Abstract
The enhancing effects of the complement system for humoral immunity have primarily focused upon the recognition of complement-bound foreign antigens by a co-receptor complex of the antigen-specific B cell receptor (BCR) and complement receptor 2 (Cr2). In vivo experiments using Cr2 gene deficient mice (which lack the expression of both the Cr1 and Cr2 proteins) do demonstrate depressed humoral responses to immunization but cannot be used to define specific contributions of the singular Cr1 or Cr2 proteins on B cell functions. To study the effect of a Cr2 deficiency in a Cr1 sufficient environment we created a mouse line in which the alternative splice site required for the expression of the Cr2 isoform was removed. This mouse line, Cr2KO, still expressed Cr1 on B cells but was deficient for the full length Cr2 protein. Surprisingly a new alternative splice within the Cr2 gene created a truncated product that encoded a novel protein termed iCr2 that was expressed on the surface of the cells. The Cr2KO mouse thus provides a new model system for the analysis of Cr1 and Cr2 functions in the immune response of the mouse.
Keywords: Alternative splicing, B cells, Complement, Complement receptors, Follicular dendritic cells
Introduction
The enhancing effects of complement for humoral immunity have been apparent since the foundational studies of Pepys (1974), and the identification of the enhancement of B cell activation through complement receptor (CR) 2 binding of complement bound antigen (Carter et al., 1988). This well accepted model of CR2 function relies initially on activation of the complement system by one of the three pathways: classical, alternative, or mannose-binding lectin. All three pathways lead to cleavage and deposition of complement component 3 (C3) fragments on antigen. The iC3b and C3d(g) fragments maintain a covalent bond with the antigen and act to direct the antigen to Cr2/CR2 via the iC3b/C3d(g) binding site on the proteins (Molina et al., 1995; Pramoonjago et al., 1993). (Cr1/Cr2 versus CR1/CR2 will be used here to clearly differentiate the mouse from the human proteins, respectively). In vitro experiments extensively defined the enhancement of B cell responses by CR2 (Carter et al., 1988; Matsumoto et al., 1991, 1993) and the overlapping binding, co-association and co-activation characteristics for Cr2 (Krop et al., 1996; Molina et al., 1994).
Cr2 null mouse lines (Cr1/2KO) have further supported the hypotheses that Cr2 gene products expressed by B cells and follicular dendritic cells (FDC) are critical for the detection of antigen and the generation of optimal antibody responses (Haas et al., 2002; Molina et al., 1996). The mouse Cr2 gene, unlike the human, creates both the Cr1 and Cr2 proteins from a single gene. The same signal sequence – encoding exon is alternatively spliced to either the domain encoding the first short consensus repeat (SCR) domain of the gene (thus generating the 190,000 Da Cr1 protein) or the exon encoding the seventh and eighth SCR (see Fig. 1a) domains of the Cr2 gene, which represent the most N-terminal coding sequences of the mature Cr2 protein (145,000 Da). The sequence of the Cr2 protein is thus fully included within that of the Cr1 protein. Recently we have found that this alternative splicing pattern is unique to B cells in that murine FDCs express only Cr1 protein from Cr2 gene transcripts (Donius et al., 2013).
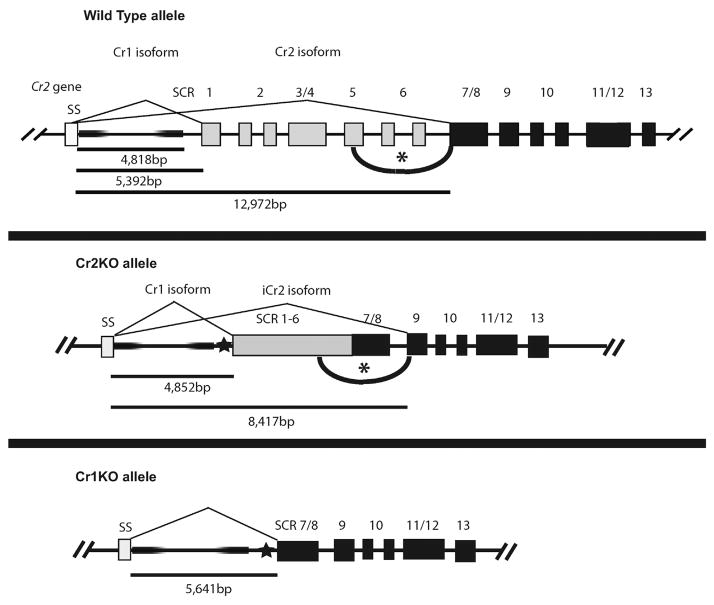
Fig. 1.
Schematic diagram and confirmation of Cr2 alternative splice site disruption. (a) The exons of the Cr2 gene are shown with numbers defining the number of the short consensus repeats (SCR) encoded within each. The initial exon denoted SS encodes the signal sequence. The Cr1 isoform is generated via the inclusion of all exons during splicing while the Cr2 isoform is generated by alternative splicing that excludes the exons encoding SCRs 1–6. The construct was generated from a cDNA version of the Cr2 gene with the addition of a pACN neomycin selection marker. Arrows represent the approximate location of restriction sites used for cloning and mapping of homologous sequence used for recombination. Triangles represent the approximate location of BamHI and MscI restriction sites utilized to Southern blot for determination of proper homologous recombination. (b) Representative Southern blots are shown for proper 5′ and 3′ homologous recombination. BamHI restriction digestion of wild type (WT) genomic DNA exhibits a 19 kb band detected by a radiolabeled probe. In the Cr2KO condition the fragment is reduced to 6 kb. Homologous recombination of the 3′ region was determined by probing for a size reduction from 15 kb to 13 kb. (c) Western blot analysis of Cr2 gene products from WT and Cr2KO mice, as well as mice heterozygous for the Cr2KO insertion.
The extensive research on the Cr1/2KO mice however has not fully discriminated the roles of the Cr1 and Cr2 proteins. The 6 N-terminal SCR domains of the Cr1 protein (which are not included in the Cr2 protein) can act as a co-factor in the regulation of C3 convertase stability and function (Molina et al., 1994), and recent studies have suggested that Cr1 may be important in regulating complement activation in the immune microenvironment (Jacobson et al., 2008; Seregin et al., 2009). To define the functions of the Cr1 protein we have recently created and described a mouse (Cr1KO) that exclusively expresses Cr2 protein on the surface of B cells and lacks expression of either protein on the surface of FDCs.
