Abstract
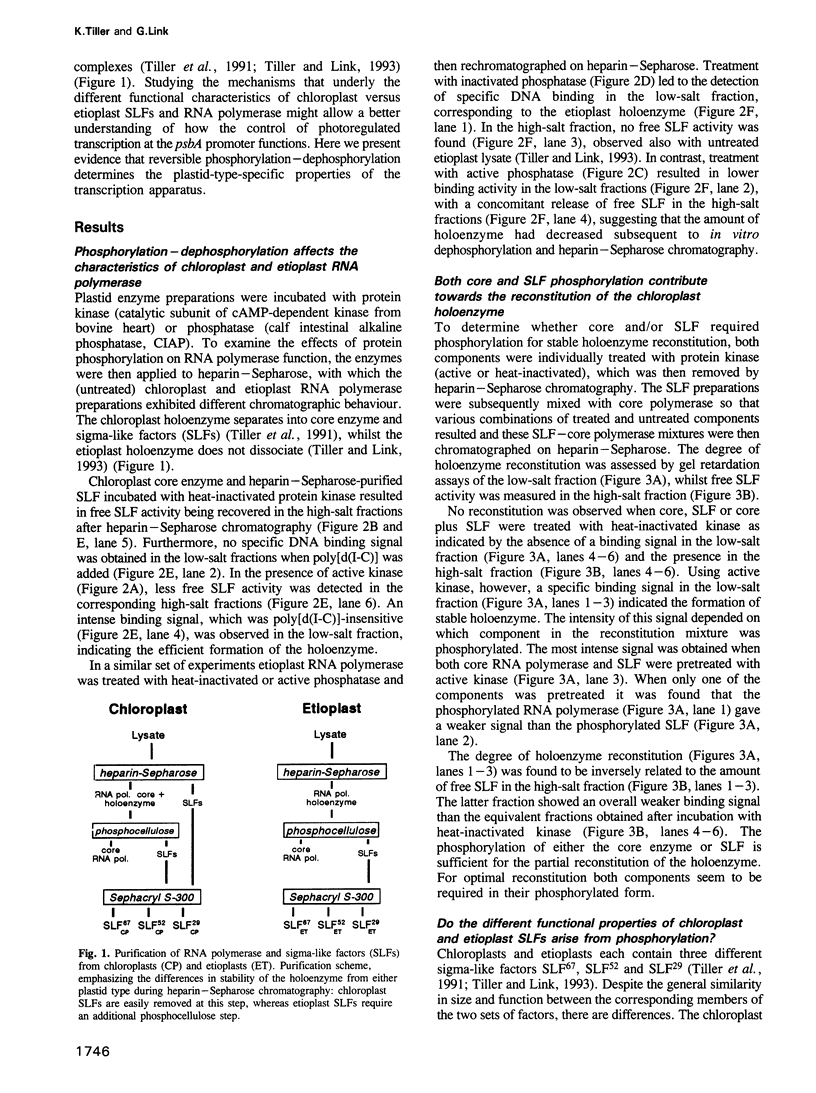
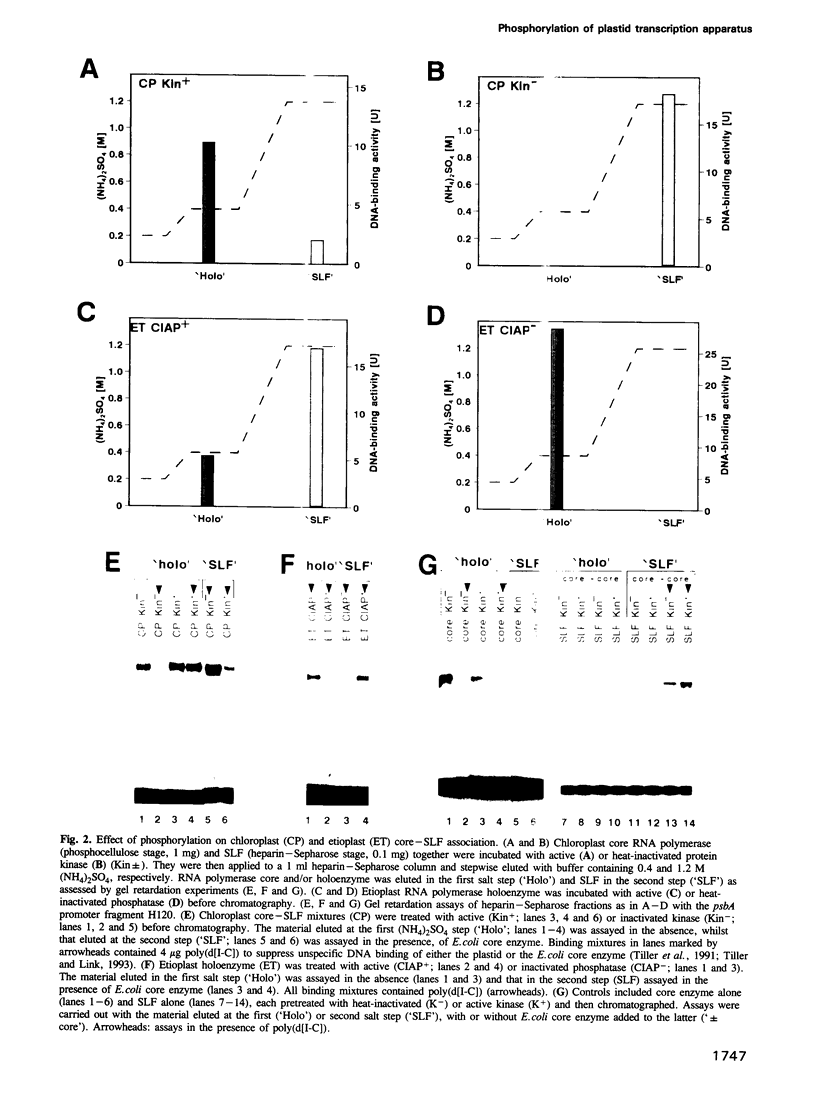
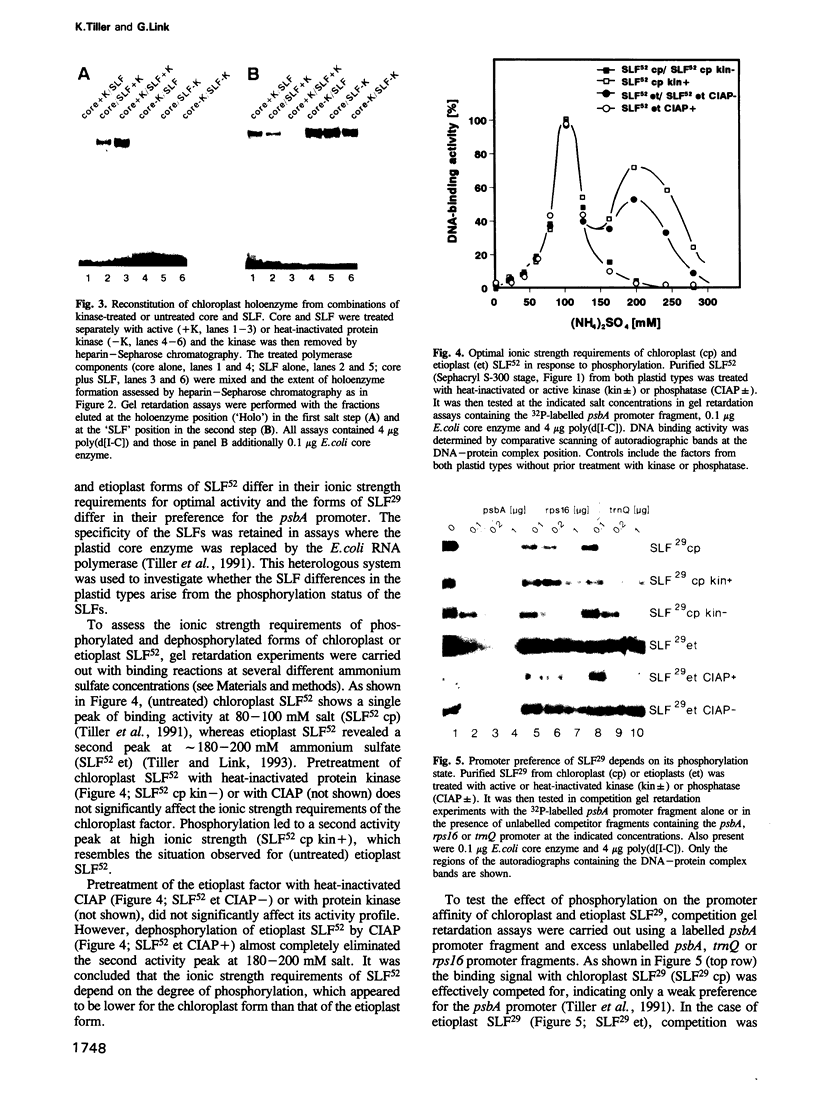
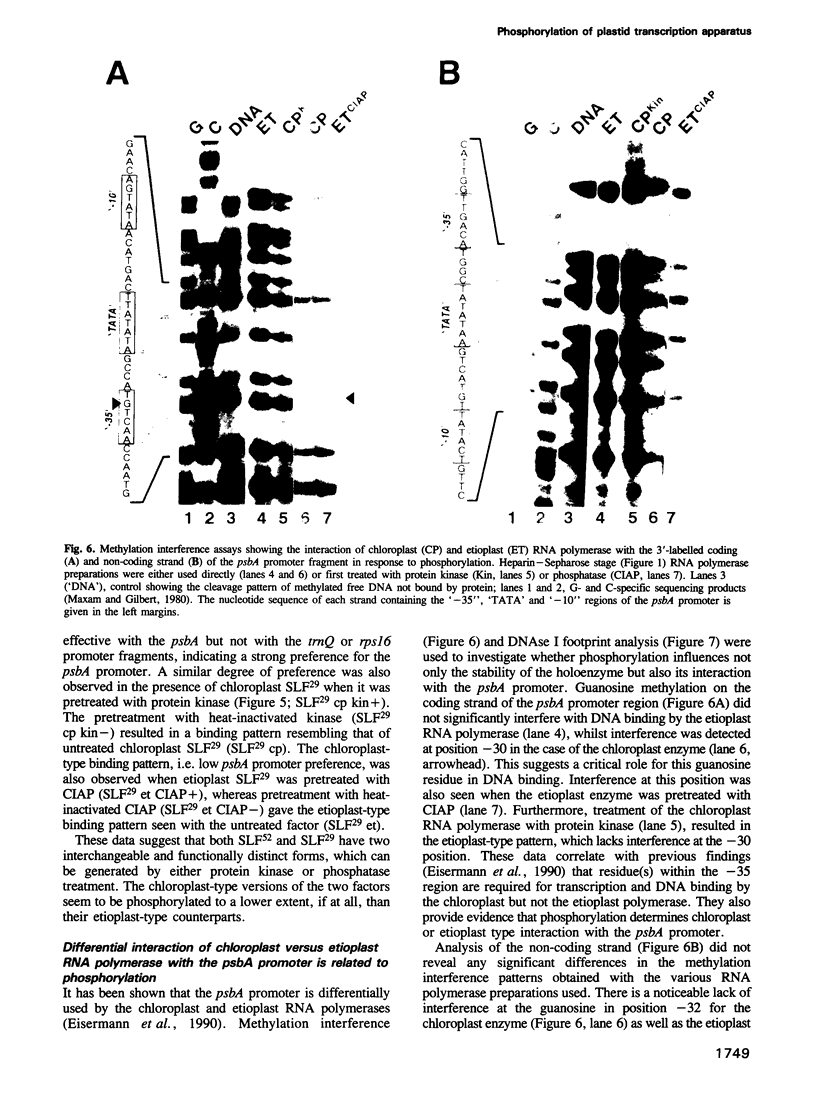
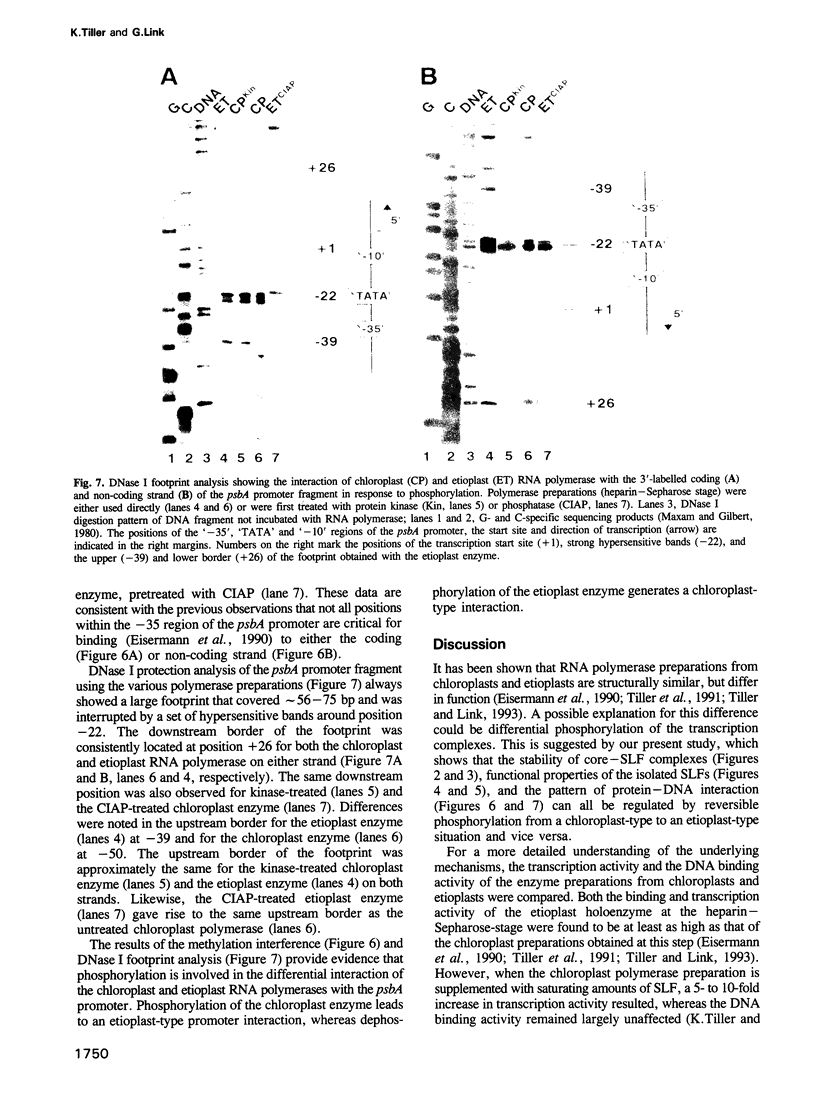
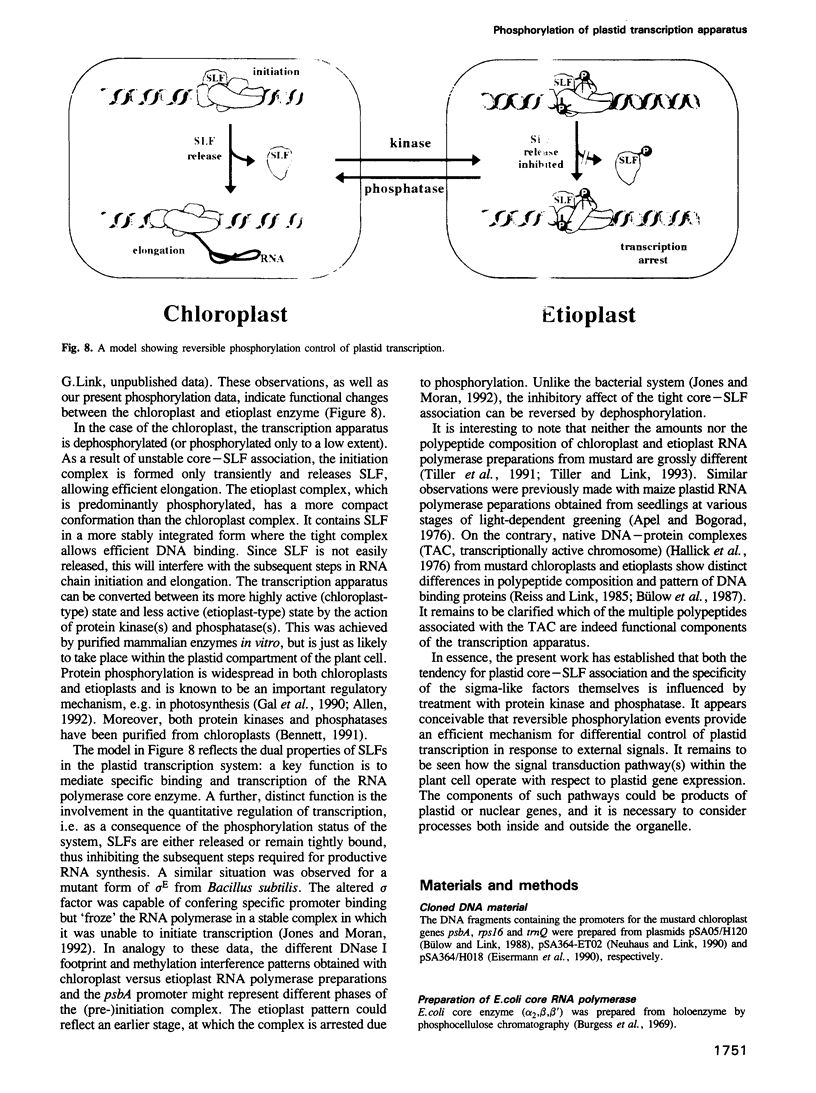
Chloroplast and etioplast RNA polymerase preparations each consist of a multi-subunit core and a set of three sigma-like transcription factors, SLF67, SLF52 and SLF29. Despite this structural similarity, the enzymes from either plastid type are functionally distinct, as is reflected by their different promoter usage and the tight core-SLF association in the etioplast but not the chloroplast holoenzyme. We tested whether these differences are related to phosphorylation. Treatment of the chloroplast enzyme with protein kinase converted it to an etioplast-type form and vice versa, treatment of the etioplast enzyme with phosphatase generated chloroplast-type properties. Although both the core enzyme and the SLF polypeptides were phosphorylation targets, only the SLFs seem to confer plastid-type-specific DNA binding characteristics. Methylation interference and DNase I footprint patterns in the psbA promoter region were found to correlate with the phosphorylation state of the chloroplast and etioplast enzymes.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Allen J. F. How does protein phosphorylation regulate photosynthesis? Trends Biochem Sci. 1992 Jan;17(1):12–17. doi: 10.1016/0968-0004(92)90418-9. [DOI] [PubMed] [Google Scholar]
