Abstract
Analyses of human evolution are fundamental to understand the current gradients of human diversity. In this concern, genetic samples collected from current populations together with archaeological data are the most important resources to study human evolution. However, they are often insufficient to properly evaluate a variety of evolutionary scenarios, leading to continuous debates and discussions. A commonly applied strategy consists of the use of computer simulations based on, as realistic as possible, evolutionary models, to evaluate alternative evolutionary scenarios through statistical correlations with the real data. Computer simulations can also be applied to estimate evolutionary parameters or to study the role of each parameter on the evolutionary process. Here we review the mainly used methods and evolutionary frameworks to perform realistic spatially explicit computer simulations of human evolution. Although we focus on human evolution, most of the methods and software we describe can also be used to study other species. We also describe the importance of considering spatially explicit models to better mimic human evolutionary scenarios based on a variety of phenomena such as range expansions, range shifts, range contractions, sex-biased dispersal, long-distance dispersal or admixtures of populations. We finally discuss future implementations to improve current spatially explicit simulations and their derived applications in human evolution.
Keywords: Demographic models, Human evolution, Human landscape genetics, Molecular evolution, Range expansion, Spatially explicit simulation.
INTRODUCTION
The evolutionary history of humans has been largely studied in order to shed light on where and when the first humans colonized the world and how such a colonization took place. Indeed, knowledge about current human genetic variation may help to understand human diseases, for example those presenting variable behaviour among ethnic groups [e.g., 1-3]. Fortunately, genetic signatures from past human evolutionary processes are still present in current humans, and together with archaeological records, may allow us to study human evolution. However, the interpretation of such genetic signatures (e.g., assign a genetic feature to a particular ancestral event) is not straightforward. For instance, different ancestral events might produce a similar genetic effect or a combination of events might lead to complex genetic information. These uncertainties can be especially noted in the literature of human evolution by continuous discussions [e.g., 4, 5]. Below we briefly describe some interesting current topics of debate:
Geographic origin of modern humans. The origins of the modern Homo sapiens remain unclear. It is widely assumed that modern humans originated in Central or South Africa, which is indeed supported by archeological data [e.g., 6-8]. However, other geographic origins have been proposed, for instance, North Africa [e.g., 9, 10] and even multiregional origins through a worldwide gradual transition from earlier humans [11, 12].
Geographic out-of-Africa migration routes. Another interesting topic is the out-of-Africa migration corridors from where modern humans started the colonization of the world at approximately 125-100 kya [5, 13]. Here there are two main routes under discussion. The first one is the traditionally considered route through the Nile Valley and the North of present Egypt [e.g., 14, 15]. The second route is through the Bab-el-Mandeb Strait towards present South Arabia [e.g., 16, 17], whose sea level may have been much lower at the time of the migration [13]. Of course, another possibility is the consideration of both migration routes [e.g., 18, 19].
Principal component analysis of European human genetic diversity gradients. The colonization of Europe by modern humans was initially studied by Cavalli-Sforza et al. [20, 21]. They proposed a demic diffusion (DD) scenario based on a progressive introgression of genes from the local populations (hunter-gatherers, Paleolithic) to the invading populations (farmers, Neolithic) that may have generated a gradient of allele frequencies along the expanding axis [21-23]. Cavalli-Sforza et al. represented these gradients by using principal component analysis (PCA) and interpreted the resulting principal components (PCs) as past migration events [20, 21]. Nevertheless, there is a controversy in this interpretation because the PCs may have arisen from isolation-by-distance scenarios at equilibrium, without requiring any expansion [4, 24, 25].
Colonization of the Americas. It is accepted that the Americas were colonized through several waves beginning 16.5 kya, by crossing the present Bering Strait [26-28], which could be transited at that time as a consequence of the last glacial maximum (LGM) [28, 29]. A proposed scenario considers an initial Pacific coastline migration due to the impediment of Canadian ice sheets that were formed during the LGM [28]. This scenario could explain the early Colombian settlement found by Hellenthal et al. [30] where independent sources of ancestry for Northern and Southern Americans are suggested. By contrast, other studies propose a series of waves where the Americas were colonized from North to South and therefore, the ancestry of South American inhabitants can be related to North America ancestral populations [e.g., 26, 31].
