311. [DOI] [PubMed] [Google Scholar]
- 2.Raff M. C., Barres B. A., Burne J. F., Coles H. S., Ishizaki Y., Jacobson M. D. Programmed cell death and the control of cell survival: lessons from the nervous system. Science. (1993);262:695–700. doi: 10.1126/science.8235590. [DOI] [PubMed] [Google Scholar]
- 3.Alnemri E. S., Livingston D. J., Nicholson D. W., Salvesen G., Thornberry. N. A., Wong W. W., Yuan J. Human ICE/CED-3 protease nomenclature. Cell. (1996);87:171. doi: 10.1016/S0092-8674(00)81334-3. [DOI] [PubMed] [Google Scholar]
- 4.Lavrik I. N., Golks A., Krammer P. H. Caspases: pharmacological manipulation of cell death. J. Clin. Invest. (2005);115:2665–2672. doi: 10.1172/JCI26252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Thornberry N. A., Rano T. A., Peterson E. P., Rasper D. M., Timkey T., Garcia-Calvo M., Houtzaqer V. M., Nordstrom P. A., Roy S., Vaillancourt J. P., Chapman K. T., Nicholson D. W. A combinatorial approach defines specificities of members of the caspase family and granzyme B. Functional relationships established for key mediators of apoptosis. J. Biol. Chem. (1997);272:17907–17911. doi: 10.1074/jbc.272.29.17907. [DOI] [PubMed] [Google Scholar]
- 6.Nicholson D. W. Caspase structure, proteolytic substrates, and function during apoptotic cell death. Cell Death Differ. (1999);6:1028–1042. doi: 10.1038/sj.cdd.4400598. [DOI] [PubMed] [Google Scholar]
- 7.Peter M. E., Krammer P. H. The CD95 (APO-1/Fas) DISC and beyond. Cell Death Differ. (2003);10:26–35. doi: 10.1038/sj.cdd.4401186. [DOI] [PubMed] [Google Scholar]
- 8.Lee E., Seo J., Jeong M., Lee S., Song J. The roles of FADD in extrinsic apoptosis and necrosis. BMB Rep. (2012);45:496–508. doi: 10.5483/BMBRep.2012.45.9.186. [DOI] [PubMed] [Google Scholar]
- 9.Kwon K. B., Kim E. K., Shin B. C., Seo E. A., Yang J. Y., Ryu D. G. Herba houttuyniae extract induces apoptotic death of human promyelocytic leukemia cells via caspase activation accompanied by dissipation of mitochondrial membrane potential and cytochrome c release. Exp. Mol. Med. (2003);35:91–97. doi: 10.1038/emm.2003.13. [DOI] [PubMed] [Google Scholar]
- 10.Thornberry N. A., Lazebnik Y. Caspases: enemies within. Science. (1998);281:1312–1316. doi: 10.1126/science.281.5381.1312. [DOI] [PubMed] [Google Scholar]
- 11.Lamkanfi M., Kanneqanti T. D. Caspase-7: a protease involved in apoptosis and inflammation. Int. J. Biochem. Cell Biol. (2010);42:21–24. doi: 10.1016/j.biocel.2009.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee A. Y., Park B. C., Jang M., Cho S., Lee D. H., Lee S. C., Myung P. K., Park S. G. Identification of caspase-3 degradome by two-dimensional gel electrophoresis and matrix-assisted laser desorption/ionization-time of flight analysis. Proteomics. (2004);4:3429–3436. doi: 10.1002/pmic.200400979. [DOI] [PubMed] [Google Scholar]
- 13.Jang M., Park B. C., Kang S., Lee D. H., Cho S., Lee S. C., Bae K. H., Park S. G. Mining of caspase-7 substrates using a degradomic approach. Mol. Cells. (2008);26:152–157. [PubMed] [Google Scholar]
- 14.Crawford E. D., Wells J. A. Caspase substrates and cellular remodeling. Annu. Rev. Biochem. (2011);80:1055–1087. doi: 10.1146/annurev-biochem-061809-121639. [DOI] [PubMed] [Google Scholar]
- 15.Lamkanfi M., Declercq W., Kalai M., Saelens X., Vandenabeele P. Alice in caspase land. A phylogenetic analysis of caspases from worm to man. Cell Death Differ. (2002);9:358–361. doi: 10.1038/sj.cdd.4400989. [DOI] [PubMed] [Google Scholar]
- 16.Takahashi A., Alnemri E. S., Lazebnik Y. A., Fernandes-Alnemri T., Litwack G., Moir R. D., Goldman R. D., Poirier G. G., Kaufmann S. H., Earnshaw W. C. Cleavage of lamin A by Mch2α but not CPP32: Multiple interleukin 1β-converting enzyme-related proteases with distinct substrate recognition properties are active in apoptosis. Proc. Natl. Acad. Sci. U. S. A. (1996);93:8395–8400. doi: 10.1073/pnas.93.16.8395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Galande S., Dickinson L. A., Mian I. S., Sikorska M., Kohwi-Shigematsu T. SATB1 cleavage by caspase 6 disrupts PDZ domain-mediated dimerization, causing detachment from chromatin early in T-cell apoptosis. Mol. Cell Biol. (2001);21:5591–5604. doi: 10.1128/MCB.21.16.5591-5604.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Graham R. K., Deng Y., Slow E. J., Haigh B., Bissada N., Lu G., Pearson J., Shehadeh J., Bertram L., Murphy Z., Warby S. C., Doty C. N., Roy S., Wellington C. L., Leavitt B. R., Raymond L. A., Nicholson D. W., Hayden M. R. Cleavage at the caspase-6 site is required for neuronal dysfunction and degeneration due to mutant huntingtin. Cell. (2006);125:1179–1191. doi: 10.1016/j.cell.2006.04.026. [DOI] [PubMed] [Google Scholar]
