Abstract
Local translation of nuclear-encoded mitochondrial mRNAs is essential for mitochondrial activity, yet there is little insight into the role that axonal trafficking of these transcripts play in neuronal function and behavior. Previously, we identified a 38 nucleotide stem-loop structure (zipcode) in the 3′ untranslated region of the Cytochrome C oxidase IV (COXIV) mRNA that directs the transport of a reporter mRNA to the axon of superior cervical ganglion neurons (SCG). Over-expression of a chimeric reporter mRNA with the COXIV zipcode competed with the axonal trafficking of endogenous COXIV mRNA, and led to attenuated axon growth in SCG neurons. Here, we show that exogenous expression of the COXIV zipcode in cultured SCG neurons also results in the reduction of local ATP levels and increases levels of reactive oxygen species (ROS) in the axon. We took advantage of this “competition” phenotype to investigate the in vivo significance of axonal transport of COXIV mRNA. Towards this end, we generated transgenic mice expressing a fluorescent reporter fused to COXIV zipcode under a forebrain-specific promoter. Immunohistological analyses and RT-PCR analyses of RNA from the transgenic mouse brain showed expression of the reporter in the deep layer neurons in the pre-frontal and frontal cortex. Consistent with the in vitro studies, we observed increased ROS levels in neurons of these transgenic animals. A battery of behavioral tests on transgenic mice expressing the COXIV zipcode revealed an “anxiety-like” behavioral phenotype, suggesting an important role for axonal trafficking of nuclear-encoded mitochondrial mRNAs in neuronal physiology and animal behavior.
Keywords: Axonal mRNAs, mitochondrial mRNAs, intra-axonal protein synthesis, energy metabolism, ROS generation, mouse behavior
INTRODUCTION
Intracellular transport and local protein synthesis of mRNAs to the distal structural/functional domains of the neuron function to regulate protein content in these subcellular compartments in response to specific stimuli or environmental and/or developmental cues (Jung et al., 2012). It is now well-established that local translation of the heterogeneous population of mRNAs in the axon is critical for axonal function, maintenance and repair (Kaplan et al., 2009; Gumy et al., 2010; Yoon et al., 2012; Kar et al., 2013).
A unique feature of all axonal mRNA profiling studies conducted to date is the presence of a large number of nuclear-encoded mitochondrial mRNAs. Local translation of nuclear-encoded mitochondrial mRNAs such as Cytochrome C oxidase IV (COXIV), and ATP synthase 9 (ATP5G1), that encode key subunits of the oxidative phosphorylation complexes, play an important role in the regulation of local axonal energy metabolism, function and growth (Hillefors et al., 2007; Aschrafi et al., 2008; Natera-Naranjo et al., 2012; Aschrafi et al., 2012). Disruption of axonal COXIV or ATP5G1 expression leads to compromised mitochondrial membrane potential, decreased ATP levels and generation of reactive oxygen species (ROS) in the axon (Hillefors et al., 2007; Natera-Naranjo et al., 2012). However, little is known about the physiological or behavioral significance of the subcellular localization and local translation of nuclear-encoded mitochondrial mRNAs. In a previous study, we reported the identification of a putative 38bp stem-loop structure (“zipcode”) in the 3′ untranslated region (3′UTR) of the COXIV transcript that directs the axonal localization of the COXIV mRNA in superior cervical ganglion (SCG) neurons. Introduction of a chimeric reporter mRNA containing the COXIV zipcode in SCG neurons resulted in decreases in endogenous axonal COXIV mRNA levels, as well as attenuation of axon growth (Aschrafi et al., 2010). Based on these finding, we hypothesized that the effects observed on axonal COXIV mRNA levels and axon growth were, in part, due to the competitive inhibition of endogenous COXIV mRNA trafficking to the axon.
In this work, we continue to investigate the physiological and behavioral consequences of exogenous COXIV zipcode expression on neuronal function. Here, we show that over-expression of COXIV zipcode in SCG neurons leads to a decrease in endogenous COXIV mRNA levels and reduction in local ATP levels in the axon, as well as increased ROS production. In addition, we tested the hypothesis that dysregulated trafficking of nuclear-encoded mitochondrial mRNAs, induced by over-expression of the COXIV zipcode, can influence animal behavior. Towards this end, we generated transgenic mice that express the COXIV zipcode in a subset of forebrain neurons and compared the behavior of these mice with their non-transgenic littermates. Results of these preliminary studies suggest the presence of an “anxiety-like” phenotype in COXIV zipcode over-expressing transgenic mice, and raises the possibility that axonal trafficking of nuclear-encoded mitochondrial mRNAs plays an important role in brain function and animal behavior.
METHODS
Neuronal Cell Cultures
SCGs were obtained from 3-day-old Harlan Sprague Dawley rats of either sex. Neurons were dissociated using Miltenyi Biotec gentle MACS Dissociator and Neuronal Tissue Dissociation Kit according to the manufacturer’s protocol. Dissociated primary neurons were plated into the center compartment of a three-compartmented Campenot culture chamber (Hillefors et al., 2007) (Figure 1 A). Cells were grown in serum-free Neurobasal medium (Invitrogen) containing 50 ng/ml nerve growth factor (NGF), 20 mM potassium chloride (KCl), and 20 U/ml penicillin and 20 mg/ml streptomycin (Hyclone) for 2–7 d before use. The culture medium was replaced every 3–4 d. Two days after plating, 50 μM 5-fluoro-2′-deoxyuridine was added to the culture medium to inhibit the growth of non-neuronal cells and remained in the medium for the duration of the experiments. Culture media also contained NGF at all times. Phase-contrast microscopy and 5-bromodeoxyuridine staining was used to check for the presence of neuronal soma and non-neuronal cells in the side compartments which contained the distal axons used in the experiments. In addition, expression of γ-actin mRNA was used to assess the purity of the RNA prepared from the distal axons.
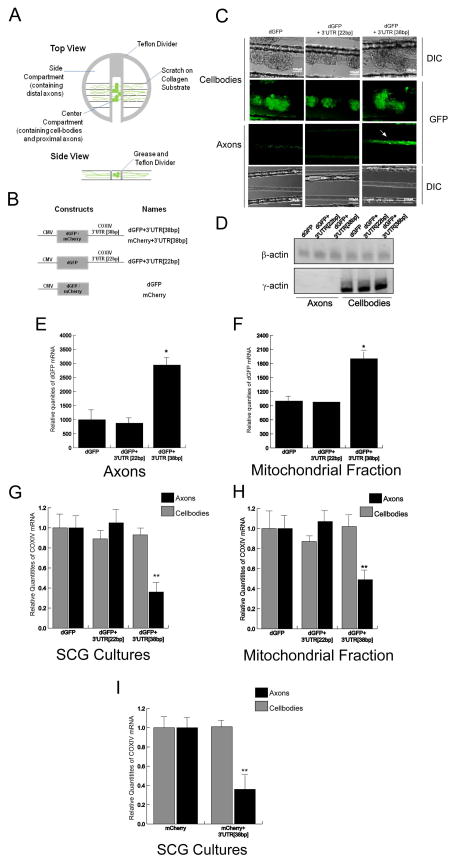
Figure 1.
Over-expression of reporter mRNA containing the 38bp COXIV zipcode inhibits the axonal localization and mitochondrial targeting of nuclear-encoded mitochondrial mRNAs. (A) Schematic diagram showing the compartmented Campenot cell culture chambers (upper panel). Lower panel shows a single track in the bottom of the culture dish indicating the location of cell-bodies and proximal axons in the center compartment and distal axons in the side compartments. (B) Overview of constructs used in this study. Schematic representation of reporter gene plasmids carrying the dGFP cDNA followed by either the final 38bp or 22bp COXIV 3′UTR (dGFP+3′UTR [38bp] and dGFP+3′UTR [22bp]) or mCherry cDNA followed by final 38bp of COXIV 3′UTR(mCherry+3′UTR [38bp]). (C) Representative DIC and fluorescence images showing expression of dGFP constructs in the transfected cell-bodies and distal axons. The arrow shows reporter gene expression in axons of cell-bodies transfected with the chimeric construct that contained the 38bp zipcode. No reporter gene expression was observed in the distal axons of neurons transfected with either a plasmid containing the GFP alone or the dGFP+3′UTR[22bp] constructs. (D) Levels of γ-actin mRNA was used to assess the purity of the RNA prepared from the distal axons located in the side compartments of the Campenot cell culture chambers. (E) Quantitation of dGFP mRNA levels in distal axons of SCG neurons were determined by qRT-PCR 24 h after transfection with plasmid DNA. The dGFP mRNA levels were expressed relative to βactin transcript. (F) SCG neurons (6DIV) were transfected with dGFP constructs and total RNA was prepared from a mitochondrial fraction isolated from distal axons using a mitochondrial isolation kit (Pierce). Quantitation of dGFP mRNA levels from total mitochondrial RNA was determined by qRT-PCR 24 h after DNA transfection. The dGFP mRNA levels were expressed relative to COXII mRNA. (G) Quantitation of COXIV mRNA levels in transfected cell-bodies and distal axons of SCG neurons was determined by qRT-PCR 24 h after DNA transfection. COXIV mRNA levels are expressed relative to β-actin mRNA. (H) Quantitation of COXIV mRNA levels from total mitochondrial mRNA was determined by qRT-PCR 24 h after DNA transfection. COXIV mRNA levels are expressed relative to COXII mRNA. (I) mCherry constructs containing the 38bp COXIV zipcode or 22bp COXIV zipcode or mCherry alone were transfected into 6DIV SCG neurons. After transfection (24 h) COXIV mRNA levels were assessed by qRT-PCR. COXIV mRNA levels were expressed relative to β-actin mRNAs. Error bars represent SEM for n=3 samples. Two-way ANOVA, Posthoc Tukey’s test *,p<0.05, **, p<0.01.