To define the specific functions of the Cr2 protein in the immune response of the mouse we created an additional mouse line that specifically lacks the Cr2 protein but still produces Cr1. This mouse, dubbed Cr2KO, was created by homologous recombination within the Cr2 gene with a construct that removed the alternative splice junction utilized in B cells to create the Cr2 protein. Analysis of mice generated from this recombination demonstrated expression of the full length Cr1 protein on B cells but the loss of Cr1 expression (and Cr2 gene expression) on FDCs suggesting the disruption of a FDC-specific transcriptional enhancer site within the Cr2 gene. Intriguingly, the removal of the native alternative splice site in the Cr2KO construct resulted in the utilization of an internal cryptic splice site, generating a truncated Cr2 gene product, termed iCr2, that lacks the iC3b/C3d(g) binding sites present on the normal Cr2 protein. The Cr2KO animals thus represent a novel animal model with which to functionally analyze the role of the Cr1 protein on the surface of B cells in the absence of functional Cr2.
Materials and methods
Generation of the Cr2KO mouse
The pKS+ Bluescript vector was used as a backbone for the creation of the 28 kb construct. Using primers (#4151 5′-GTGGTCCTTATTTCTAGGTCAGTGTAAGTTGCTGC-3′ and #4152 5′-GTTTTAATTTCCTACTTACCACTCTCACAGACTGGCAG-3′) a 1250 bp cDNA version of the exons for SCRs 1–8 of Cr1 was amplified using the long distance high-fidelity DNA polymerase Platinum Pfx (Invitrogen, Carlsbad, CA). The amplicon was additionally modified by PCR utilizing primers with overhang sequences for the restriction fragments XhoI and SbfI, 5′ and 3′, respectively. This new SCRs 1–8 fragment was cloned and ligated via XhoI and SbfI into the mouse Cr2 gene NotI-XhoI restriction fragment and 3′ to the SbfI-SalI restriction fragment (see Fig. 1a). The XhoI site of the resulting SCR1-8 construct with homology arms was utilized for insertion of the germline self deleting neomycin resistance construct pACN (Bunting et al., 1999). The entire construct was flanked by the TK1TK2 thymidine kinase containing negative selection construct. Electroporation of the Cr2KO vector into mouse strain 129-derived embryonic stem cells as well as blastocyst injection and implantation for the generation of chimeras was performed by the University of Utah Transgenic and Gene Targeting Mouse Core. Before blastocyst injection mouse embryonic stem cell clones were screened for homologous recombination by Southern blot. Restriction digestion of genomic DNA using BamHI produced a reduction in size from 19 kb for wildtype (WT) to 6 kb for proper homologous recombination of the 5′ homology arm of the vector (see Fig. 1b). The MscI digestion was used to probe for a reduction from 15 kb to 13 kb upon homologous recombination of the 3′ arm. Germline transmission of the Cr2KO allele was identified by tail or ear biopsies using PCR to generate a 193 bp product (primers: #4934 Fwd 5′-CTCCATGGTTCTGTGCATATAAACACGGGTATCG-3′ and #4937 Rev 5′-CCATTGGAGATGGCTGGAGGTGACTCACAAGG-3′). The WT condition was delineated using PCR to identify a 329 bp product (primers: #4935 Fwd 5′-GTAAATACAAAAGCACAGTCTCTTTGACTTGC-3′ and #4936 Rev 5′-GATACAATACATGTAGCAGACGAGTCACCAA-3′). All Cr2KO and WT mice as well as mice heterozygous between WT and Cr2KO were mixed background C57BL/6 and 129 mice. Cr1/2KO mice were raised on site and are progeny of the Cr2−/− described previously (Haas et al., 2002). Experiments were performed using 8–16 week old mice unless otherwise noted. All mice were housed at the Comparative Medicine Center (University of Utah Health Sciences Center) in accordance with the National Institutes of Health Guidelines on the Care and Use of Laboratory Animals.
Follicular dendritic cell preparation
To analyze follicular dendritic cell transcript and protein content the hematopoietic compartment of mice was ablated via irradiation of mice with 900 cGy two days prior to spleen isolation. The stroma of the spleen was isolated by straining the spleen through 100 μm nylon mesh. RNA was isolated as previously described (Zabel et al., 1999). Briefly, the splenic stroma from irradiated mice was homogenized in 4 M guanidine isocyathionate solution and centrifuged overnight at 30,000 rpm on a 5.7 M CsCl gradient. The pellet was resuspended in nuclease free water and isolated by ethanol precipitation. The precipitate was resuspended in nuclease free water and cDNA was generated. Protein isolation was performed as described for immunoprecipitation by lysing the stroma in 0.5% NP40 lysis buffer. Lysates were centrifuged for 10 min at 14,000 rpm and 4 °C to remove nuclei and other insoluble material. Samples were then boiled in SDS-PAGE sample buffer and assayed by western blot.
Western blot analysis
Proteins were separated on TGX SDS-PAGE gels (Bio-Rad, Hercules, CA) and transferred to PVDF membrane. Membranes were blocked in 5% milk 0.2% Tween-20 in 1× TBS. Detection antibodies used were as follows: goat anti-Cr1/2 (Santa Cruz #m-19, Dallas, TX), rabbit anti-actin (Sigma–Aldrich #A2066, St. Louis, MO), rabbit anti-MEK1/2 (ERK1/2)(Cell Signaling #9122S, Danvers, MA), and rabbit anti-CD19 (Santa Cruz Biotechnology #sc-8500-R, Dallas, TX). Actin, ERK1/2, and CD19 were all visualized with HRP-conjugated secondary donkey anti-rabbit IgG (Jackson Immunoresearch #711-035-152, West Grove, PA). The goat anti-Cr1/2 was visualized using an HRP-conjugated bovine anti-goat IgG (Jackson Immunoresearch #805-035-180, West Grove, PA).
Cr1/2 immunoprecipitation
After preparation of single cell suspensions of murine splenocytes and lysis of red blood cells with ACK lysis buffer, live cells were counted by trypan blue and 5.5 × 107 cells were collected. Cells were resuspended in 1 mL NP40 lysis buffer (0.5% NP40, 50 mM Tris–HCl (pH 7.5), 150 mM NaCl, 0.5 mM EDTA, and protease inhibitors (Complete Ultra protease inhibitor cocktail, Roche, Foster City, CA). Cells were incubated in lysis buffer for 30 min at 4 °C with rotation. Lysates were then precleared by the addition of 60 μL of a 10% suspension of protein G agarose beads (P7700, Sigma, St. Louis, MO) in PBS followed by 15 min incubation at 4 °C with rotation, centrifugation at 3000 rpm for 10 min at 4 °C, and transfer of the supernatant to a fresh 1.5 mL tube. Aliquots containing approximately 5.5 × 106 cell equivalents were taken, 1.5 μg of antibody for precipitation were added, and samples were incubated overnight at 4 °C with rotation. Precipitation of immunoconjugates was accomplished by the addition of 60 μL of 10% protein-G agarose bead suspension, incubation for 30 min at 4 °C with rotation, and centrifugation at 3000 rpm for 10 min. The beads were then washed three times with buffer containing 10 mM Tris–HCL buffer (pH 7.5), 0.1 mM EDTA and 0.05% NP40. Pellets were resuspended in 50 μL of 2× SDS-PAGE sample buffer and boiled for 5 min. Samples were then centrifuged at 14,000 rpm for 5 min at 4 °C, and supernatant was transferred to a fresh tube and stored at −80 °C until western blot analysis.
