Abstract
Background
Cetuximab, a monoclonal blocking antibody against the epidermal growth factor receptor EGFR, has been approved for the treatment of squamous cell carcinomas of the head and neck (HNSCC). However, only few patients display long-term responses, prompting the search for cetuximab resistance mechanisms and new therapeutic options enhancing cetuximab effectiveness.
Methods
Cetuximab-sensitive HNSCC cells were retro-engineered to express PIK3CA and RAS oncogenes. These cells and HNSCC cells harboring endogenous PIK3CA and RAS oncogenes were xenografted into mice (n = 10 per group) and studied for their biochemical, antitumor, antiangiogenic, and antilymphangiogenic responses to cetuximab and mTOR targeting agents. All P values are two-sided.
Results
Cetuximab treatment of PIK3CA- and RAS-expressing HNSCC xenografts promoted an initial antitumor response, but all tumors relapsed within few weeks. In these tumors, cetuximab did not decrease the activity of mTOR, a downstream signaling target of EGFR, PIK3CA, and RAS. The combined administration of cetuximab and mTOR inhibitors exerted a remarkably increased antitumor activity, particularly in HNSCC cells that are resistant to cetuximab as a single agent. Indeed, cotargeting mTOR together with cetuximab caused a rapid tumor collapse of both PIK3CA- and RAS-expressing HNSCC xenografts (P < .001), concomitant with reduced proliferation (P < .001) and lymphangiogenesis (P < .001).
Conclusion
The presence of PIK3CA and RAS mutations and other alterations affecting the mTOR pathway activity in HNSCC could be exploited to predict the potential resistance to cetuximab, and to select the patients that may benefit the most from the concomitant administration of cetuximab and PI3K and/or mTOR inhibitors as a precision molecular therapeutic option for HNSCC patients.
Squamous cell carcinomas of head and neck (HNSCC), which arise in the oral cavity, oropharynx, larynx, and hypopharynx, are a major public health concern. New therapeutic strategies to prevent and treat HNSCC patients are urgently needed. The epidermal growth factor receptor (EGFR) is overexpressed in up to 90% of HNSCC lesions (1,2) and is associated with unfavorable clinical outcome (3,4). Anti-EGFR targeted therapies have been shown to be effective in a variety of preclinical HNSCC models (5–8). Furthermore, in seminal clinical studies, cetuximab, a humanized IgG1 monoclonal antibody against the EGFR extracellular domain, prolonged the median overall survival and reduced disease progression in advanced HNSCC patients as part of combination therapies with radiation and chemotherapy (9,10). Based on these findings, cetuximab gained approval from the US Food and Drug Administration for use together with radiation or as a single agent in patients that failed to respond to platinum-based therapy, and for recurrent or metastatic HNSCC in combination with standard chemotherapy (10).
However, the overall increased response of adding cetuximab to radiation and/or chemotherapy is approximately 10% to 20% (9,10), much lower than initially expected considering the high level of EGFR expression in HNSCC. Recent studies have identified multiple mechanisms of resistance to cetuximab, including EGFR mutations, overexpression of EGFR ligands, amplification or transactivation of HER family members or the MET receptor, and deregulated EGFR recycling (11–17), all of which could explain the intrinsic or acquired resistance to cetuximab in the clinic.
EGFR regulates multiple intracellular signaling circuits, including the JAK/STAT3, RAS/MAPK, and PI3K/AKT/mTOR pathways (18–20). Among them, recent findings indicate that multiple genetic and epigenetic alterations converge on the persistent activation of PI3K/AKT/mTOR signaling in most HNSCC lesions (21–24). Thus, we asked here whether genetic alterations causing PI3K/AKT/mTOR activation can promote cetuximab resistance in HNSCC, and if so whether pharmacological inhibition of this signaling pathway represents a suitable target to prevent or overcome cetuximab resistance in HNSCC.
Methods
Cell Lines, Tissue Culture, Lentivirus, Reagents, and Tissue and Immunoblot Analysis
Cell lines, cell culture conditions, and procedures are described in detail in the supplementary Materials and Methods (available online). Briefly, Cal27 cells stably expressing an activated allele of PIK3CA and RAS were generated by infection with pLESIP HA-PIK3CA H1047A or pLESIP GFP-KRAS G12V lentiviruses. Cetuximab solution was purchased from Imclone LLC (Bridgewater, NJ). Rapamcyin and Rad001 were from LC Laboratories (Woburn, MA). All other chemicals and reagents were from Sigma-Aldrich (St. Louis, MO) unless indicated.
In Vivo Mouse Experiments and Analysis
All the mice studies were carried out according to National Institutes of Health (NIH) approved protocols (ASP # 10–569 and 13–695) in compliance with the NIH Guide for the Care and Use of Laboratory Mice. To establish tumor xenografts, cells were transplanted into the flanks of athymic nude mice (female, four to six weeks old, obtained from NCI/Frederick, MD), and when the tumor volume reached approximately 200mm3, the mice were randomized into groups and treated by intraperitoneal injection (ip) with cetuximab (40mg/kg, three times a week), rapamycin and Rad001 (5mg/kg/day), or control diluent (10 mice per group). The mice were euthanized at the indicated time points and tumors isolated for histologic and immunohistochemical evaluation.
Tumor bearing mice were randomized into groups and treated by intraperitoneal injection (ip) with cetuximab, rapamycin and Rad001, or control diluent, killed at the indicated time points, and tumors isolated for histologic and immunohistochemical evaluation.
Statistical Analysis
Data analysis was performed with GraphPad Prism version 6 for Windows (GraphPad Software, San Diego, CA). The differences between experimental groups in tumor weight, proliferation, and microvessel density, as well as viability were analyzed using the nonparametric Kruskall-Wallis test, with multiple comparisons to the control group. The Dunn’s correction was used to adjust for multiple comparisons. P values were adjusted correspondingly. P values of less than .05 were considered statistically significant. The tumor growth curves were compared by the longitudinal data analysis method. All statistical tests were two-sided.
Results
Effect of mTOR Inhibitors on the Response to Cetuximab in HNSCC Tumor Xenografts
To begin exploring mechanisms involved in intrinsic cetuximab resistance in HNSCC, we initially screened multiple HNSCC-derived xenograft models for their cetuximab sensitivity. Among the most sensitive we chose Cal27 HNSCC cells, a representative and frequently utilized HNSCC cell line that forms well-differentiated squamous carcinoma lesions in athymic nude mice (25,26). Cetuximab was very effective in preventing the growth and even causing the regression of established Cal27 HNSCC tumors (Figure 1, A–C). mTOR inhibitors such as rapamycin and Rad001 are widely used in the clinic for multiple diseases, and their efficacy and safety for the treatment of HNSCC patients is under current evaluation in multiple clinical trials (27–32).