- Apel K., Bogorad L. Light-induced increase in the activity of maize plastid DNA-dependent RNA polymerase. Eur J Biochem. 1976 Aug 16;67(2):615–620. doi: 10.1111/j.1432-1033.1976.tb10727.x. [DOI] [PubMed] [Google Scholar]
- Bedbrook J. R., Link G., Coen D. M., Bogorad L. Maize plastid gene expressed during photoregulated development. Proc Natl Acad Sci U S A. 1978 Jul;75(7):3060–3064. doi: 10.1073/pnas.75.7.3060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bottomley W., Smith H. J., Bogorad L. RNA polymerases of maize: partial purification and properties of the chloroplast enzyme. Proc Natl Acad Sci U S A. 1971 Oct;68(10):2412–2416. doi: 10.1073/pnas.68.10.2412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess R. R., Travers A. A., Dunn J. J., Bautz E. K. Factor stimulating transcription by RNA polymerase. Nature. 1969 Jan 4;221(5175):43–46. doi: 10.1038/221043a0. [DOI] [PubMed] [Google Scholar]
- Corden J. L. Tails of RNA polymerase II. Trends Biochem Sci. 1990 Oct;15(10):383–387. doi: 10.1016/0968-0004(90)90236-5. [DOI] [PubMed] [Google Scholar]