Immunohistochemistry of splenic cross-sections
Sectioning and immunohistochemistry were performed as described previously (Donius et al., 2013). Briefly, OCT (Sakura Finetek USA, Torrance, CA) embedded spleens were sectioned on a cryostat at 10–12 μm. Cr1 was detected using the biotinylated anti-Cr1 antibody clone 8C12 and Cr1/2 were detected using the biotinylated anti-Cr1/2 antibody clone 7E9. Streptavidin conjugated horseradish peroxidase (HRP) was used to detect the antibodies and was visualized using 3,3′-diaminobenzidine (DAB) (Vector Laboratories, Burlingame, CA).
Biotinylation of cell surface proteins
Total mouse splenocytes were collected and washed three times with PBS (pH 8.0). Cells were then resuspended at 2.5 × 107 cells/mL in 2.5 mM EZ-Link Sulfo-NHS-Biotin (Thermo Scientific, #21217, Waltham, MA) in PBS (pH 8.0) and incubated at 4 °C for 1 h with rotation. Unreacted biotinylation reagent was quenched and removed by three washes with 100 mM glycine in PBS (pH 8.0). Labeled cells were then lysed at 5.5 × 107 cells/mL 0.5% NP40 lysis buffer for 30 min at 4 °C with rotation. For every 1.5 mL of cell lysate, 1 mL of avidin–sepharose slurry (Protein Mods, #AVG, Madison, WI) was washed three times with 1% NP40 in PBS (pH 7.2). After removal of the last wash cell lysates were added to settled avidin sepharose gel and incubated at room temperature for 1 h with rotation. Avidin sepharose gel was transferred to a fresh micro-centrifuge tube and washed five times with 1% NP40 in PBS (pH 7.2). Settled avidin sepharose gel was resuspended in a volume of 4× SDS-PAGE sample buffer approximately equal to the volume of gel, boiled for 7 min, and placed on ice for 7 min. Samples were centrifuged at 14,000 rpm for 10 min at 4 °C, and supernatant was transferred to a fresh tube and stored at −80 °C until western blot analysis. Enrichment for cell-surface proteins was confirmed by blotting for ERK1/2.
Characterization of the iCr2 splice site using 32P-labeled PCR
Detection of small PCR amplicons was achieved by incorporation of 32P-labeled dCTP (Tan and Weis, 1992). Reaction mixes containing 0.01 mCi of 32P-labeled dCTP, 0.32 mM dNTP, MgCl2 buffer (Biofire, Salt Lake City, UT), 0.5 μg each primer, and 0.75 U of Taq polymerase were brought to 10 μl with deionized H2O and loaded into glass capillaries. Primer pairs were as follows: primer pair #1, Fwd #43 5′-ATGGGATCCTTGGGTTCGCTC-3′ with Rev #4975 5′-GTATGACTTCCATTACGAACAG-3′; primer pair #2, Fwd #43 with Rev #4974 5′-TATGAATCGTTGGAAGTGATGGGC-3′; and primer pair #3, #43 with Rev #45 Rev #45 5′-GCTAGGTGAACAAGTGTACCT-3′ (see Fig. 2a and b). Capillary tube ends were sealed by melting and rapid amplification was performed using an air thermocycler for 16–25 cycles. PCR reactions were heated to >95 °C in formamide containing loading buffer and immediately loaded onto polyacrylamide sequencing gel under denaturing conditions. The gel was transferred to chromatography paper, dried, and exposed to X-ray film overnight.
Fig. 2.
Cr2KO mice produce iCr2, an N-terminal truncated version of Cr2. (a) Schematic of PCR products from Cr2 gene transcripts (WT) and iCr2 transcripts (Cr2KO) utilizing a primer specific for the signal sequence (SS) with those from exon sequences encoding SCR 7/8 (primer set #1), SCR 10a (primer set #2) or SCR 11 (primer set #3). Cr1 encoding transcripts would not be identified with these primer sets due to the large size of the product. (b) The primer sets #1, #2, and #3 would predict PCR fragments of 866 bp, 487 bp, and 162 bp fragments, respectively, in WT samples while a PCR fragment of 476 bp from primer set #1 and 97 bp from primer set #2 would be predicted from Cr2KO cDNA samples. No PCR product from primer set #3 would be expected with the Cr2KO cDNA samples. (c) Radiographic detection of PCR amplified WT and Cr2KO cDNA samples using 32P-labeled dCTP resulted in bands corresponding to the sizes predicted in (a) and (b). Molecular weight markers are noted on the left side of the image.
Quantitative PCR
Quantitative PCR was done as described previously (Donius et al., 2013). Briefly, cDNA generated from total spleen or spleen stroma from irradiated mice was amplified in a SYBar green (Invitrogen, Carlsbad, CA) containing reaction mix and the measured Δ Ct was utilized to generate a transcript number relative to β-actin ΔCt. Primers used for Cr1, Cr1/2, and β-actin were as previously described (Debnath et al., 2007; Donius et al., 2013; Roundy et al., 2009).
Flow cytometry
Flow cytometric analysis was performed as previously described (Donius et al., 2013). Briefly, splenocytes were isolated by straining spleens through 100 μm nylon mesh into ice cold FACS buffer (0.5% BSA, 2 mM EDTA in PBS). Bone marrow from mice was isolated by flushing tibias and femurs with ice cold 0.5% BSA in PBS and straining through a 100 μm strainer and processed as below. Red blood cells were lysed with ACK buffer and the remaining cells were incubated in antibody mix. Antibodies used were as follows: eBioscience (San Diego, CA) monoclonals anti-B220-PerCP/Cy5.5 (RA3-6B2), anti-CD19-PE/Cy7 and PE (clone 1D3), anti-Cr1-biotin (clone 8C12), anti-Cr1/2-PE/Cy7 (clone 8D9), anti-Cr1/2-FITC (clone 7G6), anti-IgM-PE (clone eB121-15F9), and anti-CD40-FITC (clone HM40-3). Additionally the following BioLegend (San Diego, CA) antibodies were used anti-IgD-AlexaFluor647 (clone 11-26c.2a), anti-CD23-biotin (cloneB3B4), CD43-PerCP/Cy5.5 clone 1B11 and purified anti-CD16/32 as Fc block. Secondary incubations were performed with streptavidin-APC whenever biotinylated antibodies were used. Final samples were resuspended in 500 μl of FACS buffer and stained with 1.5 μl 1 mM DAPI for live/dead discrimination. All samples were analyzed on a FACSCantoII (BD Biosciences, San Jose, CA) and data was analyzed using FlowJo version 8.8.7 (Tree Star Inc., Ashland, OR).