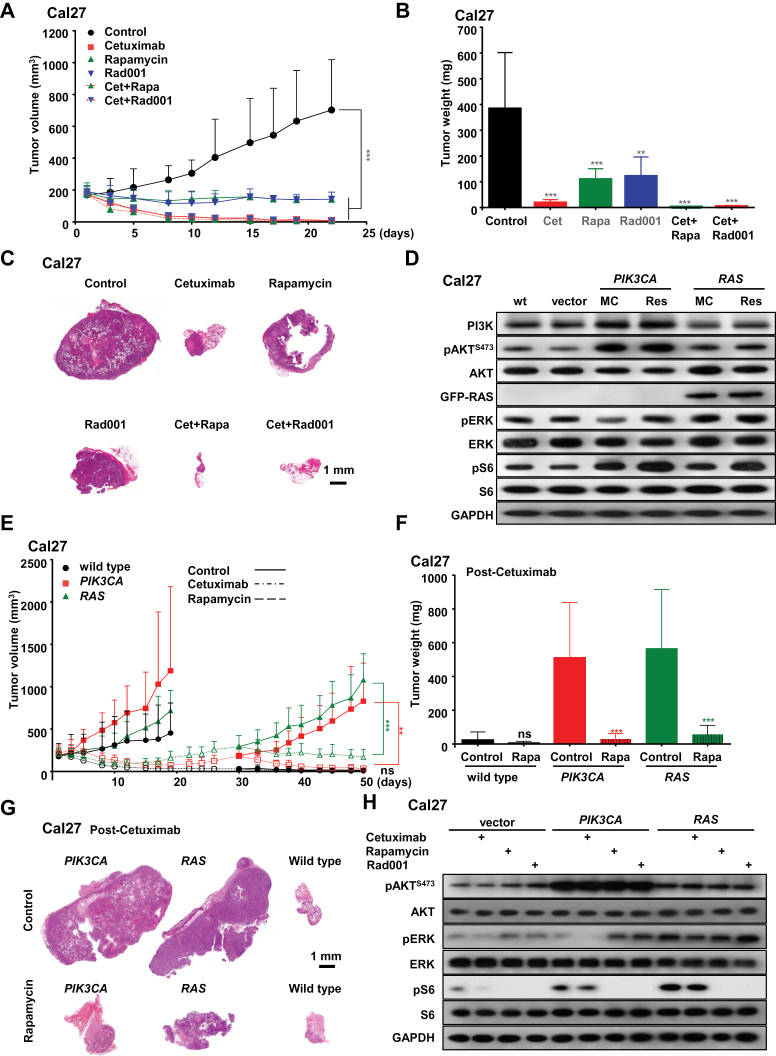
Figure 1.
Combined activity of cetuximab and mTOR inhibitors in HNSCC tumor xenografts. A) Antitumor efficacy of cetuximab, rapamycin, Rad001, and cetuximab combined with rapamycin or Rad001. Athymic nude mice were transplanted with Cal27 cells. Treatment was initiated when the tumor volume reached approximately 200mm3. The tumor growth curves were compared by the longitudinal data analysis method (two-sided). B) Tumor weights at the end of the single agent and combined treatments in panel A. C) Representative low magnification histological sections form each treatment group in panel A. Scale bars represent 1mm. D) Western blot analysis of unregulated signaling events in HNSCC cells. Mass cultures (MC) of wild-type Cal27 cells (wt), and Cal27 cells infected with empty lentiviral virus (vector), or expressing PIK3CA H1047R (PIK3CA) or RAS G12V (RAS), were serum-starved overnight, and lysates were analyzed for PI3K, pAKT, AKT, Ras, pERK, ERK, pS6, S6, and GAPDH, as indicated. Cetuximab-resistant PIK3CA and RAS cells isolated from control-treated tumors in panel E were processed in parallel Cal27 cells infected with lentiviruses encoding PIK3CA H1047R or RAS G12V mutations. Cells were transplanted into athymic nude mice, and, when they reached approximately 200mm3, mice were treated with vehicle control or cetuximab for approximately 20 days until tumors relapsed approximately to their initial size (30 days). Cetuximab was then discontinued, as indicated, and mice were divided into two groups and subsequently treated with vehicle control or rapamycin. F) Tumor weights at the end of the single agent and combined treatments in panel E. G) Representative histological tissue sections from each treatment group in panel E. Scale bars represent 1mm. H) Cal27 cell infected with control lentivirus (vector) and cetuximab-resistant PIK3CA and RAS were starved overnight, and treated with cetuximab (20 μg/mL), rapamycin (100nM), and Rad001 (100nM), and lysates were analyzed for pAKT, AKT, pERK, ERK, pS6, S6, and GAPDH, as indicated. In each case, error bars represent standard deviation, and *P < .05, ** P < .01, *** P < .001 when compared with the treatment control group; n = 10 per group.
We initially compared the effectiveness of cetuximab and rapamycin or Rad001 in these experimental HNSCC lesions and explored whether mTOR inhibition could increase the efficacy or interfere with the response to cetuximab. Regarding the latter, there is an increased interest in the immune modulatory effects of cetuximab (33), and hence it is possible that the immune suppressive activity of rapamycin may compromise cetuximab action. As reported, mTOR inhibition was quite effective in halting tumor growth (P < .001) (23,34–36), albeit in these cells cetuximab also elicited a pronounced response (P < .001) (Figure 1, A and B). When analyzing the final tumor weight, Rad001 was slightly less effective than rapamycin (P < .003 and P < .001, respectively). Remarkably, the combination of cetuximab and mTOR inhibition was highly effective (P < .001), but it was not statistically significantly better than cetuximab because of the potent activity of cetuximab alone in this model. Nonetheless, a potential increased response could be appreciated by the limited residual tumor size at the end of the combined treatment (Figure 1, A–C). Thus, combining mTOR inhibition with cetuximab does not interfere with the response to cetuximab, and it may even increase the overall efficacy of the antitumor treatment.
Effect of the Expression of PIK3CA and RAS Mutations in HNSCC on the Response to mTOR Inhibitors and Cetuximab
HNSCC lesions harbor activating mutations in PIK3CA (8% to 13%) (24,37–41) encoding the catalytic PI3K-α subunit and oncogenic mutants of the HRAS (4% to 9%) (24,41,42) or KRAS (3% to 7%) genes (41,43–45), collectively referred herein as RAS. As PIK3CA and RAS mutants stimulate mTOR downstream of EGFR, we asked whether their expression is sufficient to confer cetuximab resistance. For these studies, we genetically engineered Cal27 cells to express activating PIK3CA H1047R or KRAS G12V mutations, the latter as a GFP fusion protein to distinguish it from the endogenous gene product. Mass cultures of cells (MC) expressed increased levels of PI3K and RAS, respectively (Figure 1D). As expected, both PIK3CA and RAS increased phospho-AKT (pAKTS473) and phospho-S6 (pS6) levels, and RAS increased phospho-ERK (pERK) with limited variations in the levels of S6, AKT, and ERK proteins. PIK3CA- and RAS-expressing tumors grow statistically significantly faster than the wild-type HNSCC xenografts. Unexpectedly, Cal27 tumors expressing PIK3CA and RAS were initially sensitive to cetuximab (P < .002 and P < .001, respectively) (Figure 1E); however they relapsed within one month to their pretreatment size.