- Datta N., Cashmore A. R. Binding of a pea nuclear protein to promoters of certain photoregulated genes is modulated by phosphorylation. Plant Cell. 1989 Nov;1(11):1069–1077. doi: 10.1105/tpc.1.11.1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng X. W., Gruissem W. Control of plastid gene expression during development: the limited role of transcriptional regulation. Cell. 1987 May 8;49(3):379–387. doi: 10.1016/0092-8674(87)90290-x. [DOI] [PubMed] [Google Scholar]
- Eisermann A., Tiller K., Link G. In vitro transcription and DNA binding characteristics of chloroplast and etioplast extracts from mustard (Sinapis alba) indicate differential usage of the psbA promoter. EMBO J. 1990 Dec;9(12):3981–3987. doi: 10.1002/j.1460-2075.1990.tb07619.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fried M., Crothers D. M. Equilibria and kinetics of lac repressor-operator interactions by polyacrylamide gel electrophoresis. Nucleic Acids Res. 1981 Dec 11;9(23):6505–6525. doi: 10.1093/nar/9.23.6505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gal A., Hauska G., Herrmann R., Ohad I. Interaction between light harvesting chlorophyll-a/b protein (LHCII) kinase and cytochrome b6/f complex. In vitro control of kinase activity. J Biol Chem. 1990 Nov 15;265(32):19742–19749. [PubMed] [Google Scholar]