Statistical analysis
Statistical analysis and graphs were generated using Prism version 5.0c (GraphPad Software Inc., San Diego, CA).
Results
Creation of a Cr2 but not Cr1 deficient mouse
A Cr2 deficient, Cr1 sufficient, mouse in which Cr1 is expressed in a cell and stage specific manner has long been sought as a model for investigating the role of complement augmentation of B cell responses. The challenge in creating such a mouse was to leave the transcriptional control apparatus intact but to deprive the Cr2 gene transcripts of the option to create the Cr2 protein-specific alternative splice. To block the generation of transcripts encoding the Cr2 protein but not those encoding Cr1, we devised a strategy by which the exons encoding SCR domains 1 through 8 were fused (Fig. 1a). Normally B cells process Cr2 gene transcripts to splice the domain encoding the signal sequence (SS) to the domain encoding SCR1 (and all of the remaining exons) to create the Cr1 protein. The B cell alternative splice of the Cr2 gene transcript splices the SS-encoding exon to that encoding the SCR7/8 (and all of the remaining exons) to create the Cr2 protein. We proposed that removing this alternative splice site (just upstream of the exon encoding SCR7/8) would block the production of the Cr2 protein and instead force all of the Cr2 gene transcripts to encode the Cr1 protein. A construct containing a cDNA fusion of the SCRs 1–8 (thus removing the alternative splice junction) was ligated to the testes specific self-deleting pACN neomycin resistance gene and flanked by Cr2 homologous DNA (Fig. 1a). To reduce the odds of non-homologous recombination a gancyclovir sensitivity gene (thymidine kinase) was also attached. Homologous recombination was utilized to target the construct to the Cr2 gene of mouse strain 129 embryonic stem (ES) cells. The resulting neomycin resistant and gancyclovir insensitive clones were screened by Southern blot for the predicted insertion of novel BamHI and MscI restriction sites resulting in 19–6 kb and 15–13 kb size reductions in Cr2 restriction digest fragments, respectively (Fig. 1a and b). A single ES line was used to generate the Cr2KO mouse line.
To test the validity of our strategy we obtained protein lysates from splenocytes of our newly generated Cr2KO mice, as well as WT and heterozygous littermates. Western blot analysis of these lysates revealed that Cr2 was no longer present in the Cr2KO samples and that Cr1 expression was maintained. Surprisingly Cr2KO and heterozygous mice when compared to WT littermates expressed reduced quantities of Cr1 and produced a new protein that is detected by antibody against Cr1/2 (Fig. 1c) This new protein, termed incomplete Cr2 (iCr2), was found to be about 15,000 Da smaller than WT Cr2.
Defining the new iCr2 protein
The splenic lysates from the Cr2KO animal demonstrated two proteins, one identical in size to Cr1 and a second (iCr2) that was smaller than the WT Cr2 protein suggesting that iCr2 could be a new alternatively spliced product from the Cr2 gene transcripts. The decrease in size of the iCr2 product, compared to WT Cr2 protein, was about that predicted from the loss of two SCR domains. To identify the precise exon order in the Cr2 gene transcripts that could encode this iCr2 protein, we designed primers embedded within the exons corresponding SCR7/8, SCR10a, and SCR11 to pair with a primer located in the signal sequence (SS) encoding exon (Fig. 2a). These primer pairs are identified here as #1, #2, and #3, respectively. Primer pair #1, #2, and #3 produce 866 bp, 487 bp, and 162 bp PCR products from WT Cr2 cDNA (Fig. 2b). We predicted that if the new iCr2 product was excluding the 400 bp exon encoding SCR7/8 and splicing directly from the SS exon to the SCR9 exon then pair #1 would generate a 476 bp product, #2 would generate a 97 bp product, and pair #3 would not generate a product (Fig. 2b). In order to ensure resolution of small PCR products we performed PCR with 32P-labeled nucleotides and resolved the reactions on polyacrylamide sequencing gels. PCR of cDNA generated from Cr2KO splenocyte derived RNA using these primers revealed products 400 bp smaller than those found in WT mice (Fig. 2c). Predictably primer pair #3 did not produce a product in Cr2KO cDNA. Sanger sequencing of a product generated from primer pair #2 further demonstrated that splicing exactly joins the SS exon to the exon encoding SCR9 (data not shown). These data indicate that the iCr2 protein consists of sequences including SCR9 and greater. Therefore, this protein would not possess the C3d(g) binding site encoded within the SCR7/8 domains (Molina et al., 1994) but would include those C-terminal sequences required for membrane localization and association with CD19.
Molina et al. previously demonstrated that C3d binds to murine Cr2 within SCR 7/8 and also that the anti-Cr1/2 antibody 7G6 competitively binds to this site (Molina et al., 1994). Another anti-Cr2 antibody, 7E9, binds both Cr1 and Cr2 but does not compete for C3d binding, which suggests that it recognizes an epitope found in SCR9 or greater. Both antibodies also bind to the Cr1 protein while a third antibody, 8C12, is specific for the N-terminal sequences found only in Cr1. These three monoclonal antibodies are all of rat derivation. To evaluate the potential of these antibodies to recognize the iCr2 protein, we performed immunoprecipitations with them followed by western blot using a pan-Cr1/2 reactive goat polyclonal antibody. As shown (Fig. 3a), the 7E9 antibody can precipitate both the iCr2 and Cr1 proteins from the Cr2KO splenic lysates (and the Cr1 and Cr2 proteins from the WT lysate). Additionally, the 8C12 antibody only recognizes the Cr1 protein in both the WT and Cr2KO lysates, confirming that the Cr2KO line does continue to produce the Cr1 protein. Alternatively, the 7G6 antibody (Fig. 3b) does not precipitate the iCr2 protein but does precipitate Cr1 and WT Cr2, which supports the transcript/sequence data described above. These data indicate that the iCr2 protein does not possess the sequences encoding SCR7/8 which encode the iC3b/C3d(g) binding site.
Fig. 3.
Elucidation of structural features of the iCr2 protein by immunoprecipitation and biotin labeling of cell-surface proteins. (a) Western blot analysis of proteins precipitated from strained spleen lysates with antibodies against regions common to Cr1 and Cr2 (7E9) and the cofactor domain unique to Cr1 (8C12). IC, isotype control antibody. (b) Western blot analysis of proteins precipitated from strained spleen lysates with antibody (7G6) against the C3d(g) binding region of Cr1 and Cr2. This region remains in Cr1 produced by the Cr2KO mouse but is absent in the iCr2 protein. IC, isotype control antibody. (c) Western blot analysis of cell-surface proteins identified by biotin-labeling and precipitated with avidin–sepharose. Enrichment for cell-surface proteins was confirmed by immunoblotting with antibody against the cytoplasmic protein ERK1/2.