To explore whether targeting mTOR can be used in these cetuximab-resistant recurrent tumors, we halted cetuximab treatment and randomized the mice into control and rapamycin treated groups. Statistically significant differences in tumor volume were readily seen in just few days, with a dramatic tumor reduction in PIK3CA tumors and growth inhibition in RAS tumors (P < .001) (Figure 1, F and G). Cells were isolated from the initial cetuximab-resistant Cal27 PIK3CA and RAS tumors, referred as “Res” in Figure 1D. These cells did not express higher levels of PI3K and RAS oncoproteins or increased pAKT and pERK with respect to the initial mass cultures, but exhibited elevated mTOR activity, as judged by pS6 expression (Figure 1D), suggesting that cetuximab resistance may be associated with an increased ability of the cells to activate mTOR more efficiently, particularly the TORC1 complex that phosphorylates S6 (46,47). The mechanism underlying this selective increase in mTOR activity is still unclear and under current investigation. In these cetuximab-resistant cells, cetuximab still decreased pERK in PIK3CA, and to a much lesser extent in RAS-expressing cells, but had limited effect diminishing mTOR activity, which remained highly sensitive to rapamycin and Rad001 (Figure 1H).
Impact of mTOR Inhibition in PIK3CA and RAS-Induced Cetuximab Resistant Tumors
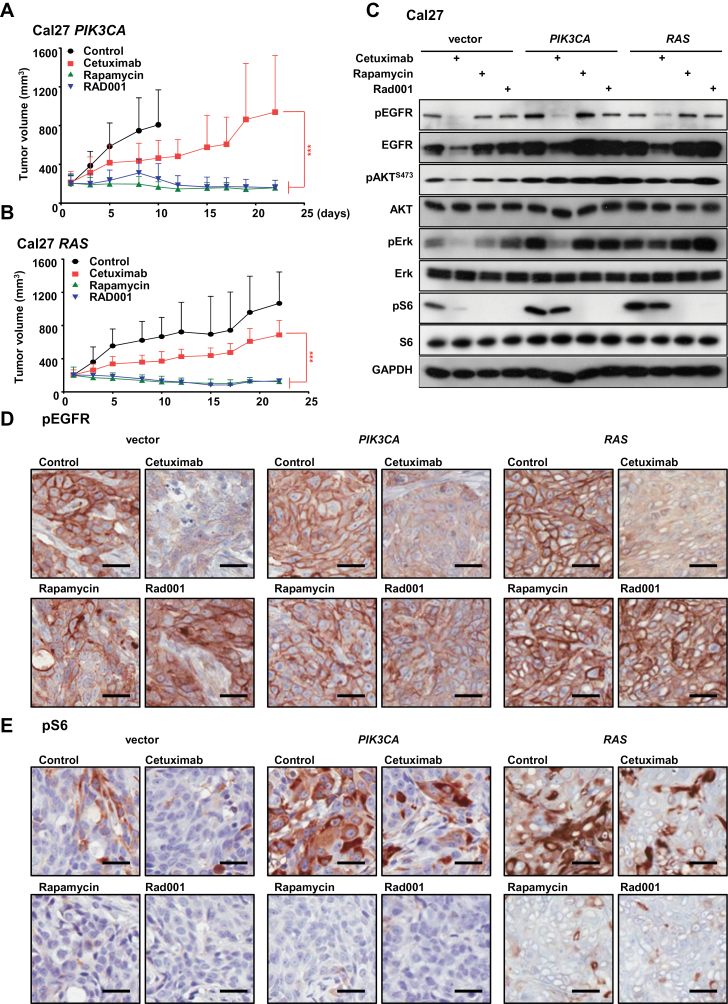
To challenge more directly whether mTOR inhibition could overcome cetuximab resistance, we used Cal27 PIK3CA- and RAS-resistant cultures (referred to as “Res” in Figure 1D). Cetuximab still showed some effect in these tumors, particularly in PIK3CA-expressing xenografts, but both PIK3CA and RAS HNSCC lesions were much more sensitive to rapamycin or Rad001 as single agents (P < .001) (Figure 2, A and B). Cetuximab decreased EGFR levels, and consequently pEGFR, pERK, pAKT, and pS6 in lysates of vector tumor controls (Figure 2, C–E). Basal EGFR and pEGFR levels were increased in PIK3CA- and RAS-expressing tumors, but both were decreased by cetuximab in tissue lysates and by immunostaining for pEGFR in tissue biopsies. Cetuximab also decreased pERK in PIK3CA cells, which may explain some of its residual antitumor activity, but failed to inhibit mTOR, while it did not inhibit ERK or mTOR in RAS-expressing cells. Rapamcyin and Rad001 instead decreased mTOR activity in all tumors treated.
Figure 2.
Effect of mTOR inhibition on PIK3CA- and RAS-induced cetuximab resistance. A and B) The cetuximab-resistant Cal27 cells expressing PIK3CA and RAS (from Figure 1E) were transplanted into athymic mice. Mice were treated with cetuximab, rapamycin, or Rad001 as a single agent as indicated. *** P < .001 when comparing the rapamycin and Rad001 treated groups with cetuximab-treated mice (n = 10 per group). The tumor growth curves were compared by the longitudinal data analysis method (two-sided). For all panels, error bars represent standard deviation. C) Tumor-bearing mice were treated with cetuximab, rapamycin, or Rad001 for four days (short-term treatment) and tumor lysates were analyzed for pEGFR, EGFR, pAKT, AKT, pERK, ERK, pS6, S6, and GAPDH, as indicated. D and E) Representative immunohistochemical analysis of pEGFR and pS6 in the short-term treatment groups from panel C. Brown chromogen deposition reflects the immunoreactivity; hematoxylin was used as a nuclear counterstain (blue). Scale bars represent 25 µm.