Transfection of neurons
The dGFP and mCherry chimeric gene constructs were transfected into the center compartment of the culture dishes (7 DIV) using Xfect transfection reagent (Clontech) according to manufacturers protocol. At least three plates were transfected for each condition and all experiments were repeated three times.
Quantitation of mRNA levels
mRNA levels were determined by qRT-PCR in total RNA samples prepared from SCG axons and parental cell soma using the Trizol reagent (Invitrogen) and the Direct-zol™ RNA MiniPrep Kit (Zymo Research) according to the manufacturer’s protocol. Total RNA was incubated with RNase-free DNaseI to remove trace DNA contamination. The total RNA sample was used for reverse transcription with qScript cDNA SuperMix (Quanta Biosciences) and qPCR analyses was conducted using gene-specific primers for COXIV, β-actin and γ-actin (Qiagen). The relative levels of the COXIV transcript were normalized to β-actin to provide an internal control for reverse transcription and axonal density. RNA values are expressed relative to control by the comparative threshold (CT) method (Schmittgen and Livak, 2008).
ATP and ROS measurements
ATP levels were assessed by luminescence using the CellTiter-Glo Luminescent Cell Viability Assay (Promega) according to the manufacturer’s instruction. Briefly, SCG neurons in the center compartment were transfected with dGFP-COXIV zipcode or the dGFP control constructs. After transfection (48 h), the distal axons were independently harvested in 100 μl of the CellTiter-Glo reagent, and the luciferase luminescence was then recorded by a GloMax-96 Microplate Luminometer (Promega) with an integration time of 1 s per well.
Reactive oxygen species (ROS) were detected using Image-iT™ Live Green Reactive Oxygen Species Detection Kit (Invitrogen). After transfection of the dGFP-COXIV zipcode and dGFP control expression constructs, cultures were incubated in Hank’s balanced salt solution with calcium and magnesium (HBSS/Ca/Mg) containing 5-(and-6)-carboxy-2′,7′-dichlorodihydrofluorescein diacetate (H2DCFDA, 25 μM) for 30 min at 37°C. Cultures were washed thrice with HBSS/Ca/Mg and fluorescence was visualized according to the manufacturer’s recommendation, using an EVOS inverted microscope (Advanced Microscopy Group). Images were deconvoluted and fluorescence quantitated using the Deconvolution Lab plugin (Vonesch and Unser, 2008) and ImageJ software (NIH).
We used MitoSOX Red (Invitrogen) to determine relative mitochondrial ROS levels. After COXIV zipcode and dGFP control expression vector transfection (24 h), cultures were incubated for 10 min at 37°C in the cell culture medium containing MitoSOX Red (4 μM). After incubation in MitoSOX Red, cultures were washed thrice with culture medium and MitoSOX Red fluorescence was measured according to the manufacturer’s recommendations.
All fluorescence quantification used original unprocessed image data, with no pixels at zero intensity or saturated. Fluorescence levels were quantified using NIH ImageJ software. For quantification of fluorescence, the boundary of the axon bundle was selected by the drawing/selection tool, and the measurement function of the analysis tool was used to measure the area, integrated density, and the mean gray value for each bundle. The integrated density value was normalized for the area by dividing by the area of the selected bundle. To determine the background, several regions near the bundle were selected and the mean fluorescence of background reading was obtained. The relative fluorescence intensity for each bundle was determined as the difference between normalized integrated density value and the product of mean background fluorescence intensity and the area of the selected bundle. All axon bundles in each culture dish were measured, and statistical comparisons were made by measuring at least 35–40 axon bundles per treatment.
Plasmid construction
The construction of the dGFP plasmids has been described previously (Aschrafi et al., 2010). The mCherry+3′UTR[38bp] construct was made by annealing and ligating the sense and antisense strands of 38 nucleotide oligomer of COXIV 3′UTR into the SalI and SacII sites following the mCherry open-reading-frame (ORF) of a pmCherry-C1 expression vector (Clontech). The clones were verified by sequencing and restriction digest analysis.
For the transgenic mouse construct, PCR primers specific for mCherry ORF and COXIV 3′UTR were used to amplify the mCherry+3′UTR[38bp] cassette. The mCherry+3′UTR [38bp] fragment was cloned into the NotI site of the pMM403 vector (Mayford et al., 1996), which inserts the mCherry+3′UTR[38bp] (COXIV zipcode) cassette downstream of the 8.5kb CaMKIIα promoter. The pMM403- mCherry+3′UTR [38bp] COXIV zipcode clones were verified by nucleic acid sequencing.
Animals
The Not1 fragment of a pMM403- mCherry+3′UTR [38bp] COXIV zipcode construct was isolated and injected into pronuclei of C57BL/6N fertilized oocytes. On the next day, embryos that had developed to the 2-cell stage were transferred to the oviducts of pseudo pregnant female recipients. The genotype of the resulting pups was determined using PCR of genomic DNA extracted from the tail of each mouse using transgene specific primers. After genotyping, the founder transgenic animals and their progeny were crossed with wild type C57BL/6N mates for at least four generations before use in behavioral experiments. All subjects were maintained under 12:12 h dark: light cycle with food and water ad libitum. Animal handling and use was in accordance with a protocol approved by the Animal Care and Use Committee of the National institutes of Health. All expression and behavioral studies employed heterozygous transgenic animals and their non-transgenic littermates.
Characterization of transgenic cassette expression
mCherry+3′UTR[38bp] COXIV zipcode cassette expression was evaluated in adult mice, both by immunohistochemical imaging of mCherry expression and RT-PCR analyses. Analysis of transgene-derived mCherry fluorescence in whole brain slices isolated from the founder lines revealed relatively low levels of mCherry fluorescence in the brain of transgenic mice (data not shown). However, immunostaining of transgenic brain slices with a mCherry-specific dsRED antibody produced a robust and reproducible mCherry expression pattern. Hence, we used immunostaining with the mCherry-specific-dsRED antibody to monitor the expression of the COXIV zipcode in transgenic animals. Seven independent founder lines were generated out of which 3-lines were used for expression and behavior analysis based on their transgene expression pattern.
For visualization of transgene expression in the brain, adult transgenic and non-transgenic littermate mice were perfused with 4% paraformaldehyde in phosphate buffered saline. The brain were coronally sectioned on a vibratome (30-μm thickness) and immunostained with dsRED antibody (Clontech) and visualized by Alexa488 to detect mCherry expression. All sections were counterstained with 4′,6-diamidino-2-phenylindole (DAPI) to label nuclei. Six coronal sections from each animal were observed under a confocal microscope (prefrontal cortex (PFC) bregma −2.10 and −1.54; frontal (M1) cortex −2.10 and −1.54). The merged confocal images show single optical sections collected with the pinhole set to 5 Airy Unit (10 micron section) for the red channel and adjusted to give the same optical slice thickness in the other channels. Acquisition settings were kept constant between specimens, and comparable optical sections were imaged. Fluorescence imaging was conducted on an inverted Zeiss Axiovert microscope with a xenon-lamp. Confocal imaging was conducted on a Zeiss LSM510 confocal microscope.
For PCR analyses, PFC, frontal cortex (M1), hippocampus (hip) and cerebellum (cb) from COXIV zipcode transgenic mice and their non-transgenic littermates were dissected and total RNA was extracted using Trizol reagent (Invitrogen) and the Direct-zol™ RNA MiniPrep Kit (Zymo Research) according to manufacturers protocol. RNA concentration and quality were examined by NanoDrop (Thermo Fisher Scientific). Four micrograms of RNA was used for reverse transcription with qScript cDNA SuperMix (Quanta Biosciences). One μl cDNA was used for PCR. GAPDH and β-actin primers used as internal positive controls were obtained from Qiagen. The transgene specific primers used in the analysis were: Forward primer: 5′ gccccaagctcgtcagtc 3′; Reverse primer: 5′ gacctcagcgtcgtagtggc 3′. GAPDH and β-actin mRNA levels were used as controls.
ROS Detection
In vivo detection of ROS was performed as described by Behrens et al., (2007). Briefly, two serial intraperitoneal (IP) injections of freshly prepared dihydroethidium (DHE, 27 mg/kg; Invitrogen) were given at 30-min intervals. Eighteen hours later, mice were perfused with 4% paraformaldehyde in phosphate-buffered saline. Brains were coronally-sectioned on a vibratome (30-μm thickness) and counterstained with DAPI. Six coronal sections from each animal were observed under a fluorescence microscope. Red fluorescence intensity of oxidized DHE was quantified with Image-J (NIH) after converting fluorescence images to gray scale. Relative fluorescence intensity from PFC or FC area was normalized to the average intensity value in the surrounding regions.
Mouse Behavior
All behavioral tasks were performed with male transgenic mice and age-matched non-transgenic littermates at 8–13 weeks after birth. Procedures were approved by the NIH Animal Care and Use Committee. After weaning, same-sex mice of mixed genotype were housed in groups of 3–5 and maintained in a 12 h light/dark cycle (light from 6 am to 6 pm). Food and water were available to the animals ad libitum. All tests, with the exception of the Y-maze, were performed during the light cycle. Testing was conducted and scored by an investigator blind to genotype.
Open-field Test
Animal activity was monitored over a 30 min period in a 40 × 40 cm Versamax test chamber (Accuscan Instruments) with a light level of 50 lux. Activity was measured by photobeam breaks and was analyzed every 3 min.
Elevated Plus-maze
“Anxiety-like” behavior was analyzed using the Elevated Plus-maze. The maze apparatus consists of two 30 × 5 cm closed arms with 16 cm high opaque walls and two 30 × 5 cm open-arms with 3 mm high ledges. The test was performed in a room homogenously lighted at 100 lux. The mouse was placed in the center of the apparatus and movement and position of the mouse was tracked for 5 min by automated tracking software (ANY-maze, Stoelting Co)
Light-Dark Test
The mouse was placed into the light side of a plexiglass light/dark box with the dark side separated from the light with a dark divider. “Anxiety-like” behavior was measured using ANY-maze automatic tracking software over a 8 min period.