If found on the membrane, the iCr2 protein would be predicted to contain all of the Cr2 protein sequences C-terminal of SCR9 including those sequences that facilitate co-association of the Cr1/Cr2 proteins with CD19. To determine if the iCr2 protein is indeed found at appropriate levels on the cell surface, we pre-formed surface biotinylation analysis of WT and Cr2KO splenocytes. Enrichment of the Cr1 and iCr2 proteins from Cr2KO splenocytes was comparable to the identification of Cr1 and Cr2 from WT cells (Fig. 3c). The intracellular protein ERK1/2 was not detected in the biotinylated fraction when compared to an equivalent exposure of total cell lysate. Additionally no Cr1, Cr2, or iCr2 was detected in western blot analysis of the non-biotinylated fractions (data not shown) suggesting that the vast majority of the Cr1, Cr2 and iCr2 proteins are localized on the surface of the expressing cells.
Characterization of B cell protein expression in the Cr2KO animal
Western blot and surface biotinylation analysis described above indicates that splenocytes (i.e., B cells) from the Cr2KO animal express both Cr1 and iCr2 on the cell surface. To compare the relative levels of expression of these proteins to Cr1 and Cr2 expression on WT splenic B cells, we isolated splenocytes from WT and Cr2KO animals and conducted flow cytometric analysis using antibodies specific for Cr1 (8C12) and for Cr1 and Cr2 (7G6, 8D9). Flow cytometric analysis of B220+ (B cells) cells demonstrated that the geometric mean fluorescent intensity (gMFI) of Cr1 expression in the Cr2KO animals (identified with the 8C12 and 7G6 antibodies) was lower than Cr1 expressed by WT B cells (defined by 8C12 staining) (Fig. 4a).
Fig. 4.
Surface expression of Cr1, Cr2, CD19, CD40 and CD23 (FcεR2a) on B cells is modulated in Cr2KO mice. (a) Representative histograms of Cr1 (antibody clone: 8C12), Cr1/2 (7G6), Cr1/2 (8D9), CD19, CD23, and CD40 expression on B220+ cells from the spleens of WT (gray line), Cr2KO (black line), and Cr1/2KO mice (gray fill). (b) The average geometric mean fluorescent intensity (gMFI) is graphed for B220+ cells from WT, Cr2KO, and Cr1/2KO mice represented in (a). Significance determined by the t-test shown between bars by black lines and denoted by asterisks (*p < 0.05, **p < 0.01, ***p < 0.001, and ns = not significant). Data representative of 2–3 independent analyses and n = 5–9 for all graphs.
The specific binding site of antibody 8D9 on the Cr1/Cr2 proteins is not known. However, staining of the Cr2KO B cells with the 8D9 antibody could be predicted to be additive of both Cr1 and iCr2, increasing the gMFI to near WT levels. The 8D9 antibody demonstrated approximately equal staining to that of the 7G6 and 8C12 antibodies (Cr1-specific for the Cr2KO cells) suggesting the epitope recognized by 8D9 is also altered by the loss of SCR7/8 in the iCr2 protein. Additional analysis of important receptors on B cells in the Cr2KO mice demonstrated identical gMFI staining for CD40 and CD23 (FcεR2α) compared to WT. The levels of CD19 expression, however are slightly elevated compared to WT but intermediate to the Cr1/2KO animal as previously described (Haas et al., 2009). The quantified gMFI of multiple similar flow cytometric analyses are shown in Fig. 4b.
Characterization of marrow and splenic B cell populations in the Cr2KO animal
To test the effect that Cr2 specific deletion has on B cell development, we assessed the subsets of marrow, splenic and peritoneal B cells. Pro-B, Pre-B, immature B and mature B cells from the mature Cr2KO animal were compared with those of WT (Fig. 5a). There were no demonstrable differences in B cell development between WT and Cr2KO mice. To determine if the expression of Cr1 by maturing B cells was altered in the Cr2KO mice, the cell surface expression of Cr1 was evaluated for the marrow B cell populations (Fig. 5b). Similar to the WT, the Cr2KO mice primarily demonstrated Cr1 expression on the mature B cells within the marrow compartment with a lesser level of expression on the immature B cells, again similar to that seen in WT mice. Therefore the stage specific expression of Cr1 was not altered from that of WT in the Cr2KO animal.
Fig. 5.
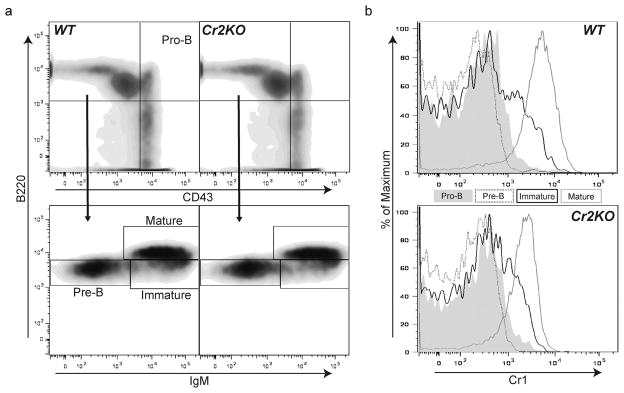
Progression of marrow B cell development in the Cr2KO animal. (a) Cell surface staining strategy for identifying Pro-B, Pre-B, immature B and mature B cells in adult WT and Cr2KO animals. Pro-B cells are identified as CD43hi and B220+ (a, top panel) and the CD43lo/− subset was further delineated by expression of IgM and B220. Pre-B cells IgMlo B220lo, immature B cells IgMhi B220lo, and mature B cells IgMhi B220hi are shown (a, bottom panel). No marked differences were identified between the bone marrow B cell subsets of Cr2KO mice compared to WT bone marrow B cell subsets. (b) Cr1 expression in maturing and mature marrow B cells from WT and Cr2KO animals was determined using the sorting gates shown in (a). Both Cr2KO and WT mice exhibit the same timing of expression of Cr1 at the immature B cell stage with all mature B cells expressing Cr1. Cr1 expression in the Cr2KO is reduced compared to WT. Data representative of n = 3 littermates.
Peritoneal B1 populations have been shown to be altered in the Cr1/2KO animals but not in the Cr1KO mouse (Ahearn et al., 1996; Donius et al., 2013). We examined B1a and B1b populations in the Cr2KO mice and found that they were not significantly different than WT (data not shown). Similarly, Cr1 expression was evident on the B1 populations of the Cr2KO mice but at levels reduced comparably to those of the other B cell populations in the Cr2KO animal (data not shown). Therefore the deletion of Cr2 in the Cr2KO animal did not alter the numbers or type of peritoneal B1 cells.