- 19.Pellegrini L., Passer B. J., Tabaton M., Ganjei J. K., D’Adamio L. Alternative, non-secretase processing of Alzheimer's beta-amyloid precursor protein during apoptosis by caspase-6 and -8. J. Biol.Chem. (1999);274:21011–21016. doi: 10.1074/jbc.274.30.21011. [DOI] [PubMed] [Google Scholar]
- 20.van de Craen M., de Jonghe C., van den Brande I., Declercq W., van Gassen G., van Criekinge W., Vanderhoeven I., Fiers W., van Broeckhoven C., Hendriks L., Vandenabeele P. Identification of caspases that cleave presenilin-1 and presenilin-2. Five presenilin-1 (PS1) mutations do not alter the sensitivity of PS1 to caspases. FEBS Lett. (1999);445:149–154. doi: 10.1016/S0014-5793(99)00108-8. [DOI] [PubMed] [Google Scholar]
- 21.Giaime E., Sunyach C., Druon C., Scarzello S., Robert G., Grosso S., Auberger P., Goldberg M. S., Shen J., Heutink P., Pouysségur J., Paqés G., Checler F., Alves da Costa C. Loss of function of DJ-1 triggered by Parkinson's disease-associated mutation is due to proteolytic resistance to caspase-6. Cell Death Differ. (2010);17:158–169. doi: 10.1038/cdd.2009.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Inoue S., Browne G., Melino G., Cohen G. M. Ordering of caspases in cells undergoing apoptosis by the intrinsic pathway. Cell Death Differ. (2009);16:1053–1061. doi: 10.1038/cdd.2009.29. [DOI] [PubMed] [Google Scholar]
- 23.Allsopp T. E., McLuckie J., Kerr L. E., Macleod M., Sharkey J., Kelly J. S. Caspase 6 activity initiates caspase 3 activation in cerebellar granule cell apoptosis. Cell Death Differ. (2000);7:984–993. doi: 10.1038/sj.cdd.4400733. [DOI] [PubMed] [Google Scholar]
- 24.Cowling V., Downward J. Caspase-6 is the direct activator of caspase-8 in the cytochrome c-induced apoptosis pathway: absolute requirement for removal of caspase-6 prodomain. Cell Death Differ. (2002);9:1046–1056. doi: 10.1038/sj.cdd.4401065. [DOI] [PubMed] [Google Scholar]
- 25.Son Y. K., Hong D. H., Kim D., Firth A. L., Park W. S. Direct effect of protein kinase C inhibitors on cardiovascular ion channels. BMB Rep. (2011);44:559–565. doi: 10.5483/BMBRep.2011.44.9.559. [DOI] [PubMed] [Google Scholar]
- 26.Slee E. A., Adrain C., Martin S. J. Executioner caspase-3, -6, and -7 perform distinct, non-redundant roles during the demolition phase of apoptosis. J. Biol. Chem. (2001);276:7320–7326. doi: 10.1074/jbc.M008363200. [DOI] [PubMed] [Google Scholar]
- 27.Germain M., Affar E. B., D’Amours D., Dixit V. M., Salvesen G. S., Poirier G. G. Cleavage of automodified poly(ADP-ribose) polymerase during apoptosis. Evidence for involvement of caspase-7. J. Biol. Chem. (1999);274:28379–28384. doi: 10.1074/jbc.274.40.28379. [DOI] [PubMed] [Google Scholar]
- 28.Li M., Chen D., Shiloh A., Lou J., Nikolaev A. Y., Qin J., Gu W. Deubiquitination of p53 by HAUSP is an important pathway for p53 stabilization. Nature. (2002);416:648–653. doi: 10.1038/nature737. [DOI] [PubMed] [Google Scholar]
- 29.Vugmeyster Y., Borodovsky A., Maurice M. M., Maehr R., Furman M. H., Ploegh H. L. The ubiquitin-proteosome pathway in thymocyte apoptosis: caspase-dependent processing of the deubiquitinating enzyme USP (HAUSP). Mol. Immunol. (2002);39:431–441. doi: 10.1016/S0161-5890(02)00123-2. [DOI] [PubMed] [Google Scholar]
- 30.Brady S. T. A novel brain ATPase with properties expected for the fast axonal transport motor. Nature. (1985);317:73–75. doi: 10.1038/317073a0. [DOI] [PubMed] [Google Scholar]
- 31.Machleidt T., Geller P., Schwandner R., Scherer G., Kroënke M. Caspase 7-induced cleavage of kinectin in apoptotic cells. FEBS Lett. (1998);436:51–54. doi: 10.1016/S0014-5793(98)01095-3. [DOI] [PubMed] [Google Scholar]
- 32.Lüthi A. U., Martin S. J. The CASBAH: a searchable database of caspase substrates. Cell Death. Differ. (2007);14:641–650. doi: 10.1038/sj.cdd.4402103. [DOI] [PubMed] [Google Scholar]
- 33.Igarashi Y., Eroshkin A., Gramatikova S., Gramatikoff K., Zhang Y., Smith J. W., Osterman A. L., Godzik A. CutDB: a proteolytic event database. Nucleic Acids Res. (2007);35:D546–549. doi: 10.1093/nar/gkl813. [DOI] [PMC free article] [PubMed] [Google Scholar]