Y-Maze Spontaneous Alternation Test
Mice were placed in one arm of a Y-maze facing the center and allowed to explore for 8 min while video was recorded and alternations were scored by hand. The Y-Maze apparatus is a transparent plexiglass apparatus with three arms of equal length (40 cm long × 4.5 cm wide × 12 cm high) each situated 120° from the others. Entries were scored by hand over the 8 min test-period based on entries over 1/3rd of the way into an arm. Alternation was considered consecutive entries into sequential arms without repeated entries (ie: ABC or CBA, but not BAB). Percent of spontaneous alternation was then calculated as actual alternations divided by total possible alternations (total entries minus two). Testing was performed during the first three hours of the dark cycle (6–9 pm) and the light in the room was at 30 lux.
Cued Fear Conditioning
Cued fear conditioning was performed in a dark room in two conditioning chambers with internal lighting. The chambers have plexiglass front and back, aluminum walls, and measure 36 × 21 × 20 cm (Med Associates). The chamber floor consists of steel rods 3.2 mm in diameter placed 7.9 mm apart.
The tests were carried out over three consecutive days. On day one, the mice were habituated to the chamber for 5 min. The chambers were cleaned with 70% ethanol before the habituation and between mice. On day two, the mice were put into the chamber and movement was recorded with FreezeFrame software (Actimetrics). Video was recorded for 3 min before the onset of an 85 db, 28 sec tone (conditioned stimulus, CS). Following the 28 sec tone, a footshock (unconditioned stimulus, US) of 0.8 mA was delivered through the grid floor. Video was recorded for 1 min after the footshock to determine freezing behavior during training. On the third day of testing, the chamber was cleaned with Windex and white plastic covers were inserted over the floor and on the walls to change the context of the testing environment. Video was recorded for 3 min prior to the onset of a 28 sec, 85 decibel tone (CS). Freezing behavior was recorded and monitored for 1 min following the tone.
Forced Swim Test
Mice were placed in a plastic cylinder 24 cm high and 18 cm in diameter filled with water (21–25° C). Swimming was recorded on video and scored by hand from video. Percentage of time immobile was calculated over the course of a 10 min trial.
Rotarod
Balance and motor coordination were evaluated using an Accurotor accelerating rotarod (AccuScan Instruments). Four animals were tested concurrently in four separate compartments, each measuring 11 cm wide on a rod 3 cm in diameter, elevated 37 cm off of the ground. Initial velocity was 4 rpm and the rod was gradually accelerated at a rate of 20 rpm/min to a maximum of 40 rpm. Latency to fall off the rod during a 2 min test period was measured.
Statistics
Quantitative data are presented as mean ± SEM. Differences between groups were assessed with a Students t test (two groups), one-way analysis of variance followed by post hoc tests for multiple group comparisons.
RESULTS
Exogenous expression of COXIV zipcode decreases the relative abundance of endogenous COXIV transcripts in the axon
In a previous study, we reported that a 38bp stem-loop structure present in the 3′UTR of COXIV mRNA (“zipcode”) contained the requisite sequence information necessary to transport a reporter mRNA to the distal axons of SCG neurons (Aschrafi et al., 2010). To confirm this finding, neurons and proximal axons located in the center compartment of Campenot culture chambers were transfected with a construct expressing the destabilized GFP (dGFP) reporter with the 38bp COXIV zipcode fused to the 3′-terminus. We also transfected SCG neurons with a dGFP construct containing a partial deletion of the COXIV zipcode (22bp). This truncated 22bp COXIV zipcode lacked the necessary information to traffic the reporter mRNA to axons (Aschrafi et al., 2010). The dGFP plasmid alone was used as a negative control (Figure 1B). As shown in Figure 1C (upper panel), fluorescence imaging of the central compartment of the Campenot chamber 24 h after transfection showed comparable transfection efficiency in the soma and proximal axons for all three dGFP expression plasmids. Consistent with our previous studies, we observed dGFP signal in the distal axons of neurons expressing the reporter gene construct containing the 38bp COXIV zipcode, while very low levels of axonal dGFP fluorescence was detected in distal axons of neurons transfected with the truncated 22bp COXIV zipcode construct and dGFP alone (Figure 1C). Consistent with our earlier reports, qRT-PCR analyses from total RNA isolated from distal axons 24 h after transfection showed a significant increase in dGFP mRNA levels in axons of SCG cultures transfected with the dGFP construct containing the 38bp COXIV zipcode as compared to either the truncated 22bp COXIV zipcode construct or dGFP alone (Figure 1E). The expression of γ-actin mRNA was used as a control to assess the purity of the axonal mRNA preparations. The lack of γ-actin transcript in the side compartment of the Campenot culture chambers attests to the purity of the RNA preparations obtained from the distal axons (Figure 1D). Additionally, similar to our previous results, we also observed a significant increase in the levels of dGFP mRNA in the mitochondrial fractions isolated from distal axons of SCG cultures transfected with the dGFP construct containing the 38bp COXIV zipcode as compared to either the truncated 22bp COXIV zipcode construct or GFP alone (Figure 1F). These results indicate the full-length 38bp COXIV zipcode is necessary and sufficient to direct the trafficking of a reporter mRNA to the axon in SCG neurons.
To assess the effect of COXIV zipcode over-expression on axonal COXIV mRNA levels, we performed a qRT-PCR analysis on total RNA isolated from the distal axons of transfected neurons situated in the side compartments of the Campenot culture chambers, 24 h after transfection with either dGFP+3′UTR[38bp], dGFP+3′UTR[22bp], or dGFP expression constructs. As shown in Figure 1G, the expression of either dGFP or the 22bp COXIV zipcode deletion (dGFP+3′UTR[22bp]) construct in SCG neurons showed minimal effects on axonal COXIV transcript levels. In contrast, a 50–60% decrease in endogenous COXIV mRNA levels was observed in the distal axons of neurons transfected with the dGFP+3′UTR [38bp] construct. No significant change in the somal levels of COXIV mRNA was observed after dGFP+3′UTR[38bp] expression suggesting that over-expression of COXIV zipcode had minimal effect on total COXIV mRNA levels in the neuron (Figure 1G). These results suggest that reporter mRNA containing the COXIV zipcode competes with the endogenous COXIV transcript for trafficking to the axon.
Previous studies in yeast and mammalian cells suggested that trafficking of specific nuclear-encoded mRNAs to the vicinity of the mitochondria required specific cis-regulatory elements present in the 3′UTR of the transcripts (Corral-Debrinski et al., 1999, 2000). Additionally, our previous results showed that the 38bp COXIV zipcode could target a reporter gene transcript to the mitochondrial fraction isolated from distal axons (Aschrafi et al., 2010, and Figure 1F). To investigate the affect of COXIV zipcode expression on trafficking of COXIV mRNA to the axonal mitochondrial fraction, we quantified the levels of COXIV mRNA in mitochondrial fractions derived from SCG cell-bodies and distal axons 24 h after transfection with either dGFP+3′UTR [38bp], dGFP+3′UTR [22bp], or dGFP expression constructs. The relative abundance of COXIV mRNA in the mitochondrial fractions was determined by qRT-PCR using the COXII gene, a mitochondrial DNA-encoded gene, as an internal control. Results of the experiments revealed that over-expression of the COXIV zipcode in SCG neurons led to a 50% decrease in the relative abundance of the COXIV transcript in the axonal mitochondrial fraction: no such affect was observed in neurons transfected with either the 22bp zipcode deletion construct or dGFP alone (Figure 1H). In addition, no change in relative abundance of COXIV mRNA was observed in the mitochondrial fractions derived from cell-bodies expressing the COXIV zipcode (Figure 1H). Taken together, these results suggest that over-expression of the COXIV zipcode not only blocks the axonal transport of endogenous COXIV, but also inhibits its localization to the mitochondria in SCG axons.
To test the hypothesis that the disrupted trafficking COXIV mRNA to the axon is independent of the reporter mRNA used in the study, we performed a qRT-PCR analysis on total RNA isolated from soma and distal axons of neurons transfected with either mCherry+3′UTR [38bp], or native mCherry expression constructs. Consistent with the results obtained from dGFP reporter constructs, exogenous over-expression of mCherry+3′UTR [38bp] led to a decrease in axonal levels of COXIV mRNA, as compared to mCherry alone. Similar to our previous results obtained with the dGFP reporter the relative abundance of COXIV mRNA in transfected cell-bodies also remained unchanged after expression of either mCherry+3′UTR [38bp], or mCherry construct (Figure 1I). These results establish that the effects on axonal trafficking of COXIV transcript is independent of the reporter used in the assay and depends solely on the presence of a functional full-length COXIV zipcode sequence.