- Gilmartin P. M., Sarokin L., Memelink J., Chua N. H. Molecular light switches for plant genes. Plant Cell. 1990 May;2(5):369–378. doi: 10.1105/tpc.2.5.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilfoyle T. J. A protein kinase from wheat germ that phosphorylates the largest subunit of RNA polymerase II. Plant Cell. 1989 Aug;1(8):827–836. doi: 10.1105/tpc.1.8.827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallick R. B., Lipper C., Richards O. C., Rutter W. J. Isolation of a transcriptionally active chromosome from chloroplasts of Euglena gracilis. Biochemistry. 1976 Jul 13;15(14):3039–3045. doi: 10.1021/bi00659a016. [DOI] [PubMed] [Google Scholar]
- Helmann J. D., Chamberlin M. J. Structure and function of bacterial sigma factors. Annu Rev Biochem. 1988;57:839–872. doi: 10.1146/annurev.bi.57.070188.004203. [DOI] [PubMed] [Google Scholar]
- Jones C. H., Moran C. P., Jr Mutant sigma factor blocks transition between promoter binding and initiation of transcription. Proc Natl Acad Sci U S A. 1992 Mar 1;89(5):1958–1962. doi: 10.1073/pnas.89.5.1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein R. R., Mullet J. E. Light-induced transcription of chloroplast genes. psbA transcription is differentially enhanced in illuminated barley. J Biol Chem. 1990 Feb 5;265(4):1895–1902. [PubMed] [Google Scholar]