Combined Treatment with Cetuximab and mTOR Inhibitors of HNSCC Cells Engineered to Express PIK3CA and RAS
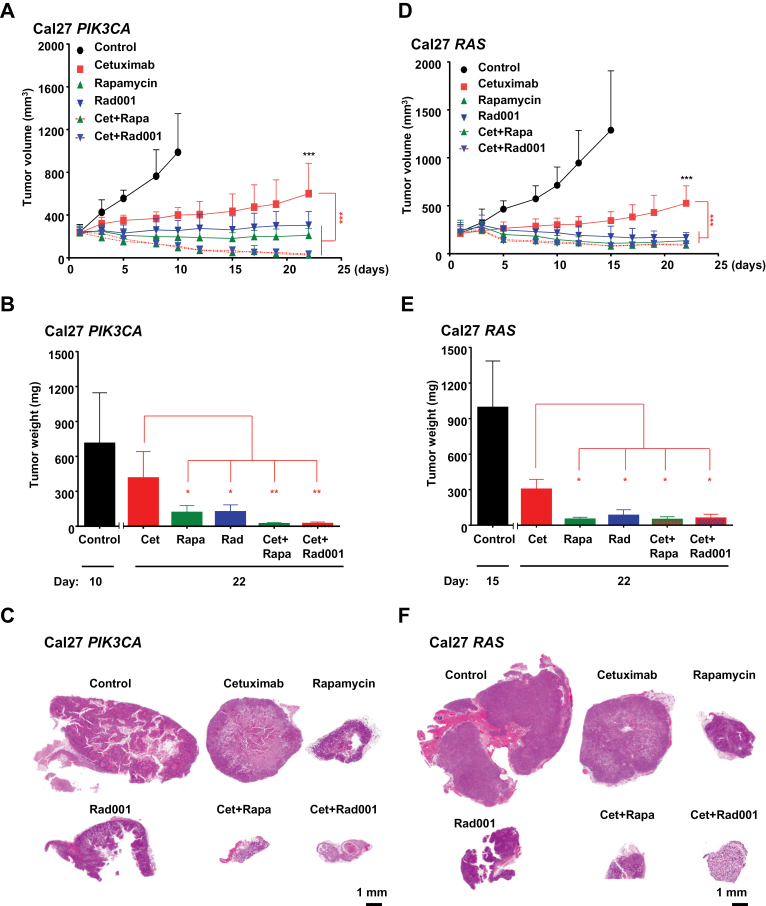
Given that mTOR inhibitors do not diminish the response of wild-type Cal27 HNSCC to cetuximab, we next investigated the consequences of combining cetuximab with rapamycin or Rad001 instead of using the mTOR inhibitors after the relapse of cetuximab-resistant cells. The impact of the drug combination was remarkable. mTOR inhibition together with cetuximab caused a rapid tumor collapse (P < .001), with nearly nonexistent residual tumor masses at the end of the treatment (Figure 3, A–F). This potentiation was also recapitulated in vitro using small molecule inhibitors. PI3K- and RAS-expressing cells exhibited reduced sensitivity to the EGFR tyrosine kinase inhibitor (TKI) gefitinib, aligned with recent reports (Supplementary Figure 1, available online) (48). The concomitant blockade of mTOR restored the sensitivity to gefitinib in these HNSCC cells in vitro, thus independently of the tumor microenvironment and the potential immune response elicited by cetuximab (Supplementary Figure 1, available online).
Figure 3.
Sensitivity of HNSCC cells engineered to express PIK3CA and RAS to combined treatment with cetuximab and mTOR inhibitors. A and D) The cetuximab-resistant PIK3CA and RAS Cal27 cells were transplanted into athymic mice and mice were treated with cetuximab, rapamycin, Rad001, and cetuximab combined with rapamycin or Rad001 as indicated. B and E) Tumor weights at the end of the single agent and combined treatments in panels A and D, respectively. B: Cet vs Rapa: P < .04; Cet vs Rad001: P < .02; Cet vs Cet+Rapa: P < .004; Cet vs Cet+Rad001: P < .006; E: Cet vs Rapa P < .03; Cet vs Rad001 P < .03; Cet vs Cet+Rapa: P < .02; Cet vs Cet+Rad001 P < .03). C and F) Representative histological sections from each treatment group in panels A and D. Scale bars represent 1mm. (***P < .001 when compared with the control-treated group [black stars], or when comparing with the mice treated with cetuximab as a single agent [red stars]; n = 10 per group. The tumor growth curves were compared by the longitudinal data analysis method (two-sided).
Impact of EGFR and mTOR Cotargeting on Tumor Relapse After Cetuximab Treatment in HNSCC Cells Harboring Endogenous PIK3CA and RAS Oncogenes
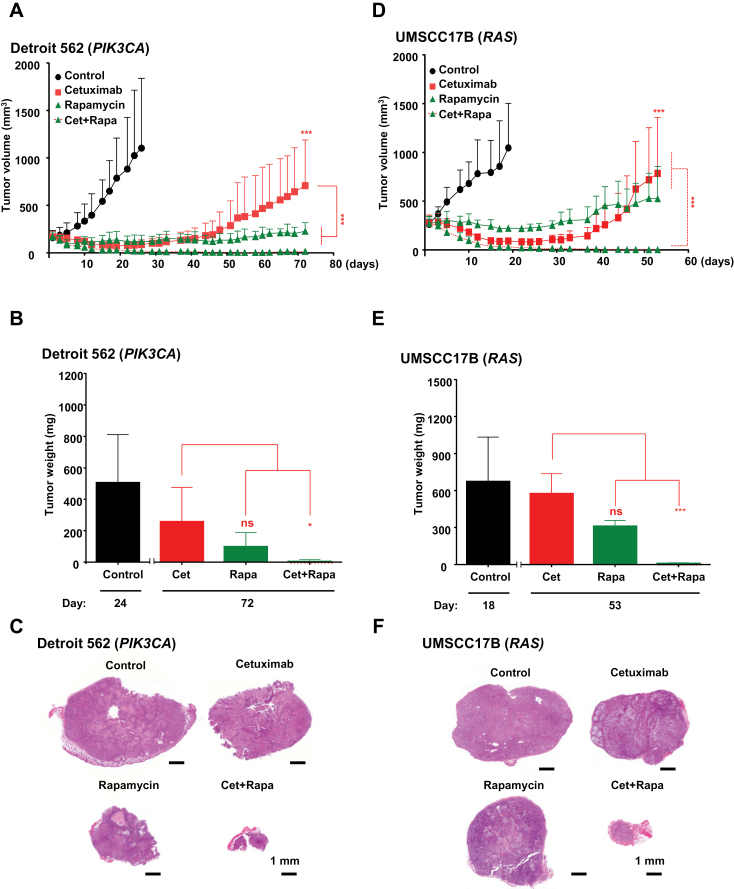
We next investigated whether the combination of cetuximab and the mTOR inhibitor rapamycin was also effective in HNSCC tumors harboring activating mutations of endogenous PIK3CA and RAS. For these studies, we used Detroit 562 cells exhibiting a PIK3CA H1047R mutation (38) and UMSCC17B, which has a HRAS Q61L mutation (not shown). Both PIK3CA- and RAS-harboring tumors were initially sensitive to cetuximab when compared with the control-treated groups (P < .001), but both tumors relapsed after one month, and mice succumbed to disease in two months (Figure 4). These PI3KCA- and RAS-expressing tumors were sensitive to rapamycin as a single agent, but the drug combination groups showed a remarkably statistically significant tumor volume decrease for both PIK3CA- (P < .03) and RAS-expressing (P < .001) tumors when compared with the cetuximab and rapamycin alone groups (Figure 4). Indeed, residual disease after the combined treatment was negligible.
Figure 4.
Effect of EGFR and mTOR cotargeting on tumor relapse after cetuximab treatment in HNSCC cells harboring PIK3CA and RAS oncogenes. A and D) Detroit 562 cells (harboring PIK3CA H1047R mutations) and UMSCC17B cells (expressing HRAS Q61L mutant oncogene) were transplanted into athymic mice, and mice were treated with cetuximab or rapamycin alone, or cetuximab together with rapamycin, as indicated. B and E) Tumor weights in each group at the end of the treatments in panels A and D, respectively. C and F) Representative histological sections for each treatment group in figures A and D, respectively. Scale bars represent 1mm. (***P < .001 comparing the treatment groups with the control [black stars], or comparing with the cetuximab treated group [red stars], n = 10 per group, * P < .04). The tumor growth curves were compared by the longitudinal data analysis method (two-sided).