Depletion of local COXIV mRNA affects axonal mitochondrial function and alters ROS levels in the axon
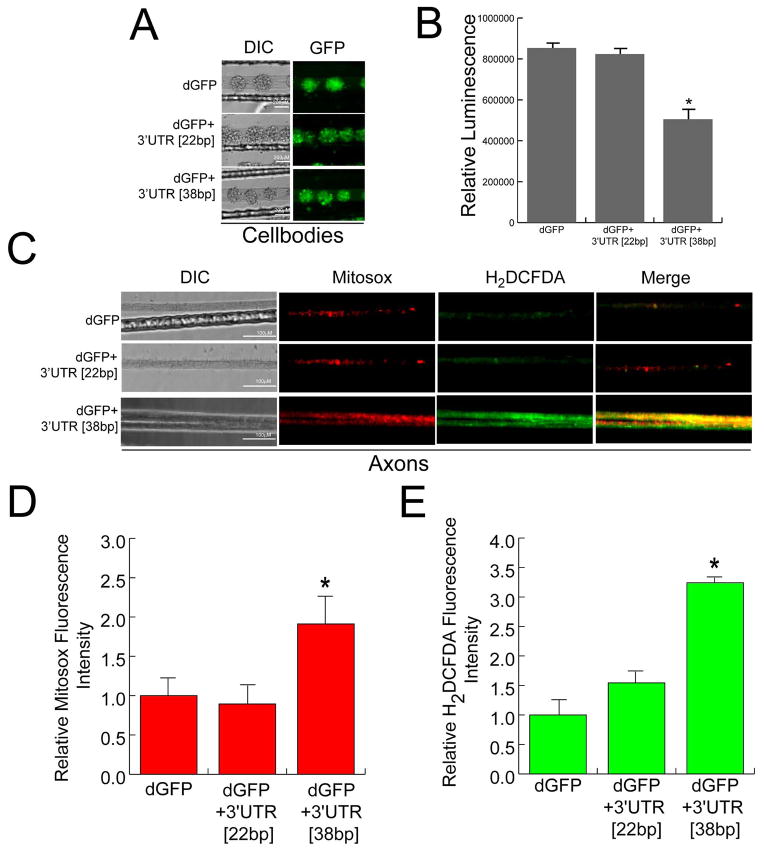
To examine the functional significance of disrupted axonal trafficking of COXIV mRNA, we evaluated the effects of over-expression of the COXIV zipcode on local ATP levels and axonal ROS. In previous studies, we had shown that axonal translation of COXIV in the axons is important for maintaining axonal mitochondrial function and growth. In this regard, the silencing of axonal COXIV expression using small interfering RNAs (siRNA) was shown to affect axonal ATP production and altered ROS production in the axon (Aschrafi et al., 2008; Aschrafi et al., 2012). In the present study, we measured ATP levels in the distal axons, 48 h after transfection of the parental soma with dGFP+3′UTR [38bp], dGFP+3′UTR [22bp], or dGFP constructs. Comparable levels of dGFP expression were observed in the soma for each construct after transfection as assessed by fluorescence microscopy (Figure 2A). The results from the luminometric ATP assay showed a 40% decrease in total axonal ATP levels in the distal axons of neurons transfected with the dGFP+3′UTR [38bp] construct as compared to dGFP+3′UTR [22bp], or dGFP expressing neurons (Figure 2B). No statistically significant difference in cell-body ATP levels were observed in these experiments (data not shown).
Figure 2.
Over-expression of dGFP mRNA containing the 38bp COXIV zipcode decreases local ATP levels in axons and alters axonal ROS production. SCG neuron cell-bodies (6DIV) were transfected with dGFP+3′UTR [38bp], dGFP+3′UTR[22bp], or dGFP constructs (3ug each). (A) Representative DIC and fluorescence images showing the expression of dGFP contructs in the transfected cell-bodies. (B) Total axonal ATP levels were measured in distal axons 48 h after transfection of SCG neurons using the CellTiter-Glo Luminescent Cell Viability Assay from Promega (see Methods). Values are plotted as relative luminescence units. (C) Intra-axonal reactive oxygen species (ROS) levels were measured in distal axons 24 h after transfection of cell-bodies by fluorescence microscopy using Carboxy-H2DCFDA (green) and Mitosox (red). Fluorescence intensity was quantified using ImageJ, and fluorescence levels for Mitosox (D) and Carboxy-H2DCFDA (E) are indicated as relative fluorescence units (RFU). Data are the mean ± SEM from the measurement of 35–45 axons. One-way ANOVA, *, p < 0.01.
Alteration in mitochondrial function that affect oxidative phoshorylation often results in the generation of reactive oxygen species (ROS) (for review, see Patten et al., 2010), raising the possibility that downregulation of local levels of COXIV could induce ROS formation in the axon. To examine if decreases in relative abundance of COXIV mRNA in the axon affects axonal ROS levels, we transfected central compartments of 7-day-old neuronal cultures with dGFP+3′UTR [38bp], dGFP+3′UTR [22bp], or dGFP expression plasmids. Forty eight hours after transfection, ROS levels were assessed with H2-DCFDA and MitoSOX, fluorescent indicators of total and mitochondrial-generated ROS (i.e., superoxide), respectively. As shown in Figure 2C, little fluorescence was observed in distal axons of soma transfected with either the dGFP control, or the dGFP+3′UTR [22bp] deletion construct with either dye. In contrast, there was a marked elevation in ROS levels in axons whose soma were transfected with dGFP+3′UTR [38bp] (Figure 2C). A partial overlap between H2-DCFDA and MitoSOX staining was observed in the merge shown in Figure 2C. It bears mention here, that we did observe a small increase in H2-DCFDA and MitoSOX levels in the cell-bodies of SCG neurons transfected with dGFP+3′UTR [38bp] construct as compared to those transfected with dGFP+3′UTR [22bp], or dGFP plasmids, however this increase in ROS levels was not statistically significant (data not shown). Quantitation of the fluorescent signal in distal axons from cultures transfected with the dGFP+3′UTR[38bp] or controls revealed that mitochondrial-generated ROS was elevated nearly 2.5-fold (MitoSOX, Figure 2D), while total ROS levels increased 3.0–3.5 -fold (H2-DCFDA, Figure 2E) as compared to cultures expressing the dGFP+3′UTR[22bp] deletion construct or the dGFP control plasmid.
Taken together, these results suggest that dysregulated trafficking of endogenous COXIV mRNA to the axon has a detrimental effect on mitochondrial activity in the axon which can subsequently lead to alterations in axonal ROS levels.
COXIV zipcode is expressed in neurons situated in the deep layers of the prefrontal and frontal cortices of transgenic animals
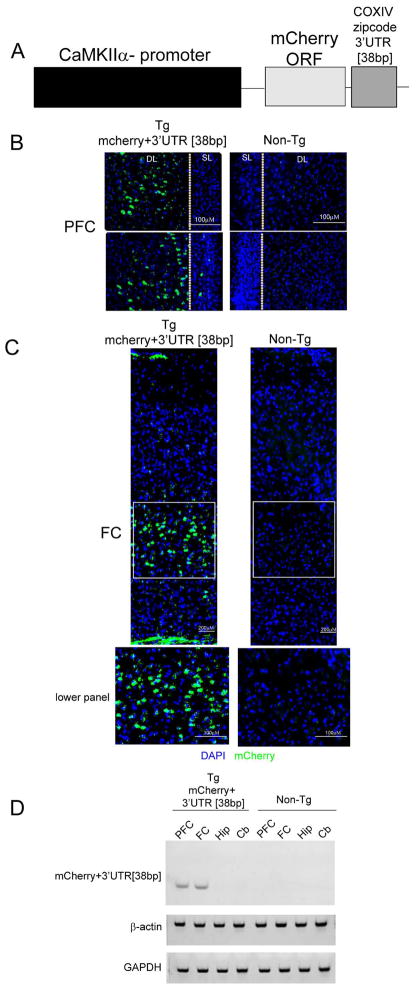
To determine the effect of altering the trafficking of nuclear-encoded mitochondrial mRNAs on brain ROS levels and animal behavior, we expressed the COXIV zipcode specifically in forebrain neurons of transgenic mice. To facilitate studies of transgene expression, we inserted the COXIV zipcode downstream of a mCherry ORF and used mCherry expression as a surrogate marker for the expression of the COXIV zipcode in the transgenic mouse brain (Figure 3A). The expression of this mCherry+3′UTR [38bp] (COXIV zipcode) cassette was driven by the CaMKIIα promoter to target expression of the transgene to the forebrain, more specifically in the excitatory neurons of the forebrain (Mayford et al., 1996).
Figure 3.
Transgenic mice express the COXIV zipcode in the frontal and pre-frontal cortex. (A) Schematic diagram showing the chimeric transgenic expression construct used to generate the transgenic animals. The construct contains the mCherry open-reading-frame followed by the 38bp COXIV zipcode. The expression of this cassette is driven by the α-calmodulin kinase II promoter. The mCherry+3′UTR [38bp] transgene expression in coronal brain sections show mCherry signal in the prefrontal cortex (PFC) (B) and the frontal cortex (FC) (C) as detected by fluorescence microscopy using a dsRED antibody that was visualized by Alexa488 (Green). SL-superficial layer; DL- deep layer in the section. The sections were counterstained with 4′,6-diamidino-2-phenylindole (blue).(D) Prefrontal cortex (PFC), frontal cortex (FC), hippocampus (Hip) and cerebellum (Cb) was dissected from transgenic animals and non-transgenic littermates, and total RNA was extracted (see Methods). Following reverse transcription, transgene specific primers were used to detect the presence of exogenous mCherry+3′UTR [38bp] transcripts. GAPDH and β-actin were used as internal positive controls.
Investigation into the expression pattern of the mCherry+3′UTR [38bp] (COXIV zipcode) transgene was conducted using coronal sections derived from transgenic mice by immunostaining with the dsRED antibody to detect the presence of mCherry signal. Based on the intensity of transgene expression, three transgenic lines were selected for further characterization. The results of the immunofluorescence analyses showed that the mCherry signal was localized to neurons situated in the deep layers of the prefrontal and frontal cortices. No significant mCherry staining was observed in the superficial layers in these sections (Figure 3B and C). Magnified images of the deep layer neurons from the frontal cortex (see lower panel Figure 3C) show strong labeling in the soma surrounding the nucleus. No immunohistofluorescent signal was observed in deep layer neurons of the frontal and prefrontal cortex in non-transgenic littermates (Figure 3B and C). No significant mCherry expression was observed in the hippocampus, striatum, amygdala or cerebellar regions of the transgenic mice (data not shown).