- Klein R. R. Regulation of light-induced chloroplast transcription and translation in eight-day-old dark-grown barley seedlings. Plant Physiol. 1991 Sep;97(1):335–342. doi: 10.1104/pp.97.1.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhlemeier C. Transcriptional and post-transcriptional regulation of gene expression in plants. Plant Mol Biol. 1992 May;19(1):1–14. doi: 10.1007/978-94-011-2656-4_1. [DOI] [PubMed] [Google Scholar]
- Kung S. D., Lin C. M. Chloroplast promoters from higher plants. Nucleic Acids Res. 1985 Nov 11;13(21):7543–7549. doi: 10.1093/nar/13.21.7543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latchman D. S. Eukaryotic transcription factors. Biochem J. 1990 Sep 1;270(2):281–289. doi: 10.1042/bj2700281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Link D. P., Lantz B. M. Vasodilator response in the lower extremity induced by contrast medium. I Canine model. Acta Radiol Diagn (Stockh) 1982;23(2):81–86. doi: 10.1177/028418518202300201. [DOI] [PubMed] [Google Scholar]
- Link G. DNA sequence requirements for the accurate transcription of a protein-coding plastid gene in a plastid in vitro system from mustard (Sinapis alba L.). EMBO J. 1984 Aug;3(8):1697–1704. doi: 10.1002/j.1460-2075.1984.tb02034.