Effects of the Combination of Cetuximab and Rapamycin on PIK3CA- and RAS-Induced Cell Proliferation
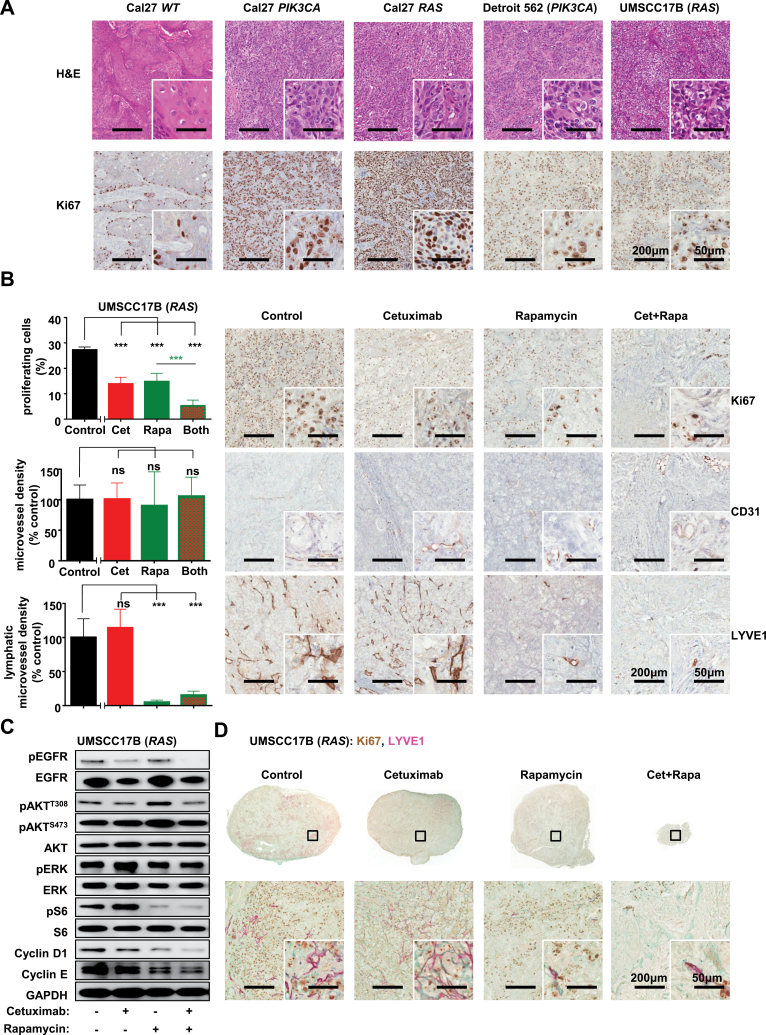
Cal27 cells engineered to express PIK3CA and RAS have distinct cell morphology, resembling tumors induced by HNSCC cells harboring endogenous PIK3CA and RAS gene mutations (Figure 5A). Immunohistochemistry (IHC) for Ki67 showed increased proliferation in tumor xenografts expressing PIK3CA and RAS genes endogenously or ectopically. To study the biological and biochemical consequences of cetuximab treatment and mTOR inhibition and their combination, we focused on the RAS-driven UMSCC17B cells and PIK3CA-driven Detroit 562 cells. Xenografts were treated with vehicle control, cetuximab, rapamycin, and cetuximab plus rapamycin. IHC for Ki67 showed decreased cell proliferation in the combination-treated group as compared with vehicle control and the cetuximab or rapamycin alone groups (P < .001). IHC for CD31 and LYVE1 showed limited changes in overall angiogenesis, but mTOR inhibitors alone or in combination with cetuximab provoked a remarkable reduction in lymphangiogenesis (P < .001), aligned with prior studies from our laboratory and others (Figure 5B) (32,36). Rapamycin or cetuximab and rapamycin inhibited the mTOR pathway, while cetuximab alone was less effective. Cetuximab reduced ERK activity in HNSCC expressing active PIK3CA. Importantly, in HNSCC cells expressing mutant PIK3CA and RAS cetuximab prevented the upregulation of AKT activity caused by mTOR inhibition (Figure 5C; Supplementary Figure 2, B and D, available online). Paralleling the impact on cell proliferation, cyclin D levels were clearly decreased (Figure 5C; Supplementary Figure 2D, available online). We also investigated the proliferative potential of residual HNSCC cells after prolonged tumor treatment. Whereas many Ki67+-proliferating cells are still visible in cetuximab- and rapamycin-treated mice, very few proliferating cells are found in the residual lesion after two month of the combined treatment. Most of the histological sections revealed dense fibrous tissue infiltrated by chronic inflammatory reaction, with few or no remaining neoplastic cells and intratumor lymphatic vessels (Figure 5D; Supplementary Figure 2C, available online).
Figure 5.
Effects of the combination of cetuximab and rapamycin on tumor growth driven by PIK3CA and RAS mutations. A) Representative histological sections of tumor xenografts stained by H&E and Ki67 immunohistochemistry as a proliferative marker after transplantation of Cal27 cells, Cal27 cells expressing PIK3CA and RAS oncogenes, as well as Detroit 562 and UMSCC17B HNSCC cells into nude mice. Scale bars represent 200 µm (low magnification) or 50 µm (inset). B) Quantification (left) and representative tumor tissue sections (right) stained for Ki67, CD31, and LYVE1 by immunohistochemistry after a long-term treatment for the indicated days of UMSCC17B xenograft with control, cetuximab, rapamycin, and cetuximab combined with rapamycin, as indicated. (***P < .001 comparing the treatment groups with the control [black stars], or comparing with the indicated treated groups [green stars]; n = 4 per group). The tumor growth curves were compared by the longitudinal data analysis method (two-sided). C) As an example of naturally cetuximab-resistant HNSCC cells, UMSCC17B were transplanted into athymic mice and treated with cetuximab, rapamycin, and with cetuximab combined with rapamycin for four days (short-term treatment). Tumor lysates were analyzed for pEGFR, EGFR, pAKTT308, pAKTS473, AKT, pERK, ERK, pS6, S6, Cylin D1, Cyclin E, and GAPDH. D) Representative tumor tissue sections stained for Ki67 and LYVE1 by immunohistochemistry after a long-term treatment of UMSCC17B xenograft with control, cetuximab, rapamycin, and cetuximab combined with rapamycin, as indicated. Brown color for Ki67, red color for LYVE1.
Discussion
While cetuximab offers clinical benefits in HNSCC, its impact in advanced HNSCC patients is still quite limited, likely due to intrinsic mechanisms preventing a full beneficial response to cetuximab, or to the development of acquired resistance (9,10,17,49,50). Clearly, new strategies are needed to treat and aid in the management of HNSCC. Here, we show that genetic alterations causing PI3K/AKT/mTOR activation, such as by expression of activated PIK3CA and RAS alleles, are sufficient to prevent a sustained response to cetuximab after an initial short-lasting beneficial effect. We also show that clinically relevant mTOR targeting agents, such as rapamycin and Rad001, are effective in HNSCC lesions refractory to cetuximab. Surprisingly, the concomitant administration of cetuximab- and mTOR-targeting agents caused the rapid and complete regression of both genetically engineered and naturally occurring PIK3CA- and RAS-driven HNSCC lesions. These findings provide support to the early evaluation of the rationale combination of cetuximab and/or other EGFR-targeting agents with mTOR inhibitors for the treatment of HNSCC lesions harboring genetic alterations resulting in mTOR activation downstream from EGFR.