In addition, expression of the COXIV zipcode transgene was also assessed by RT-PCR from brain regions dissected from several brain sections namely, prefrontal cortex (PFC), frontal cortex (FC), hippocampus (Hip), and Cerebellum (Cb). Consistent with the findings obtained from the immunofluorescence analyses, the results of the RT-PCR assay revealed the presence of the mCherry+3′UTR[38bp] transcript in the prefrontal cortex and frontal cortices of transgenic mouse (Figure 3D). No mCherry+3′UTR [38bp] transcripts were observed in RNA samples isolated from either the hippocampus or cerebellum. The intensity of GAPDH and β-actin amplicons, used as loading controls, indicate that there was no significant differences in RNA amounts or quality between the various samples used in the PCR analyses (Figure 3D). No transgene-specific bands were observed in brain sections isolated from the non-trangenic littermates (Figure 3D). Taken together, these results establish that the COXIV zipcode is expressed in the deep layer neurons of the frontal and prefrontal cortex of transgenic mice. Similar transgene expression profiles were observed in all three transgenic lines that were analyzed.
Exogenous expression of COXIV zipcode in the brain of transgenic mice leads to increased cortical ROS levels
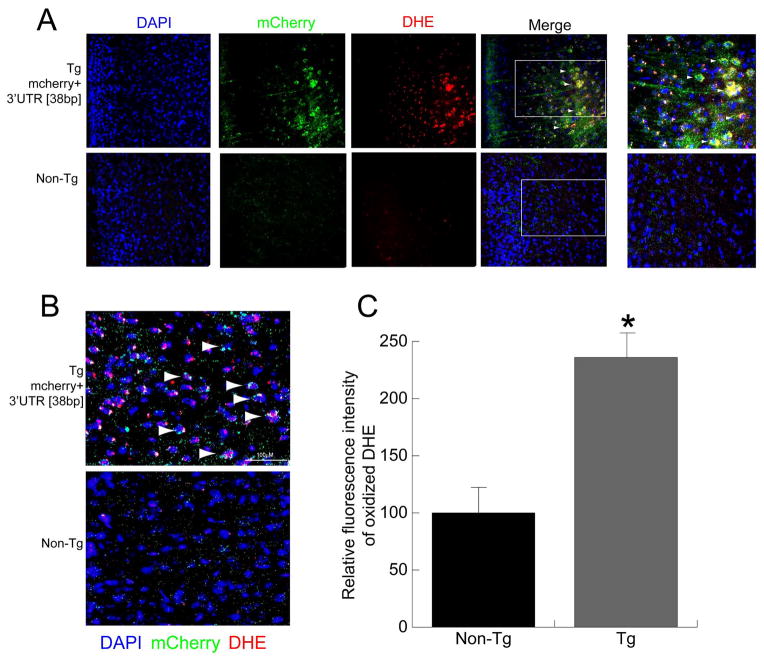
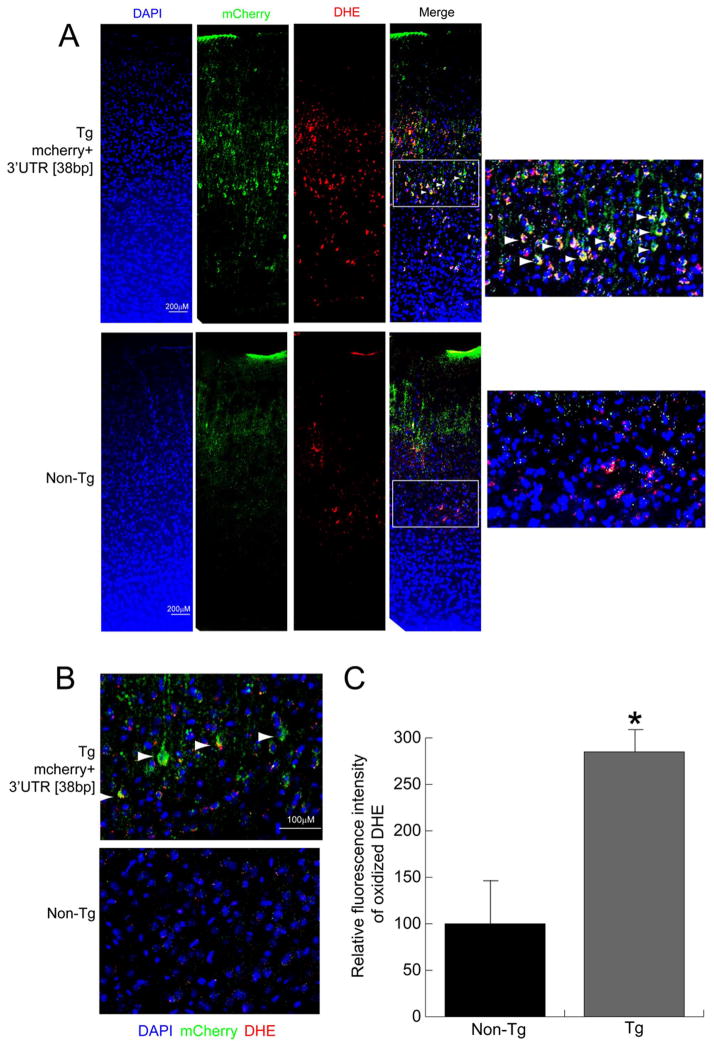
Our in vitro studies in SCG neurons over-expressing the COXIV zipcode suggested that the zipcode functions as a competitive inhibitor of axonal trafficking of endogenous COXIV transcripts. It was also shown that, decreased local COXIV mRNA levels in the axon leads to mitochondrial dysfunction and increased axonal ROS levels (Aschrafi et al., 2008, 2012 and see Figure 2). To investigate whether expression of COXIV zipcode in the deep layer neurons of the frontal and prefrontal cortices of transgenic mice leads to alterations in cortical ROS levels, brain sections from transgenic mice and non-transgenic littermates were examined after IP injection of DHE, a fluorescent indicator used to monitor cortical ROS production in vivo (Behrens et al., 2007; Jiang et al., 2013). Consistent with the results obtained in the in vitro studies, DHE staining revealed a significant increase in cortical ROS levels in the deep layer neurons of transgenic mice as compared to non-transgenic littermates (Figure 4 and 5). Magnified images of the deep layer neurons from both prefrontal and frontal cortices showed co-localization of DHE staining and dsRED immunolabeling (Figures 4 and 5, respectively). It is important to note that, similar to the mCherry expression pattern, the DHE flurorescence was also restricted to the deep layer neurons in these sections (Figures 4 and 5). Confocal microscopy of the brain sections co-stained with dsRED and DHE also showed neurons in the prefrontal and frontal cortex that manifested double-labeling with dsRED antibody and oxidized DHE staining (Figure 4B and 5B respectively). To quantify the increase in cortical ROS levels, the fluorescence intensity of the oxidized DHE signal was measured from deep layer neurons in the prefrontal and frontal cortices of COXIV transgenic and age-matched non-transgenic littermates. The relative fluorescence intensity from PFC and FC revealed an 2.5-fold increase in ROS levels in the prefrontal cortex, while approximately 3-fold increases in ROS levels were observed in deep layer neurons of the frontal cortex relative to sections from age-matched non-transgenic littermates (Figures 4C and 5C, respectively). These results suggest that similar to our in vitro results in primary SCG neurons, in vivo expression of COXIV zipcode in the deep layer neurons produced increased cortical ROS levels in the prefrontal and frontal cortices of transgenic mice. All three transgenic lines showed increased ROS levels in the deep-layer neurons expressing the transgene.
Figure 4.
Neurons expressing mCherry+3′UTR [38bp] transgene in the prefrontal cortex show elevated ROS levels. (A) Expression of the mCherry+3′UTR [38bp] in brain sections containing the prefrontal cortex from transgenic mice and non-transgenic littermates was detected by immunostaining with dsRED antibody and visualized by Alexa488 (Green) using fluorescence microscopy. The ROS levels in prefrontal cortex were visualized by oxidized DHE (red) in sections counterstained with 4′,6-diamidino-2-phenylindole (blue). (B) Confocal micrographs showing co-localization of mcherry (green) and DHE (red) fluorescence signals in the prefrontal cortex. Arrowheads show neurons that have mCherry expression (dsRED:Green), as well as elevated ROS levels (DHE: Red). The merged confocal images show a single optical section. (C) Relative ROS levels in prefrontal cortex neurons were quantified from fluorescence images and compared to the levels of ROS in non-transgenic littermates. Values represent the mean ± SEM. Students t test, *, p < 0.01.
Figure 5.
Exogenous expression of the mCherry+3′UTR [38bp] transgene in the frontal cortex leads to increased cortical ROS levels. (A) Expression of the mCherry+3′UTR [38bp] in brain sections containing the frontal cortex from transgenic mice and non-transgenic littermates was visualized by fluorescence microscopy using Alexa488 (Green) after immunostaining with dsRED antibody. The ROS levels in the frontal cortex were visualized by oxidized DHE (red) in sections counterstained with 4′,6-diamidino-2-phenylindole (blue). (B) Confocal micrographs showing neurons in the frontal cortex that were double-labeled with mCherry (green) and DHE (red). Arrowheads show neurons that show mCherry expression (dsRED:Green), as well as elevated ROS levels (DHE: Red). The merged confocal images show a single optical section. (C) Relative ROS levels in the frontal cortex neurons were quantified from fluorescence images and compared to the ROS levels of non-transgenic littermates. Values represent the mean ± SEM. Students t test, *, p < 0.01.