Admixture of Human populations. The admixture of different ancestral human populations is an interesting topic of debate. There is some evidence of admixture between Paleolithic and archaic humans (0.5-2.1%) [32-34] but the genomic distributions of such an admixture are still lacking [see for a review, 35]. On the other hand, the amount of admixture between Paleolithic and Neolithic populations is highly debated and current estimates are described between 20 and 80% depending on the applied methods and data [see e.g. 36-40].
A strategy to help with the above debates consists of the application of computer simulations. In general, computer simulations aim to mimic real world processes and present a variety of applications [see the reviews, 41-45]. Simulations allow for the study of evolutionary aspects that may alter entire processes or enable the understanding of complex systems that are analytically intractable [46]. As noted in [41], computer simulations are widely applied in population genetics for hypothesis testing [e.g., 47-50], to validate and compare analytical frameworks [e.g., 51, 52], to study interactions among evolutionary forces [e.g., 49, 53], or to estimate evolutionary parameters [e.g., 54, 55]. The choice of an appropriate simulator is fundamental because generally simulations should be as realistic as possible to mimic real-world scenarios of population genetics [56-58]. Computer simulations using spatially explicit models can be useful to analyze the influence of habitat on organism evolution at different spatial and temporal scales [59]. In human evolution, spatially explicit simulations have provided important advances to the current understanding of genetic diversity through the estimation of evolutionary parameters and through hypothesis testing of alternative evolutionary models.
This study provides an overview of spatially explicit models and the derived simulation frameworks that are commonly applied to study human evolution. The implemented evolutionary scenarios, with their associated advantages and limitations, are discussed. Then, we describe a variety of human evolution studies based on spatially explicit computer simulations. Finally, we conclude with a discussion on the importance of considering more rational evolutionary scenarios that would help to simulate a more realistic human evolution and generate more accurate inferences. We also discuss the direct incorporation of spatially explicit simulations on analytical methods like the approximate Bayesian computation approach.
SPATIAL AND TEMPORAL SIMULATIONS
Two main approaches are commonly used in population and landscape genetics to simulate evolutionary histories, the coalescent (backward-time) and the forward in time (forward-time). The latter approach includes the spatially explicit models. Below we describe briefly the main particularities of both approaches.
Coalescent Simulations
The coalescent [60] describes the genealogical history of a sample of alleles from the present to a single ancestral copy [see reviews, 61, 62]. Interestingly, it only simulates the backwards in time evolution of a sample and therefore, coalescent simulations are frequently computationally faster than other methods based on the evolution of the whole population (see later). Currently, the coalescent can only simulate a few population genetics models such as demographics [e.g., 63], population history and migration [e.g., 64, 65], gene flow and recombination [e.g., 66, 67] and selection [e.g., 68, 69]. Coalescent simulations can be used in human evolutionary studies [e.g., 50, 70]. In fact, the coalescent is especially interesting when extensive simulations are required (i.e., in analyses based on the approximate Bayesian computation approach) [e.g., 50, 70]. Nevertheless, forward-time simulations can be much more realistic to mimic human evolution due to the consideration of a wide variety of evolutionary processes (see following subsections).
Forward-time Simulations
The forward-time approach evolves the whole population from the past to the present [see reviews, 45, 71, 72]. As a consequence, this approach considers all the ancestral information of the population allowing for individual-individual interactions [e.g., 73], admixture of populations [e.g., 74], complex selection [e.g., 73, 75, 76] and complex migration models [e.g., 74, 76, 77]. Nevertheless, forward simulations are computationally slower than coalescent simulations due to the simulation of the entire population, although recent methods showed improvements in this concern [e.g., 78]. Interestingly, two recent simulators have combined both coalescent and forward-time approaches allowing fast simulation under some complex evolutionary scenarios [79, 80].
Spatially Explicit Simulations
The forward approach includes temporal and spatial (1-dimensional, 2D, or 3D)) models. In terrestrial animals like humans, it is known that 2D spatially explicit models may generate more realistic simulations than models with a lower number of dimensions [45, 59]. This improvement is probably due to the consideration of spatial constraints such as population range expansions [see for a review, 58, 81] or environmental changes [e.g., 49, 53, 58, 82]. Overall, spatially explicit models can better consider the available information and provide more realistic explanations for the observations.