B cells in the spleen can be segregated by their varied expression of IgM and IgD. By gating on total splenic B220+ cells the marginal zone and transitional 1 B cells are identified as IgDLO IgMHI cells, while follicular mature B cells are identified by the marker profile IgDHI IgMLO (Loder et al., 1999). A transitional 2 immature subset can also be identified by this strategy as IgDHI and IgMHI. Flow cytometric analysis of splenic B cells, identified by expression of the B cell marker B220, did not identify any significant effects on frequency or total number of cells from the transitional 1, transitional 2, marginal zone, or follicular B cell populations (Fig. 6a and b). Therefore, similar to the Cr1/2KO mouse, the Cr2KO mouse has normal B2 populations of splenic B cells.
Fig. 6.
Splenic B cell populations are fully represented in Cr2KO mice. (a) Representative plots of B220+ B cells from total live splenocytes isolated from WT, Cr2KO, and Cr1/2KO mice. Gates represent follicular B cells (IgDHI IgMLO), transitional 2 B cells (IgDHI IgMHI), and the marginal zone, B1, and transitional 1 B cell populations (IgDLO IgMHI). (b) The total number of cells from each population were calculated using gate percentages for B220+ and IgM IgD parameters and the initial number of splenocytes counted using trypan blue. No significant differences were seen between any genotypes for any populations by ANOVA.
Cr2 gene product expression on follicular dendritic cells of Cr2KO mice
As described above, the expression of the Cr1 protein is reduced on the surface of the Cr2KO B cells compared to WT B cells. Previously we have shown that FDCs do not produce the Cr2 protein but instead primarily express Cr1 (Donius et al., 2013). To determine if FDC expression of Cr1 was also depressed in the Cr2KO animals, we first analyzed Cr1 specific transcripts in FDCs relative to that of total spleen. Irradiation treatment of mice is very effective at removing virtually all B cells from the spleen, leaving behind the radiation-resistant stromal cells and FDCs. As shown (Fig. 7a), the level of Cr1-specific transcripts is depressed in both the total spleen samples (containing both B cells and FDCs) as well as the irradiated samples (highly enriched for FDCs) indicating that FDC expression of Cr1 in the Cr2KO mouse is also reduced. Western blot analysis of spleen lysates from these B cell depleted mice further demonstrated that Cr1 is not expressed by FDCs, at least not at a detectable level (Fig. 7b). Splenic cross-sections from WT and Cr2KO mice analyzed by immunohistochemistry using the anti-Cr1 antibody clone 8C12 labels Cr2KO spleens but in a considerably reduced pattern compared to WT (Fig. 7c).
Fig. 7.
Follicular dendritic cell Cr2 gene expression. (a) Quantitative PCR measurement of Cr1 transcripts per 1000 β-actin transcripts in cDNA samples generated from either total splenic RNA or stromal isolations from irradiated mice (B cell depleted). Black lines denote statistical analysis by the t-test. (b) Western blot analysis of Cr1, Cr2, and iCr2 in lysates isolated from spleens using the same methods as in (a). (c) Detection of Cr1 utilizing antibody clone 8C12 in splenic cross-sections from naive WT and Cr2KO mice. All images obtained at a magnification of 4×. (**p < 0.01, ***p < 0.001, and ns = not significant; total spleen graphs in (a) n = 6, n = 3 for Cr1/2KO; irradiated stromal cells n = 4).
Discussion
The generation of a mouse specifically and singularly deficient in the Cr2 protein has been a sought after tool for the study of the effects of the complement system on humoral immunity. A similar goal was advanced recently by the creation of a transgenic Cr1/2KO mouse line expressing human CR1/CD35 (Pappworth et al., 2011). However, in addition to the expression of human CR1 in a non-autochthonous environment these mice possess other disadvantages including premature expression of CR1 during B cell development, a broader cell specific expression pattern than murine Cr1, and the absence of the CD19 co-association exhibited by mouse Cr1. Therefore to create a Cr2KO animal without these limitations, we adopted the strategy of modifying the endogenous Cr2 gene via homologous recombination with a construct that would block Cr2 protein production but leave intact those coding sequences required for the Cr1 protein.
This strategy of specifically manipulating the endogenous Cr2 gene was designed to leave Cr2 under the transcriptional control of its endogenous promoter in an approach nearly identical to that taken in the creation of the Cr1KO mouse (Donius et al., 2013). Creation of the Cr2KO mouse and the biological assessments of the animal described here however, uncovered the importance of the intronic sequence and/or the alternative splice site in promoting overall expression of the Cr2 gene.
Both the Cr1KO and Cr2KO mice were designed to maintain the normal positioning of the Cr2 promoter, first exon encoding the signal sequence, and much of the first intron of the gene, especially the regions that are rich in transcription factor binding sites (Fig. 8). While the Cr1KO mouse lacks the exons encoding the Cr1-specific exons encoding SCRs 1–6 (and the intervening introns), the Cr2KO animal is primarily lacking only the intronic sequences between the exons encoding the first 8 SCR domains. There was no indication of a “new” alternative splice in the Cr1KO animal suggesting that sequences that prompt the alternative splice are somewhere within the exons encoding the first 6 SCRs of the protein. The concept that coding exons could influence such a splicing event is not new. Recent studies on alternative splicing of CD45 in T and B cells have identified specific residues within coding exons of that gene that are recognized by the HnRNP L and L-like proteins that help facilitate the complex alternative splicing of CD45 transcripts (Oberdoerffer et al., 2008; Preussner et al., 2012; Rothrock et al., 2005; Tong et al., 2005). These target sequences that facilitate constitutive and inducible alternative exon splicing are comprised of CA rich sequences that are specifically bound by members of the HnRNP L family. Exons encoding the SCRs 4–6 of the Cr2 gene would be the likely site for such sequences and CA-rich HnRNP L-binding regions can be identified in silico in these coding exons (data not shown). Future experiments using Cr2 minigenes (similar to the minigenes constructed for the alternatively spliced exons of the CD45 gene) will be required to definitively identify sites and mechanisms controlling this alternative splicing pathway.
Fig. 8.
Schematic representation of the alternative splicing of the Cr2 gene seen in WT and Cr2KO animal lines. The first intron sequence of 4818 bp in the WT Cr2 gene is present in both the Cr2KO and Cr1KO lines (denoted by shaded black box). The genomic span between the signal sequence (SS) exon and that encoding SCR1 is 5392 bp and SS to that encoding SCR7/8 is 12,972 bp in the WT gene. The star in the Cr2KO and Cr1KO alleles denotes the insertion site of the Neo gene sequence which, during germline propagation, resolves to a single Lox site. The Cr1KO allele does not demonstrate any alternative splice suggesting the alternative splicing target sequence(s) is within coding sequences for SCRs 1–6, not the promoter or first intron of the Cr2 gene. The position of the hypothetical location within the exons for SCRs 1–6 is indicated by the black arc and asterisk.