Behavioral consequences of COXIV zipcode expression in transgenic mice
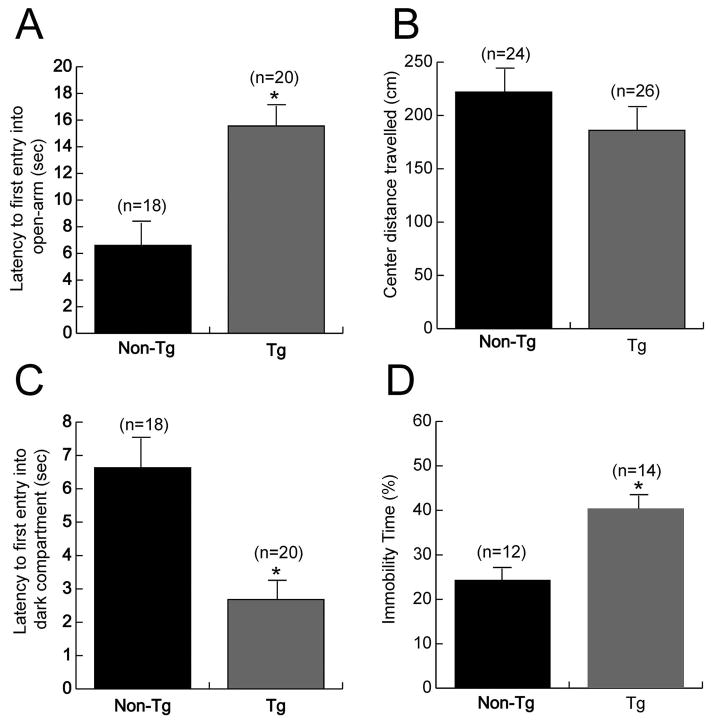
To assess the behavioral consequences of exogenous expression of the COXIV zipcode, we subjected heterozygous COXIV zipcode transgenic males (Tg) and age- and sex-matched non-transgenic littermates (Non-Tg) to a battery of behavioral tests. The transgenic animals showed no obvious impairment in basic functions, such as body weight, auditory, visual and olfactory function and motor coordination. Additionally, no significant difference in grooming behavior was observed. Also, transgenic mice and littermate controls showed no significant locomotor abnormality at 9–10 weeks of age, the time at which we started conducting the behavioral tests.
Naive transgenic mice subjected to an elevated plus-maze task showed a 2.5 fold increase in their latency to first entry into the unprotected open-arm as compared to age-matched non-transgenic littermates (Figure 6A). No significant differences were found between transgenic and control mice in the total number of entries into the open- or closed-arm, or the total time spent in the open- or closed-arm (data not shown). Spontaneous exploration in a novel open-field test showed decreased exploration of the unprotected center area of the open-field by transgenic mice as compared to age-matched non-transgenic controls; however the difference between the two groups did not reach statistical significance (Figure 6B). Additionally, no differences were observed between the transgenic and non-transgenic animals in the amount of horizontal and vertical exploration during the open-field test. A light-dark box exploration test of transgenic mice revealed a 2-fold decrease in the latency to first entry into the dark compartment after the mice were placed in the light compartment at the start of the test (Figure 6C). No significant differences were observed between transgenic and control mice in either the number of entries into the light- or dark-compartments or the time spent in each of these compartments. Taken together, the differences observed in the latencies to first entry in the elevated plus-maze and light-dark exploration, as well as the decreased exploration of the center area of the open-field suggest the presence of a higher level of “anxiety” in the COXIV zipcode expressing transgenic mice as compared to controls.
Figure 6.
Transgenic mice expressing COXIV zipcode manifest behavioral deficits. (A) Transgenic mice expressing the mCherry+3′UTR [38bp] show a significant increased latency to enter the anxiogenic open-arm in an elevated plus-maze test as compared to non-transgenic control littermates. (B) A consistent decrease in exploration of the anxiogenic center region of a novel open-field was observed in transgenic mice as compared to control littermates; however the differences did not reach statistical significance. (C) Light-dark box test showed that transgenic mice decreased latency to first entry into the dark-compartment as compared to control littermates, when the mice were placed in the light-compartment at the start of the test. (D) Transgenic mice expressing COXIV zipcode show statistically higher immobility in forced swim test as compared to non-transgenic littermates. Data are shown as mean values ± SEM. Number in parentheses: number of animals. One-way ANOVA,*, p<0.05
As depression-like behavior is often reported to be a comorbid feature of the anxiety phenotype, we assessed the presence of depression-like behavior in transgenic mice as adjudged by a forced swim test. The results of this behavioral analyses revealed that COXIV zipcode expressing transgenic mice manifest an approximately 2-fold increase in immobility time as compared to non-transgenic littermate controls (Figure 6D). The findings of the forced swim test suggest the presence of a “depression-like” behavior in the COXIV zipcode expressing transgenic mice.
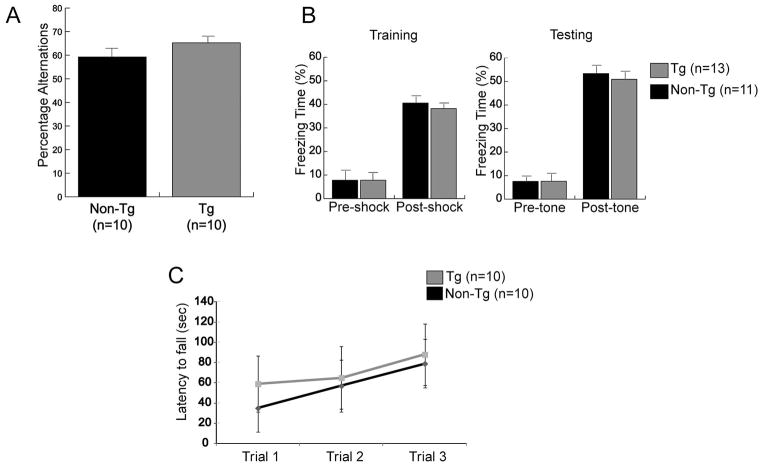
To investigate the effect of COXIV zipcode expression on short-term memory, we conducted a test of spontaneous Y-maze alternations in a spatial memory task based on the natural behavior of mice to alternate between the maze arms. Both the transgenic and non-transgenic littermate control mice showed reliable alternation in the Y-maze with greater than 50% alternations (Figure 7A). These results suggest that transgenic animal’s manifest no spatial memory deficit. A one-trial cued-fear conditioning test was also conducted to test deficits in associative learning. In the training/conditioning session both COXIV zipcode transgenics and littermate control mice showed similar freezing levels after shock presentation. No difference in freezing behavior was observed between the transgenic and control mice after tone presentation, during the testing session conducted 24 h after training (Figure 7B). These data suggest that there was no significant deficit in associative memory in the COXIV zipcode expressing transgenic mice.
Figure 7.
The behavior of transgenic and non-transgenic control mice were similar in measures of learning and memory. (A) In Y-maze spontaneous alternation task, the transgenic mice and the non-transgenic littermates showed similar alternation percentages. (B) Transgenic and control mice were subjected to cued-fear conditioning. Freezing was similar in both groups during conditioning. No significant difference was observed in freezing behavior between transgenic and control littermates after presentation of the tone during the testing phase 24 h after training. (C) Rotarod test was conducted to measure motor coordination and learning. Both transgenic and non-transgenic littermates exhibited similar motor coordination, as well as motor learning as evidenced by an increased latency to fall across trials.
To test for gross motor skills and motor coordination, we conducted rotarod training in which the animals were placed on an accelerating horizontal rod. Multiple trials were conducted to assess motor learning. In this assay, no significant difference was observed in the latency to fall between the transgenic mice and their non-transgenic littermate controls, after the first trial (Figure 7C). Additionally, repeated trials on the accelerating rotarod showed similar levels of learning in both the transgenic and control mice, as evidenced by the increase in the latency to fall-off the rotarod (Figure 7C). These results suggest that gross motor co-ordination and motor learning was intact in transgenic mice expressing the COXIV zipcode.
Combined, the results from our behavioral studies indicate that transgenic mice expressing the COXIV zipcode in the deep layer neurons of the prefrontal and frontal cortex show the presence of “anxiety-like” and “depression-like” behavioral phenotype.
DISCUSSION
Intracellular trafficking and localized translation of mRNAs within neurons provides a mechanism for rapid, and spatially regulated, activity-dependent protein synthesis in axons presynaptic nerve terminals (Goldie and Crains, 2012). Studies have shown that local expression and translation of nuclear-encoded mitochondrial mRNAs, such as COXIV and ATP5G1, are essential for regulation of local energy metabolism, growth and neuronal function (Hillefors et al., 2007; Aschrafi et al., 2008; 2012, Natera-Naranjo et al., 2012). In a recent study, we reported that axonal trafficking of COXIV mRNA was regulated by sequences present in a 38bp stem-loop structure (“zipcode”) in the 3′UTR of the transcript (Aschrafi et al., 2010). Interestingly, exogenous expression of the COXIV zipcode in SCG neurons led to decreased local levels of endogenous COXIV mRNA. Additionally, over-expression of the COXIV zipcode also resulted in attenuated axon growth (Aschrafi et al., 2010). Based on these observations, we hypothesized that these COXIV zipcode expression-mediated effects were, at least in part due to, competitive inhibition of the trafficking of the endogenous COXIV transcript. In this study, we further investigated the effects of over-expression of COXIV zipcode on brain function and behavior.
Previous studies had shown that the diminution of local expression of COXIV mRNA in the axon affected axonal mitochondrial activity and altered ROS levels (Aschrafi et al., 2008; 2012). Consistent with these reports, we observed that COXIV zipcode over-expression, also results in a reduction in axonal ATP levels, as well as increases in ROS production in the axon. The decrease in local ATP levels, and increased ROS generation in the axon, could explain the decreased axon growth observed in our earlier studies (Aschrafi et al., 2010). These observations suggest that both trafficking and local translation of nuclear-encoded mitochondrial mRNAs play an important role in maintaining and modulating mitochondrial activity and function in these neuronal domains (Gioio et al., 2001; Margeot et al., 2002; Aschrafi et al., 2010; Yoon et al., 2012). Furthermore, these findings also indicate a link between disrupted local transport and/or translation of nuclear-encoded mitochondrial mRNAs and mitochondrial dysfunction in the distal structural and functional domains of the neuron. Additional studies will be needed to define the role of trafficking of nuclear-encoded mitochondrial mRNAs in the mitochondrial dysfunction associated with various neurological diseases.