RAS mutations are predictive of cetuximab resistance in colon cancer, with some PIK3CA mutations diminishing its response (51,52). Instead, HNSCC cells expressing RAS and PIK3CA endogenously or after gene transfer still exhibited an initial, albeit limited tumor response. Thus, EGFR may promote HNSCC growth by stimulating signaling pathways in addition to PI3K and RAS, making them initially sensitive to cetuximab. This sensitivity might then be overcome by the emergence of compensatory mechanisms leading to tumor relapse. Ultimately, the presence of RAS and PIK3CA mutations or other genetic and epigenetic alterations leading to PI3K/AKT/mTOR activation, such as reduced PTEN expression, may provide an intrinsic resistance mechanism contributing to tumor recurrence after an initial short-lasting favorable response to cetuximab. We can postulate that epigenetic events or de novo mutations resulting in the activation of the PI3K/AKT/mTOR signaling circuitry might also contribute to acquired cetuximab resistance.
The frequent overreliance on the PI3K/AKT/mTOR pathway for HNSCC growth (22) may in turn represent a cancer vulnerability that can be exploited therapeutically. This includes the use of inhibitors targeting PI3K-α, AKT, and mTOR, the latter as part of its complex TORC1, using rapalogs, or TORC1 and TORC2 using novel mTOR kinase inhibitors and PI3K/TORC1/TORC2 blocking agents. Based on this widespread activation of PI3K/AKT/mTOR in HNSCC and preclinical information (21–23,25,36,53,54), multiple clinical trials targeting this pathway are now under evaluation (27–31). Specifically, our observations suggest that mTOR inhibitors will exert a beneficial response in PI3KCA-expressing tumors, particularly when combined with cetuximab up front to increase the therapeutic response and limit the risk of tumor relapse.
Interestingly, HNSCC-expressing RAS mutants ectopically were sensitive to mTOR inhibition, while those expressing endogenous RAS oncogenes where refractory. This can be explained by the fact that we expressed KRAS mutations previously reported to confer cetuximab resistance in colon cancer (52), while HNSCC lesions and derived cells most often express HRAS mutants. Different RAS protein isoforms may initiate distinct signaling events in HNSCC, being KRAS sensitive to mTOR inhibitors, whereas HRAS may not. Alternatively, HNSCC cells with endogenous HRAS mutations may exhibit other alterations resulting in insensitivity to mTOR inhibitors, a possibility that warrants further investigation. In every case, cetuximab combined with mTOR inhibitors was quite effective in HNSCC cells harboring HRAS mutations in their endogenous allele, representing the most biologically relevant situation.
The finding that we observed a complete response when combining cetuximab with mTOR inhibitors in every genetic background tested is quite remarkable. This included tumors expressing endogenous RAS and PI3KCA oncogenes, which collapsed rapidly and did not relapse even after prolonged observation. The biological and molecular bases of this increased activity are still unclear and under current investigation. At the biochemical level, cetuximab prevented the compensatory increase in AKT activity often seen after treatment with rapamycin alone, which may limit the clinical benefit of mTOR inhibitors as single agents (55,56). This may help explain the increased response to the combined treatment. At the biological level, we did not observe an overt increase in apoptosis after cetuximab treatment and/or mTOR inhibition (not shown). Preliminary results suggest that HNSCC cells exposed to the combination of cetuximab and mTOR inhibitors stop proliferating, exit the self-renewal cancer pool, and undergo rapid morphological changes consistent with an increased autophagy (57,58) and differentiation, and exhibit a remarkably reduced lymphangiogenesis. A possibility also exists that cetuximab may promote a cytotoxic immune response against EGFR-overexpressing cells, which would synergize with mTOR growth-signaling inhibition (33). Of interest, the analysis of the very limited residual lesions after the combined treatment indicates that most remaining cancer cells were no longer proliferating and that they were embedded in a highly fibrotic scaring tissue that may fail to support the potential cancer cell regrowth at the end of the treatment.
While our experimental models address PIK3CA- and RAS-induced resistance to cetuximab, multiple additional mechanisms may exist in HNSCC patients, which may need to be evaluated experimentally and in the clinical setting. Similarly, our experimental models in immunocompromised mice may not reflect fully the immune modulatory effects of cetuximab (33). On the other hand, the antilymphangiogenic activity or mTOR inhibitors combined with cetuximab in mice was remarkable. However, this activity and its potential benefits may require further evaluation in the clinic.
This raises the issue as to whether mTOR inhibitors would be best utilized as single agents after the development of cetuximab resistance or instead to explore cotargeting EGFR and mTOR as a first line of treatment. Cetuximab is frequently used in HNSCC patients concomitant with radiation and chemotherapy (9,10), which can cause DNA alterations and the emergence of cetuximab-resistant cancer cells. In this scenario, the use of a cotargeting therapeutic strategy using cetuximab and PI3K or mTOR inhibitors as a first line of treatment may instead be ideal. In addition, human papillomavirus (HPV)–positive HNSCCs frequently harbor activating PIK3CA mutations (39,59,60) and exhibit aberrant mTOR activity (23), suggesting that this combined targeted therapy might be also useful in at least a subset of HPV-associated HNSCCs. Overall, we can conclude that the evaluation of mutations in genes encoding RAS and PIK3CA and/or other genetic and epigenetic alterations affecting the activity of the PI3K/AKT/mTOR pathway may predict the potential intrinsic resistance to cetuximab. In turn, this information can help select a patient population that may benefit the most from the concomitant administration of cetuximab and PI3K and/or mTOR inhibitors as a precision molecular therapeutic option for HNSCC patients.
Funding
This work was supported by the Intramural Research Program (Z01DE00558), NIDCR, NIH, and by the 111 Project of MOE & ISTCPC (2012DFA31370), China.
The sponsoring institutions did not play a role in the study design and execution, or in the data analysis and publication.