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Mullet J. E., Klein R. R. Transcription and RNA stability are important determinants of higher plant chloroplast RNA levels. EMBO J. 1987 Jun;6(6):1571–1579. doi: 10.1002/j.1460-2075.1987.tb02402.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuhaus H., Link G. The chloroplast psbK operon from mustard (Sinapis alba L.): multiple transcripts during seedling development and evidence for divergent overlapping transcription. Curr Genet. 1990 Nov;18(4):377–383. doi: 10.1007/BF00318220. [DOI] [PubMed] [Google Scholar]
- Nickelsen J., Link G. RNA-protein interactions at transcript 3' ends and evidence for trnK-psbA cotranscription in mustard chloroplasts. Mol Gen Genet. 1991 Aug;228(1-2):89–96. doi: 10.1007/BF00282452. [DOI] [PubMed] [Google Scholar]
- Reiss T., Link G. Characterization of transcriptionally active DNA-protein complexes from chloroplasts and etioplasts of mustard (Sinapis alba L.). Eur J Biochem. 1985 Apr 15;148(2):207–212. doi: 10.1111/j.1432-1033.1985.tb08826.x. [DOI] [PubMed] [Google Scholar]
- Rodermel S. R., Bogorad L. Maize plastid photogenes: mapping and photoregulation of transcript levels during light-induced development. J Cell Biol. 1985 Feb;100(2):463–476. doi: 10.1083/jcb.100.2.