The ability to over-express the COXIV zipcode in the live animal provided us with a new experimental approach to test the functional significance of the trafficking of COXIV mRNA to the axon in vivo. In this regard, we generated transgenic mice expressing the mCherry ORF with the COXIV zipcode under the control of forebrain-specific and excitatory neuron-restricted CaMKIIα promoter in an attempt to disrupt the normal trafficking of the COXIV transcript to the axon. Immunofluorescence analyses of brain sections from transgenic animals revealed that the expression of the transgene was restricted to the deep layer neurons of the prefrontal and frontal cortices. Neurons expressing the transgene showed transgene-specific staining in the neuronal cell-bodies, as well as in the proximal neurites. Consistent with our in vitro studies, expression of the transgene in these neurons resulted in a significant increase in cortical ROS levels. These findings suggest that expression of COXIV zipcode in these neurons disrupted COXIV trafficking, resulting in decrements in mitochondrial activity and increased ROS production. Further studies are needed to accurately define the identity of the deep-layer neurons that express the transgene and delineate the affects that exogenous expression of COXIV zipcode has on the function of these neurons.
Several past studies have suggested the role for impaired mitochondrial function and increased ROS production in the pathophysiology of neurodegenerative and neuropsychiatric diseases (for review, see Federico and Radi et al., 2010; Manji and Chen et al., 2012 respectively). To assess whether our transgenic mice exhibited behavioral abnormalities, we exposed transgenic animals and non-transgenic littermates to a battery of behavioral tests. The results of these analyses demonstrated that transgenic mice expressing the COXIV zipcode showed a significant increase in latency to first enter an open-arm in an elevated plus-maze test. Consistent with this behavior, the results derived from a light-dark box test showed a decrease in latency to first entry into the dark-compartment by transgenic animals as compared to non-transgenic littermates. Both these test parameters have been interpreted to indicate anxiety-related behavior (van Gaalen and Steckler, 2000; Ionescu et al., 2012; Unemori et al., 2013). Earlier studies in rodent models of depression had reported the presence of a comorbid anxiety-like phenotype (Wang et al., 2007). Therefore, we tested the presence of a depression-like phenotype in our transgenic mice using a forced swim test. The results of this test showed a statistically significant increase in immobility in transgenic mice as compared to non-transgenic littermates, suggesting the presence of “depression-like” behavior in these transgenic animals. Previous studies in rodents have shown the involvement of the prefrontal cortex in mediating anxiety-like behavior (Stern et al., 2010; Adhikari et al., 2010, 2011; Bi et al., 2013). In addition, local drug infusion into the prefrontal cortex affects anxiety-like behavior in mice (Shah et al., 2004; Bruening et al., 2006; Stern et al., 2010; Bi et al., 2013). Interestingly, increased ROS levels in the prefrontal cortex are associated with stress-induced exacerbation of the cognitive deficits in mice showing schizophrenia-like behavioral and pathophysiological features (Belforte et al., 2010; Jiang et al., 2013). Consistent with these studies, our results suggest that perturbations in prefrontal cortical neuron activity, due to mitochondrial dysfunction as a result of COXIV zipcode expression, could lead to the observed behavioral outcomes. In light of recent reports that suggest antioxidant treatment is able to rescue behavioral impairments (Jiang et al., 2013), it would be intriguing to test if administration of antioxidant to our transgenic animals is able to normalize the behavioral anomalies observed in this study.
It is important to note that though transgene expression was detected in frontal cortices (to include the primary motor cortex M1), the open-field test showed no motor dysfunction in transgenic mice. This observation suggests that there may exist brain-region specific vulnerabilities in cortical neurons to mitochondrial dysfunction and ROS production, possibly due to differential antioxidant defense mechanisms.
In view of the fact that we observed no COXIV zipcode expression in the cerebellum, we hypothesized that transgenic mice would not manifest abnormalities in balance and motor behavior. To evaluate the viability of this hypothesis, we used the rotarod test. Consistent with this postulate, transgenic mice showed no deficiency in either motor coordination or motor learning. Similarly, consistent with the lack of expression of the transgene in the hippocampus, results from a Y maze test show no difference in the spontaneous alternations between transgenic and non-transgenic littermates. In addition, cued-fear conditioning showed no difference in cue learning between transgenic and non-transgenic littermates, which is again consistent with a lack of transgene expression in the amygdala. Taken together, these results suggest a correlation between COXIV zipcode expression and abnormalities in behavior of transgenic mice.
Previous studies that employed competitive inhibition derived from over-expression of an axonal zipcode (e.g. β-actin) suggest that the dominant negative phenotype on endogenous transcript trafficking is a result of limited availability of the cognate RNA binding protein (RBP), zipcode binding protein-1 (ZBP-1) (Donnelly et al., 2011, 2013). Additionally, over-expression of ZBP-1 partially rescued the disrupted trafficking of the endogenous β-actin transcript (Donnelly et al., 2011). Based on these studies, we can postulate that a similar molecular mechanism may be responsible for the results observed in our studies. However the cognate RBPs that interact with the COXIV-zipcode have yet to be identified. Thus, future studies will be aimed at identifying the RBPs involved in regulating COXIV trafficking, and testing if increasing the in vivo levels of these proteins will rescue the behavioral phenotypes observed in the COXIV zipcode transgenic mice.
Several studies have implicated the prefrontal cortex in higher-order cognitive and executive functions such as working memory, behavioral inhibition, and cognitive flexibility (for review, see Euston et al., 2012). Accumulating evidence also indicates that stress can profoundly alter structure and neuronal morphology in rodent PFC (Arnsten, 2009; Holmes and Wellman, 2009; Shansky and Morrison, 2009), as well as lead to exacerbation of transgenic animal behavioral phenotypes in mice (Jiang et al., 2013). Given the prefrontal expression pattern in our transgenic mice, it would be interesting to test these animals with behavioral paradigms that evaluate higher-order cognitive and executive functions. Additionally, the exposure of these transgenic mice to various stress-paradigms could reveal important environmental effects on the phenotypes manifest by COXIV zipcode expressing animals and raises the possibility that the intracellular trafficking and local translation of mRNAs in the neuron represent a new focus for the pathophysiology of neuropsychiatric diseases.
Acknowledgments
The authors work was supported by the Division of Intramural Research Programs of the National Institute of Mental Health (ZIAMH002768). We thank Noreen Gervasi for critical reading of the manuscript.
Footnotes
CONFLICT OF INTEREST
The authors declare no competing financial interests.