References
- 1. Grandis JR, Tweardy DJ. Elevated Levels of Transforming Growth Factor a and Epidermal Growth Factor Receptor Messenger RNA Are Early Markers of Carcinogenesis in Head and Neck Cancer. Cancer Res. 1993;53(15):3579–3584 [PubMed] [Google Scholar]
- 2. Kalyankrishna S, Grandis JR. Epidermal growth factor receptor biology in head and neck cancer. J Clin Oncol. 2006;24(17):2666–2672 [DOI] [PubMed] [Google Scholar]
- 3. Grandis JR, Melhem MF, Gooding WE. Levels of TGF-alfa and EGFR Protein in Head and Neck Squamous Cell Carcinoma and Patient Survival. J Natl Cancer Inst. 1998;90(11):824–832 [DOI] [PubMed] [Google Scholar]
- 4. Chung CH, Ely K, McGavran L, et al. Increased epidermal growth factor receptor gene copy number is associated with poor prognosis in head and neck squamous cell carcinomas. J Clin Oncol. 2006;24(25):4170–4176 [DOI] [PubMed] [Google Scholar]
- 5. Grandis JR, Chakraborty A, Melhem MF, Zeng Q, Tweardy DJ. Inhibition of epidermal growth factor receptor gene expression and function decreases proliferation of head and neck squamous carcinoma but not normal mucosal epithelial cells. Oncogene. 1997;15(4):409–416 [DOI] [PubMed] [Google Scholar]
- 6. Erjala K, Sundvall M, Junttila TT, et al. Signaling via ErbB2 and ErbB3 associates with resistance and epidermal growth factor receptor (EGFR) amplification with sensitivity to EGFR inhibitor gefitinib in head and neck squamous cell carcinoma cells. Clin Cancer Res. 2006;12(13):4103–4111 [DOI] [PubMed] [Google Scholar]
- 7. Nozawa H, Tadakuma T, Ono T, et al. Small interfering RNA targeting epidermal growth factor receptor enhances chemosensitivity to cisplatin, 5-fluorouracil and docetaxel in head and neck squamous cell carcinoma. Cancer Sci. 2006;97(10):1115–1124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cassell A, Grandis JR. Investigational EGFR-targeted therapy in head and neck squamous cell carcinoma. Expert Opin Investig Drugs. 2010;19(6):709–722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bonner JA, Harrari PM, J. G, et al. Radiotherapy plus Cetuximab for Squamous-Cell Carcinoma of the Head and Neck. N Engl J Med. 2006;354(6)567–578 [DOI] [PubMed] [Google Scholar]
- 10. Vormorken J B, Mesia R, Rivera F, et al. Platinum-Based Chemotherapy plus Cetuximab in Head and Neck Cancer. N Engl J Med. 2008; 359(11):1116–1127 [DOI] [PubMed] [Google Scholar]
- 11. Montagut C, Dalmases A, Bellosillo B, et al. Identification of a mutation in the extracellular domain of the Epidermal Growth Factor Receptor conferring cetuximab resistance in colorectal cancer. Nat Med. 2012;18(2):221–223 [DOI] [PubMed] [Google Scholar]
- 12. Yonesaka K, Zejnullahu K, Okamoto I, et al. Activation of ERBB2 signaling causes resistance to the EGFR-directed therapeutic antibody cetuximab. Sci Transl Med. 2011;3(99):99ra86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lu Y, Li X, Liang K, et al. Epidermal growth factor receptor (EGFR) ubiquitination as a mechanism of acquired resistance escaping treatment by the anti-EGFR monoclonal antibody cetuximab. Cancer Res. 2007;67(17):8240–8247 [DOI] [PubMed] [Google Scholar]
- 14. Benavente S, Huang S, Armstrong EA, et al. Establishment and characterization of a model of acquired resistance to epidermal growth factor receptor targeting agents in human cancer cells. Clin Cancer Res. 2009;15(5):1585–1592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Li C, Iida M, Dunn EF, et al. Nuclear EGFR contributes to acquired resistance to cetuximab. Oncogene. 2009;28(43):3801–3813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wheeler DL, Huang S, Kruser TJ, et al. Mechanisms of acquired resistance to cetuximab: role of HER (ErbB) family members. Oncogene. 2008;27(28):3944–3956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wheeler DL, Dunn EF, Harari PM. Understanding resistance to EGFR inhibitors-impact on future treatment strategies. Nat Rev Clin Oncol. 2010;7(9):493–507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Sharma SV, Bell DW, Settleman J, et al. Epidermal growth factor receptor mutations in lung cancer. Nat Rev Cancer. 2007;7(3):169–181 [DOI] [PubMed] [Google Scholar]
- 19. Tebbutt N, Pedersen MW, Johns TG. Targeting the ERBB family in cancer: couples therapy. Nat Rev Cancer. 2013;13(9):663–673 [DOI] [PubMed] [Google Scholar]
- 20. Maulik G, Shrikhand A, Kijima T, et al. Role of the hepatocyte growth factor receptor, c-Met, in oncogenesis and potential for therapeutic inhibition. Cytokine Growth Factor Rev. 2002;13(1):41–59 [DOI] [PubMed] [Google Scholar]
- 21. Amornphimoltham P, Patel V, Sodhi A, et al. Mammalian target of rapamycin, a molecular target in squamous cell carcinomas of the head and neck. Cancer Res. 2005;65(21):9953–9961 [DOI] [PubMed] [Google Scholar]
- 22. Molinolo AA, Hewitt SM, Amornphimoltham P, et al. Dissecting the Akt/mammalian target of rapamycin signaling network: emerging results from the head and neck cancer tissue array initiative. Clin Cancer Res. 2007;13(17):4964–4973 [DOI] [PubMed] [Google Scholar]
- 23. Molinolo AA, Marsh C, El Dinali M, et al. mTOR as a molecular target in HPV-associated oral and cervical squamous carcinomas. Clin Cancer Res. 2012;18(9):2558–2568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Iglesias-Bartolome R, Martin D, Gutkind JS. Exploiting the head and neck cancer oncogenome: widespread PI3K-mTOR pathway alterations and novel molecular targets. Cancer Discov. 2013;3(7):722–725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Amornphimoltham P, Patel V, Leelahavanichkul K, et al. A retroinhibition approach reveals a tumor cell-autonomous response to rapamycin in head and neck cancer. Cancer Res. 2008;68(4):1144–1153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Leelahavanichkul K, Amornphimoltham P, Molinolo AA, et al. A role for p38 MAPK in head and neck cancer cell growth and tumor-induced angiogenesis and lymphangiogenesis. Mol Oncol. 2014;8(1):105–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Massarelli E, Ginsberg LE, Lin H, et al. Phase II trial of everolimus and erlotinib in patients with platinum-resistant recurrent and/or metastatic head and neck squamous cell carcinoma. Chicago, IL: ASCO Annual Meeting; May 31-June 4, 2013. Abstract 6607. [Google Scholar]
- 28. Clark C, Shah S, Herman-Ferdinandez L, et al. Teasing out the best molecular marker in the AKT/mTOR pathway in head and neck squamous cell cancer patients. Laryngoscope. 2010;120(6):1159–1165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Nguyen SA, Walker D, Gillespie MB, et al. mTOR inhibitors and its role in the treatment of head and neck squamous cell carcinoma. Curr Treat Options Oncol. 2012;13(1):71–81 [DOI] [PubMed] [Google Scholar]
- 30. Varadarajan P, Kotsakis AP, Martin D, et al. Phase II trial of everolimus in patients with previously treated recurrent or metastatic squamous cell carcinoma of the head and neck (SCCHN). Chicago, IL: ASCO Annual Meeting; June 1–5, 2012. Abstract 5541. [Google Scholar]