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarokin L. P., Chua N. H. Binding sites for two novel phosphoproteins, 3AF5 and 3AF3, are required for rbcS-3A expression. Plant Cell. 1992 Apr;4(4):473–483. doi: 10.1105/tpc.4.4.473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiura M. The chloroplast genome. Plant Mol Biol. 1992 May;19(1):149–168. doi: 10.1007/BF00015612. [DOI] [PubMed] [Google Scholar]
- Surzycki S. J., Shellenbarger D. L. Purification and characterization of a putative sigma factor from Chalamydomonas reinhardi. Proc Natl Acad Sci U S A. 1976 Nov;73(11):3961–3965. doi: 10.1073/pnas.73.11.3961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiller K., Eisermann A., Link G. The chloroplast transcription apparatus from mustard (Sinapis alba L.). Evidence for three different transcription factors which resemble bacterial sigma factors. Eur J Biochem. 1991 May 23;198(1):93–99. doi: 10.1111/j.1432-1033.1991.tb15990.x. [DOI] [PubMed] [Google Scholar]
- Tiller K., Link G. Sigma-like transcription factors from mustard (Sinapis alba L.) etioplast are similar in size to, but functionally distinct from, their chloroplast counterparts. Plant Mol Biol. 1993 Feb;21(3):503–513. doi: 10.1007/BF00028807. [DOI] [PubMed] [Google Scholar]
- Trewavas A., Gilroy S. Signal transduction in plant cells. Trends Genet. 1991 Nov-Dec;7(11-12):356–361. doi: 10.1016/0168-9525(91)90255-o. [DOI] [PubMed] [Google Scholar]