Although our goal was to create a mouse that only expressed the Cr1 protein from native Cr2 gene transcripts, the creation of such a mouse would have provided for much higher Cr1 surface expression than WT. Instead we found the iCr2 protein was the major product from the targeted Cr2KO allele and the Cr1 protein was expressed below WT levels. The production of this iCr2 protein is nevertheless a positive outcome of the Cr2KO mouse since iCr2 occupies a B cell surface niche vacated in the absence of Cr2. The iCr2 protein lacks the sequences encoding the SCR7/8 domains which include the iC3b and C3d(g) binding site, but still maintains a membrane localization with full availability for CD19 association. The potential of iCr2 to associate with CD19 is especially important in light of the effects that complement receptor surface expression is proposed have on CD19 expression (Haas et al., 2009). Haas et al. have shown that overexpression of CD19 is correlated with reduced B cell responsiveness and is controlled by Cr1/2 expression (Haas et al., 2005). The Cr2KO mice express an intermediate level of CD19 compared to WT and the Cr1/2KO mice (Fig. 4b) suggesting that the Cr2KO mouse may avoid the complications associated with aberrant CD19 expression.
The most unexpected effect of deletion of the Cr2 alternative splice site is the reduction of Cr1 expression. This reduction is especially surprising in the FDC compartment where a strong preference for Cr1 is observed (Donius et al., 2013). Instead of remaining the same or becoming enhanced compared to WT (which would be expected since the alternative splice to create the Cr2 protein is not evident in FDCs) Cr1 was found to be reduced in FDCs at the transcriptional level and could not be detected by western blot (Fig. 7a and b). The promoter and first intronic sequences of the Cr1KO, Cr2KO and WT strains are virtually identical yet the production of Cr1 in FDCs from the Cr2KO animal is dramatically reduced.
The Cr2KO animals will provide us with a unique strain with which to trace B cell activation within germinal centers. C3KO, Cr1KO and Cr1/2KO animals are all deficient in the generation of activated germinal centers following immunization with sheep red blood cells (Donius et al., 2013; Wu et al., 2000). By creating bone marrow chimeras of Cr2KO marrow into a WT animal, we will be able to determine if B cell expression of the Cr2 protein is required for appropriate B cell activation in the germinal centers. Similarly, by creating bone marrow chimeras of WT bone marrow into Cr2KO animals we will be able to determine if the reduced level of expression of Cr1 on FDC seen in the Cr2KO mouse is still sufficient for appropriate germinal center B cell activation.
In total this work describes the generation of a mouse deficient in the Cr2 isoform of Cr2. Surprisingly, this mouse illustrates a dominant alternative splicing event that creates a novel iCr2 protein product that is expressed on the cell surface yet lacks the critical complement binding domains. The Cr2KO mice are thus functionally deficient in Cr2 but not Cr1, however the effects of hypomorphism for Cr1 expression by B cells and FDCs are yet to be determined. Intriguingly, the human CR2 homolog lacks a direct mouse Cr1 homolog, but maintains non-coding pseudoexons with sequence conservation to mouse (Holguin et al., 1990). It is tempting to speculate that these pseudoexons of the human CR2 gene contain regulatory sequences for splicing and CR2 expression. In humans CR2 produces a long and short isoform with a strong preference for the long isoform of CR2 on FDCs (Liu et al., 1997). Understanding the splicing control mechanism disrupted in the Cr2KO mouse may elucidate the mechanism behind CR2 isoform preference in humans.
Acknowledgments
The authors would like to thank the members of the Weis’ Laboratories for their insight and advice during the course of this investigation. We thank the Utah Transgenic and Knockout Mouse Core facility for helping with the creation of the Cr2KO animal. We also thank core facilities at this university for oligonucleotide preparation and DNA sequencing. This work was supported by the NIH (AI-24158 to JHW, and AI-32223 and AI-43521 to JJW), by the Weber Presidential Endowed Chair (JHW) and by the Training Program in Microbial Pathogenesis 5T32-AI-055434 (LRD).
Abbreviations
- Cr1/CR1
complement receptor 1
- Cr2/CR2
complement receptor 2
- C3
complement component 3
- BCR
B cell receptor
- s
follicular dendritic cell
- SCR
short consensus repeat
- PCR
polymerase chain reaction
- TK
thymidine kinase
- WT
wild type
- SS
signal sequence
- iCr2
incomplete Cr2
- IC
isotype control
- gMFI
geometric mean fluorescent intensity
Footnotes
Conflict of interest
None declared.
References
- Ahearn JM, Fischer MB, Croix D, Goerg S, Ma M, Xia J, Zhou X, Howard RG, Rothstein TL, Carroll MC. Disruption of the Cr2 locus results in a reduction in B-1a cells and in an impaired B cell response to T-dependent antigen. Immunity. 1996;4 (3):251–262. doi: 10.1016/s1074-7613(00)80433-1. [DOI] [PubMed] [Google Scholar]
- Bunting M, Bernstein KE, Greer JM, Capecchi MR, Thomas KR. Targeting genes for self-excision in the germ line. Genes Dev. 1999;13 (12):1524–1528. doi: 10.1101/gad.13.12.1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter RH, Spycher MO, Ng YC, Hoffman R, Fearon DT. Synergistic interaction between complement receptor type 2 and membrane IgM on B lymphocytes. J Immunol. 1988;141 (2):457–463. [PubMed] [Google Scholar]
- Debnath I, Roundy KM, Weis JJ, Weis J. Defining in vivo transcription factor complexes of the murine CD21 and CD23 genes. J Immunol. 2007;178 (11):7139–7150. doi: 10.4049/jimmunol.178.11.7139. [DOI] [PubMed] [Google Scholar]
- Donius LR, Handy JM, Weis JJ, Weis J. Optimal germinal center B cell activation and T-dependent antibody responses require expression of the mouse complement receptor Cr1. J Immunol. 2013;191 (1):434–447. doi: 10.4049/jimmunol.1203176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas KM, Hasegawa M, Steeber DA, Poe JC, Zabel MD, Bock CB, Karp DR, Briles DE, Weis J, Tedder TF. Complement receptors CD21/35 link innate and protective immunity during Streptococcus pneumoniae infection by regulating IgG3 antibody responses. Immunity. 2002;17 (6):713–723. doi: 10.1016/s1074-7613(02)00483-1. [DOI] [PubMed] [Google Scholar]