- 31. Freudlsperger C, Burnett JR, Friedman JA, et al. EGFR-PI3K-AKT-mTOR signaling in head and neck squamous cell carcinomas: attractive targets for molecular-oriented therapy. Expert Opin Ther Targets. 2011;15(1):63–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ekshyyan O, Moore-Medlin TN, Raley MC, et al. Anti-lymphangiogenic properties of mTOR inhibitors in head and neck squamous cell carcinoma experimental models. BMC Cancer. 2013;13:320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Srivastava RM, Lee SC, Andrade Filho PA, et al. Cetuximab-activated natural killer and dendritic cells collaborate to trigger tumor antigen-specific T-cell immunity in head and neck cancer patients. Clin Cancer Res. 2013;19(7):1858–1872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Czerninski R, Amornphimoltham P, Patel V, et al. Targeting mammalian target of rapamycin by rapamycin prevents tumor progression in an oral-specific chemical carcinogenesis model. Cancer Prev Res (Phila). 2009;2(1):27–36 [DOI] [PubMed] [Google Scholar]
- 35. Raimondi AR, Molinolo A, Gutkind JS. Rapamycin prevents early onset of tumorigenesis in an oral-specific K-ras and p53 two-hit carcinogenesis model. Cancer Res. 2009;69(10):4159–4166 [DOI] [PubMed] [Google Scholar]
- 36. Patel V, Marsh CA, Dorsam RT, et al. Decreased lymphangiogenesis and lymph node metastasis by mTOR inhibition in head and neck cancer. Cancer Res. 2011;71(22):7103–7112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Cohen Y, Goldenberg-Cohen N, Shalmon B, et al. Mutational analysis of PTEN/PIK3CA/AKT pathway in oral squamous cell carcinoma. Oral Oncol. 2011;47(10):946–950 [DOI] [PubMed] [Google Scholar]
- 38. Qiu W, Schonleben F, Li X, et al. PIK3CA mutations in head and neck squamous cell carcinoma. Clin Cancer Res. 2006;12(5):1441–1446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lui VW, Hedberg ML, Li H, et al. Frequent mutation of the PI3K pathway in head and neck cancer defines predictive biomarkers. Cancer Discov. 2013;3(7):761–769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kozaki K, Imoto I, Pimkhaokham A, et al. PIK3CA mutation is an oncogenic aberration at advanced stages of oral squamous cell carcinoma. Cancer Sci. 2006;97(12):1351–1358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Pickering CR, Zhang J, Yoo SY, et al. Integrative genomic characterization of oral squamous cell carcinoma identifies frequent somatic drivers. Cancer Discov. 2013;3(7):770–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Anderson JA, Irish JC, McLachlin CM, Ngan BY. H-ras oncogene mutation and human papillomavirus infection in oral carcinomas. Arch Otolaryngol Head Neck Surg. 1994;120(7):755–760 [DOI] [PubMed] [Google Scholar]
- 43. Forbes SA, Bhamra G, Bamford S, et al. The catalogue of somatic mutations in cancer (COSMIC). Curr Protoc Hum Genet. 2008;Chapter 10:Unit 10.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Bissada E, Abboud O, Abou Chacra Z, et al. Prevalence of K-RAS Codons 12 and 13 Mutations in Locally Advanced Head and Neck Squamous Cell Carcinoma and Impact on Clinical Outcomes. Int J Otolaryngol. 2013;2013:848021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Das N, Majumder J, DasGupta UB. ras Gene mutations in oral cancer in eastern India. Oral Oncol. 2000;36(1):76–80 [DOI] [PubMed] [Google Scholar]
- 46. Hay N, Sonenberg N. Upstream and downstream of mTOR. Genes Dev. 2004;18(16):1926–1945 [DOI] [PubMed] [Google Scholar]
- 47. Laplante M, Sabatini DM. mTOR signaling in growth control and disease. Cell. 2012;149(2):274–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Young NR, Liu J, Pierce C, et al. Molecular phenotype predicts sensitivity of squamous cell carcinoma of the head and neck to epidermal growth factor receptor inhibition. Mol Oncol. 2013;7(3):359–368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Bonner JA, Harari PM, Cohen RB, et al. Radiotherapy plus cetuximab for locoregionally advanced head and neck cancer: 5-year survival data from a phase 3 randomised trial, and relation between cetuximab-induced rash and survival. Lancet Oncol. 2010;11(1):21–28 [DOI] [PubMed] [Google Scholar]
- 50. Kim S, Grandis JR, Rinaldo A, et al. Emerging perspectives in epidermal growth factor receptor targeting in head and neck cancer. Head Neck. 2008;30(5):667–674 [DOI] [PubMed] [Google Scholar]
- 51. Normanno N, Tejpar S, Morgillo F, et al. Implications for KRAS status and EGFR-targeted therapies in metastatic CRC. Nat Rev Clin Oncol. 2009;6(9):519–527 [DOI] [PubMed] [Google Scholar]
- 52. Bardelli A, Siena S. Molecular mechanisms of resistance to cetuximab and panitumumab in colorectal cancer. J Clin Oncol. 2010;28(7):1254–1261 [DOI] [PubMed] [Google Scholar]
- 53. Vitale-Cross L, Molinolo AA, Martin D, et al. Metformin prevents the development of oral squamous cell carcinomas from carcinogen-induced premalignant lesions. Cancer Prev Res (Phila). 2012;5(4):562–573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Vitale-Cross L, Czerninski R, Amornphimoltham P, et al. Chemical carcinogenesis models for evaluating molecular-targeted prevention and treatment of oral cancer. Cancer Prev Res (Phila). 2009;2(5):419–422 [DOI] [PubMed] [Google Scholar]
- 55. Cloughesy TF, Yoshimoto K, Nghiemphu P, et al. Antitumor activity of rapamycin in a Phase I trial for patients with recurrent PTEN-deficient glioblastoma. PLoS Med. 2008;5(1):e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Tamburini J, Chapuis N, Bardet V, et al. Mammalian target of rapamycin (mTOR) inhibition activates phosphatidylinositol 3-kinase/Akt by up-regulating insulin-like growth factor-1 receptor signaling in acute myeloid leukemia: rationale for therapeutic inhibition of both pathways. Blood. 2008;111(1):379–382 [DOI] [PubMed] [Google Scholar]
- 57. Li X, Fan Z. The epidermal growth factor receptor antibody cetuximab induces autophagy in cancer cells by downregulating HIF-1alpha and Bcl-2 and activating the beclin 1/hVps34 complex. Cancer Res. 2010;70(14):5942–5952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Kim J, Kundu M, Viollet B, et al. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat Cell Biol. 2011;13(2):132–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Nichols AC, Palma D A, Chow W, et al. High Frequency of Activating PIK3CA Mutations in Human Papillomavirus–Positive Oropharyngeal Cancer. JAMA Otolaryngol Head Neck Surg. 2013;139:617–622 [DOI] [PubMed] [Google Scholar]
- 60. Sewell A, Brown BT, Biktasova A, et al. Reverse Phase Protein Array profiling of oropharyngeal cancer and significance of PIK3CA mutations in HPV-associated head and neck cancer. Clin Cancer Res. 2014; 20(9):2300–2311 [DOI] [PMC free article] [PubMed] [Google Scholar]