References
- Adhikari A, Topiwala MA, Gordon JA. Synchronized activity between the ventral hippocampus and the medial prefrontal cortex during anxiety. Neuron. 2010;65(2):257–69. doi: 10.1016/j.neuron.2009.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adhikari A, Topiwala MA, Gordon JA. Single units in the medial prefrontal cortex with anxiety-related firing patterns are preferentially influenced by ventral hippocampal activity. Neuron. 2011;71(5):898–910. doi: 10.1016/j.neuron.2011.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnsten AF. Stress signalling pathways that impair prefrontal cortex structure and function. Nat Rev Neurosci. 2009;10(6):410–22. doi: 10.1038/nrn2648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aschrafi A, Schwechter AD, Mameza MG, Natera-Naranjo O, Gioio AE, Kaplan BB. MicroRNA-338 regulates local cytochrome c oxidase IV mRNA levels and oxidative phosphorylation in the axons of sympathetic neurons. J Neurosci. 2008;28(47):12581–90. doi: 10.1523/JNEUROSCI.3338-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aschrafi A, Natera-Naranjo O, Gioio AE, Kaplan BB. Regulation of axonal trafficking of cytochrome c oxidase IV mRNA. Mol Cell Neurosci. 2010;43(4):422–30. doi: 10.1016/j.mcn.2010.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aschrafi A, Kar AN, Natera-Naranjo O, Macgibeny MA, Gioio AE, Kaplan BB. MicroRNA-338 regulates the axonal expression of multiple nuclear-encoded mitochondrial mRNAs encoding subunits of the oxidative phosphorylation machinery. Cell Mol Life Sci. 2012;69 (23):4017–4027. doi: 10.1007/s00018-012-1064-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behrens MM, Ali SS, Dao DN, Lucero J, Shekhtman G, Quick KL, Dugan LL. Ketamine-induced loss of phenotype of fast-spiking interneurons is mediated by NADPH-oxidase. Science. 2007;318(5856):1645–7. doi: 10.1126/science.1148045. [DOI] [PubMed] [Google Scholar]
- Behrens MM, Ali SS, Dugan LL. Interleukin-6 mediates the increase in NADPH-oxidase in the ketamine model of schizophrenia. J Neurosci. 2008;28(51):13957–66. doi: 10.1523/JNEUROSCI.4457-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belforte JE, Zsiros V, Sklar ER, Jiang Z, Yu G, Li Y, Quinlan EM, Nakazawa K. Postnatal NMDA receptor ablation in corticolimbic interneurons confers schizophrenia-like phenotypes. Nat Neurosci. 2010;13(1):76–83. doi: 10.1038/nn.2447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi LL, Wang J, Luo ZY, Chen SP, Geng F, Chen YH, Li SJ, Yuan CH, Lin S, Gao TM. Enhanced excitability in the infralimbic cortex produces anxiety-like behaviors. Neuropharmacology. 2013;72C:148–156. doi: 10.1016/j.neuropharm.2013.04.048. [DOI] [PubMed] [Google Scholar]
- Bruening S, Oh E, Hetzenauer A, Escobar-Alvarez S, Westphalen RI, Hemmings HC, Jr, Singewald N, Shippenberg T, Toth M. The anxiety-like phenotype of 5-HT receptor null mice is associated with genetic background-specific perturbations in the prefrontal cortex GABA-glutamate system. J Neurochem. 2006;99(3):892–9. doi: 10.1111/j.1471-4159.2006.04129.x. [DOI] [PubMed] [Google Scholar]
- Corral-Debrinski M, Belgareh N, Blugeon C, Claros MG, Doye V, Jacq C. Overexpression of yeast karyopherin Pse1p/Kap121p stimulates the mitochondrial import of hydrophobic proteins in vivo. Mol Microbiol. 1999;31(5):1499–511. doi: 10.1046/j.1365-2958.1999.01295.x. [DOI] [PubMed] [Google Scholar]
- Corral-Debrinski M, Blugeon C, Jacq C. In yeast, the 3′ untranslated region or the presequence of ATM1 is required for the exclusive localization of its mRNA to the vicinity of mitochondria. Mol Cell Biol. 2000;20(21):7881–92. doi: 10.1128/mcb.20.21.7881-7892.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donnelly CJ, Willis DE, Xu M, Tep C, Jiang C, Yoo S, Schanen NC, Kirn-Safran CB, van Minnen J, English A, Yoon SO, Bassell GJ, Twiss JL. Limited availability of ZBP1 restricts axonal mRNA localization and nerve regeneration capacity. EMBO J. 2011;30(22):4665–77. doi: 10.1038/emboj.2011.347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donnelly CJ, Park M, Spillane M, Yoo S, Pacheco A, Gomes C, Vuppalanchi D, McDonald M, Kim HH, Merianda TT, Gallo G, Twiss JL. Axonally synthesized β-actin and GAP-43 proteins support distinct modes of axonal growth. J Neurosci. 2013;33(8):3311–22. doi: 10.1523/JNEUROSCI.1722-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Euston DR, Gruber AJ, McNaughton BL. The role of medial prefrontal cortex in memory and decision making. Neuron. 2012;76(6):1057–70. doi: 10.1016/j.neuron.2012.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Federico A, Cardaioli E, Da Pozzo P, Formichi P, Gallus GN, Radi E. Mitochondria, oxidative stress and neurodegeneration. J Neurol Sci. 2012;322(1–2):254–62. doi: 10.1016/j.jns.2012.05.030. [DOI] [PubMed] [Google Scholar]
- van Gaalen MM, Steckler T. Behavioural analysis of four mouse strains in an anxiety test battery. Behav Brain Res. 2000;115(1):95–106. doi: 10.1016/s0166-4328(00)00240-0. [DOI] [PubMed] [Google Scholar]
- van Gaalen MM, Stenzel-Poore MP, Holsboer F, Steckler T. Effects of transgenic overproduction of CRH on anxiety-like behaviour. Eur J Neurosci. 2002;15(12):2007–15. doi: 10.1046/j.1460-9568.2002.02040.x. [DOI] [PubMed] [Google Scholar]
- Gioio AE, Eyman M, Zhang H, Lavina ZS, Giuditta A, Kaplan BB. Local synthesis of nuclear-encoded mitochondrial proteins in the presynaptic nerve terminal. J Neurosci Res. 2001;64(5):447–53. doi: 10.1002/jnr.1096. [DOI] [PubMed] [Google Scholar]
- Goldie BJ, Cairns MJ. Post-transcriptional trafficking and regulation of neuronal gene expression. Mol Neurobiol. 2012;45(1):99–108. doi: 10.1007/s12035-011-8222-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gumy LF, Yeo GS, Tung YC, Zivraj KH, Willis D, Coppola G, Lam BY, Twiss JL, Holt CE, Fawcett JW. Transcriptome analysis of embryonic and adult sensory axons reveals changes in mRNA repertoire localization. RNA. 2011;17(1):85–98. doi: 10.1261/rna.2386111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillefors M, Gioio AE, Mameza MG, Kaplan BB. Axon viability and mitochondrial function are dependent on local protein synthesis in sympathetic neurons. Cell Mol Neurobiol. 2007;27(6):701–16. doi: 10.1007/s10571-007-9148-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes A, Wellman CL. Stress-induced prefrontal reorganization and executive dysfunction in rodents. Neurosci Biobehav Rev. 2009;33(6):773–83. doi: 10.1016/j.neubiorev.2008.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ionescu IA, Dine J, Yen YC, Buell DR, Herrmann L, Holsboer F, Eder M, Landgraf R, Schmidt U. Intranasally administered neuropeptide S (NPS) exerts anxiolytic effects following internalization into NPS receptor-expressing neurons. Neuropsychopharmacology. 2012;37(6):1323–37. doi: 10.1038/npp.2011.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Z, Rompala GR, Zhang S, Cowell RM, Nakazawa K. Social isolation exacerbates schizophrenia-like phenotypes via oxidative stress in cortical interneurons. Biol Psychiatry. 2013;73(10):1024–34. doi: 10.1016/j.biopsych.2012.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung H, Yoon BC, Holt CE. Axonal mRNA localization and local protein synthesis in nervous system assembly, maintenance and repair. Nat Rev Neurosci. 2012;13(5):308–24. doi: 10.1038/nrn3210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan BB, Gioio AE, Hillefors M, Aschrafi A. Axonal protein synthesis and the regulation of local mitochondrial function. Results Probl Cell Differ. 2009;48:225–42. doi: 10.1007/400_2009_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kar AN, MacGibeny MA, Gervasi NM, Gioio AE, Kaplan BB. Intra-axonal synthesis of eukaryotic translation initiation factors regulates local protein synthesis and axon growth in rat sympathetic neurons. J Neurosci. 2013;33(17):7165–74. doi: 10.1523/JNEUROSCI.2040-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manji H, Kato T, Di Prospero NA, Ness S, Beal MF, Krams M, Chen G. Impaired mitochondrial function in psychiatric disorders. Nat Rev Neurosci. 2012;13(5):293–307. doi: 10.1038/nrn3229. [DOI] [PubMed] [Google Scholar]
- Margeot A, Blugeon C, Sylvestre J, Vialette S, Jacq C, Corral-Debrinski M. In Saccharomyces cerevisiae, ATP2 mRNA sorting to the vicinity of mitochondria is essential for respiratory function. EMBO J. 2002;21(24):6893–904. doi: 10.1093/emboj/cdf690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayford M, Bach ME, Huang YY, Wang L, Hawkins RD, Kandel ER. Control of memory formation through regulated expression of a CaMKII transgene. Science. 1996;274(5293):1678–83. doi: 10.1126/science.274.5293.1678. [DOI] [PubMed] [Google Scholar]
- Natera-Naranjo O, Kar AN, Aschrafi A, Gervasi NM, Macgibeny MA, Gioio AE, Kaplan BB. Local translation of ATP synthase subunit 9 mRNA alters ATP levels and the production of ROS in the axon. Mol Cell Neurosci. 2012;49(3):263–70. doi: 10.1016/j.mcn.2011.12.006. [DOI] [PubMed] [Google Scholar]
- Patten DA, Germain M, Kelly MA, Slack RS. Reactive oxygen species: stuck in the middle of neurodegeneration. J Alzheimers Dis. 2010;20(Suppl 2):S357–67. doi: 10.3233/JAD-2010-100498. [DOI] [PubMed] [Google Scholar]
- Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 2008;3(6):1101–8. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- Shah AA, Sjovold T, Treit D. Inactivation of the medial prefrontal cortex with the GABAA receptor agonist muscimol increases open-arm activity in the elevated plus-maze and attenuates shock-probe burying in rats. Brain Res. 2004;1028(1):112–5. doi: 10.1016/j.brainres.2004.08.061. [DOI] [PubMed] [Google Scholar]
- Shansky RM, Morrison JH. Stress-induced dendritic remodeling in the medial prefrontal cortex: effects of circuit, hormones and rest. Brain Res. 2009;1293:108–13. doi: 10.1016/j.brainres.2009.03.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern CA, Do Monte FH, Gazarini L, Carobrez AP, Bertoglio LJ. Activity in prelimbic cortex is required for adjusting the anxiety response level during the elevated plus-maze retest. Neuroscience. 2010;170(1):214–22. doi: 10.1016/j.neuroscience.2010.06.080. [DOI] [PubMed] [Google Scholar]
- Umemori J, Takao K, Koshimizu H, Hattori S, Furuse T, Wakana S, Miyakawa T. ENU-mutagenesis mice with a non-synonymous mutation in Grin1 exhibit abnormal anxiety-like behaviors, impaired fear memory, and decreased acoustic startle response. BMC Res Notes. 2013;6:203. doi: 10.1186/1756-0500-6-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vonesch C, Unser M. A fast thresholded landweber algorithm for wavelet-regularized multidimensional deconvolution. IEEE Trans Image Process. 2008;17(4):539–49. doi: 10.1109/TIP.2008.917103. [DOI] [PubMed] [Google Scholar]
- Wang J, Mack AL, Coop A, Matsumoto RR. Novel sigma (sigma) receptor agonists produce antidepressant-like effects in mice. Eur Neuropsychopharmacol. 2007;17(11):708–16. doi: 10.1016/j.euroneuro.2007.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon BC, Jung H, Dwivedy A, O’Hare CM, Zivraj KH, Holt CE. Local translation of extranuclear lamin B promotes axon maintenance. Cell. 2012;148(4):752–64. doi: 10.1016/j.cell.2011.11.064. [DOI] [PMC free article] [PubMed] [Google Scholar]