- Haas KM, Poe J, Steeber D. B-1a and B-1b cells exhibit distinct developmental requirements and have unique functional roles in innate and adaptive immunity to S. pneumoniae. Immunity. 2005;23 (1):7–18. doi: 10.1016/j.immuni.2005.04.011. [DOI] [PubMed] [Google Scholar]
- Haas KM, Poe J, Tedder TF. CD21/35 promotes protective immunity to Streptococcus pneumoniae through a complement-independent but CD19-dependent pathway that regulates PD-1 expression. J Immunol. 2009;183 (6):3661–3671. doi: 10.4049/jimmunol.0901218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holguin MH, Kurtz CB, Parker CJ, Weis JJ, Weis J. Loss of human CR1-and murine Crry-like exons in human CR2 transcripts due to CR2 gene mutations. J Immunol. 1990;145 (6):1776–1781. [PubMed] [Google Scholar]
- Jacobson A, Weis JJ, Weis J. Complement receptors 1 and 2 influence the immune environment in a B cell receptor-independent manner. J Immunol. 2008;180 (7):5057–5066. doi: 10.4049/jimmunol.180.7.5057. [DOI] [PubMed] [Google Scholar]
- Krop I, Shaffer AL, Fearon DT, Schlissel MS. The signaling activity of murine CD19 is regulated during cell development. J Immunol. 1996;157 (1):48–56. [PubMed] [Google Scholar]
- Liu YJ, Xu J, de Bouteiller O, Parham CL, Grouard G, Djossou O, de Saint-Vis B, Lebecque S, Banchereau J, Moore KW. Follicular dendritic cells specifically express the long CR2/CD21 isoform. J Exp Med. 1997;185 (1):165–170. doi: 10.1084/jem.185.1.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loder F, Mutschler B, Ray RJ, Paige CJ, Sideras P, Torres R, Lamers MC, Carsetti R. B cell development in the spleen takes place in discrete steps and is determined by the quality of B cell receptor-derived signals. J Exp Med. 1999;190 (1):75–89. doi: 10.1084/jem.190.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto A, Martin D, Carter RH, Klickstein L, Ahearn J, Fearon D. Functional dissection of the CD21/CD19/TAPA-1/Leu-13 complex of B lymphocytes. J Exp Med. 1993;178 (4):1407–1417. doi: 10.1084/jem.178.4.1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto AK, Kopicky-Burd J, Carter RH, Tuveson DA, Tedder TF, Fearon DT. Intersection of the complement and immune systems: a signal transduction complex of the B lymphocyte-containing complement receptor type 2 and CD19. J Exp Med. 1991;173 (1):55–64. doi: 10.1084/jem.173.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molina H, Holers VM, Li B, Fung Y, Mariathasan S, Goellner J, Strauss-Schoenberger J, Karr RW, Chaplin DD. Markedly impaired humoral immune response in mice deficient in complement receptors 1 and 2. Proc Natl Acad Sci USA. 1996;93 (8):3357–3361. doi: 10.1073/pnas.93.8.3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molina H, Kinoshita T, Webster CB, Holers VM. Analysis of C3b/C3d binding sites and factor I cofactor regions within mouse complement receptors 1 and 2. J Immunol. 1994;153 (2):789–795. [PubMed] [Google Scholar]
- Molina H, Perkins SJ, Guthridge J, Gorka J, Kinoshita T, Holers VM. Characterization of a complement receptor 2 (CR2 CD21) ligand binding site for C3: an initial model of ligand interaction with two linked short consensus repeat modules. J Immunol. 1995;154 (10):5426–5435. [PubMed] [Google Scholar]
- Oberdoerffer S, Moita LF, Neems D, Freitas RP, Hacohen N, Rao A. Regulation of CD45 alternative splicing by heterogeneous ribonucleoprotein, hnRNPLL. Science. 2008;321 (5889):686–691. doi: 10.1126/science.1157610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pappworth IY, Hayes C, Dimmick J, Morgan BP, Holers VM, Marchbank KJ. Mice expressing human CR1/CD35 have an enhanced humoral immune response to T-dependent antigens but fail to correct the effect of premature human CR2 expression. Immunobiology. 2011;217 (2):147–157. doi: 10.1016/j.imbio.2011.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pepys MB. Role of complement in induction of antibody production in vivo Effect of cobra factor and other C3-reactive agents on thymus-dependent and thymus-independent antibody responses. J Exp Med. 1974;140 (1):126–145. doi: 10.1084/jem.140.1.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pramoonjago P, Takeda J, Kim YU, Inoue K, Kinoshita T. Ligand specificities of mouse complement receptor types 1 (CR1) and 2 (CR2) purified from spleen cells. Int Immunol. 1993;5 (4):337–343. doi: 10.1093/intimm/5.4.337. [DOI] [PubMed] [Google Scholar]
- Preussner M, Schreiner S, Hung LH, Porstner M, Jack HM, Benes V, Ratsch G, Bindereif A. HnRNP L and L-like cooperate in multiple-exon regulation of CD45 alternative splicing. Nucleic Acids Res. 2012;40 (12):5666–5678. doi: 10.1093/nar/gks221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothrock CR, House AE, Lynch KW. HnRNP L represses exon splicing via a regulated exonic splicing silencer. EMBO J. 2005;24 (15):2792–2802. doi: 10.1038/sj.emboj.7600745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roundy KM, Weis JJ, Weis J. Deletion of putative intronic control sequences does not alter cell or stage specific expression of Cr2. Mol Immunol. 2009;47 (2):517–525. doi: 10.1016/j.molimm.2009.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seregin SS, Aldhamen YA, Appledorn DM, Schuldt NJ, McBride AJ, Bujold M, Godbehere SS, Amalfitano A. CR1/2 is an important suppressor of Adenovirus-induced innate immune responses and is required for induction of neutralizing antibodies. Gene Ther. 2009;16 (10):1245–1259. doi: 10.1038/gt.2009.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan SS, Weis JH. Development of a sensitive reverse transcriptase PCR assay, RT-RPCR, utilizing rapid cycle times. PCR Methods Appl. 1992;2 (2):137–143. doi: 10.1101/gr.2.2.137. [DOI] [PubMed] [Google Scholar]
- Tong A, Nguyen J, Lynch KW. Differential expression of CD45 isoforms is controlled by the combined activity of basal and inducible splicing-regulatory elements in each of the variable exons. J Biol Chem. 2005;280 (46):38297–38304. doi: 10.1074/jbc.M508123200. [DOI] [PubMed] [Google Scholar]
- Wu X, Jiang N, Fang YF, Xu C, Mao D, Singh J, Fu YX, Molina H. Impaired affinity maturation in Cr2−/− mice is rescued by adjuvants without improvement in germinal center development. J Immunol. 2000;165 (6):3119–3127. doi: 10.4049/jimmunol.165.6.3119. [DOI] [PubMed] [Google Scholar]
- Zabel MD, Weis JJ, Weis J. Lymphoid transcription of the murine CD21 gene is positively regulated by histone acetylation. J Immunol. 1999;163 (5):2697–2703. [PubMed] [Google Scholar]