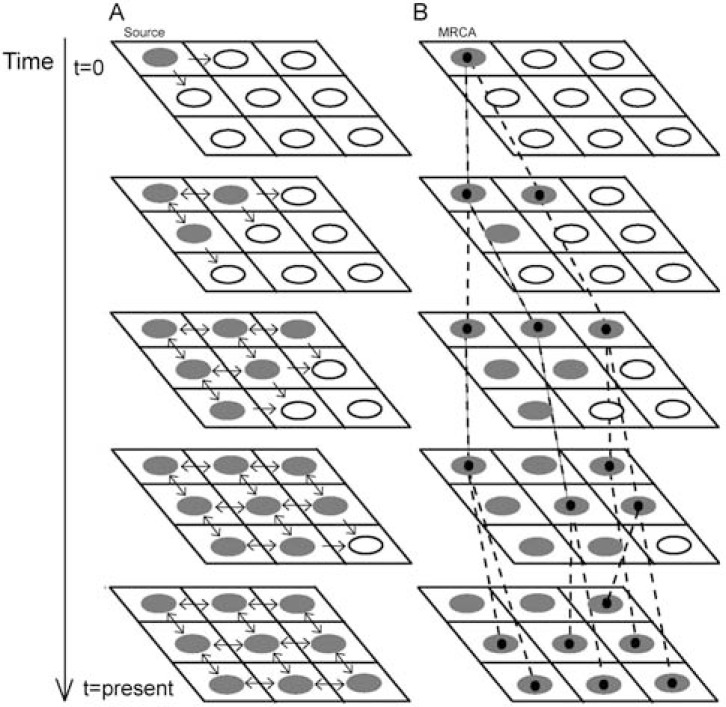
The main goal of spatially explicit models is to combine demographic and genetic processes with a given landscape map, where the landscape features may influence the evolution of the population. Real-world maps can be imported from a Geographical Information System (GIS) tool that usually can also split the map into a lattice of demes by defining a deme size [e.g., 83]. Initially, a deme is chosen to start the colonization, and migration events can occur towards the other demes under a migration model (e.g., the stepping-stone model [84]) (Fig. 1A). In addition to the migration rate, the number of emigrants and immigrants depend on the local and departure deme sizes, respectively. Intra-deme demography can be modelled by the population growth rate [e.g., 85]. A carrying capacity and friction (facility to move through) for each deme can also be considered to model the environmental conditions. All together the process can occur during a user-specified time (or number of generations), and at the end the landscape may become colonized (Fig. 1A).
Fig. (1).
Spatially explicit simulation of a range expansion according to a 2D stepping-stone migration model [84] and posterior representation of the evolutionary history of a sample. A: The colonization of the lattice starts from the upper-left deme (source) that sends migrants to its neighboring demes. Colonized demes can send/receive individuals to/from the neighboring demes while non-colonized demes can only receive individuals. B: Evolutionary history of a random sample of 7 individuals collected at the present from different demes. Going backwards in time, these individuals can reach a most recent common ancestor (MRCA), which not necessarily (but often) belongs to the source deme.
Additional evolutionary processes of general interest, which can be simulated with spatially explicit models, are described in (Table 1).
Table 1.
Evolutionary processes of general interest that can be simulated with spatially explicit models.
| Evolutionary Process | Commentary | References |
|---|---|---|
| Population range expansion | The colonization of a landscape through a spatial expansion is quite different from a pure demographic expansion and may generate particular genetic features such as sectors [107] and allele surfing (alleles riding on the wave of population range expansions [see, 143-145]). | [81] |
| Population range contractions and population range shifts | Under hard living conditions (i.e., as a consequence of a climatic change or a invasive species) a population can reduce or shift its living range. | [49] |
| Heterogeneous environment and habitat fragmentation | Habitats are frequently heterogeneous in the distribution of resources and as a consequence, they are not uniformly occupied. Indeed, habitats can be fragmented with spatial barriers leading to population fragmentation, which may result in loss of genetic diversity and sometimes may cause allopatric speciation [146-148]. | [77, 117] |
| Complex migration | Species dispersal abilities can eventually determine the fate of the populations [146, 149] and should be carefully considered. Anisotropic migration (different migration rates towards the neighboring demes), sex-biased dispersal (i.e., induced by post-marital residence rules) or long-distance dispersal (LDD) may alter the colonization process and may influence genetic diversity. For instance, anisotropic migration towards refugia areas may lead to a larger loss of genetic diversity than isotropic migration [49]. LDD often increases genetic diversity [76, 77]. | [49, 74, 76] |
| Admixed populations | Admixture between two populations may occur if both populations can interbreed [e.g., 113]. In this situations, demic diffusion can influence the spatial distribution of gene frequencies [21, 23]. | [53, 74, 105] |
When applying computer simulations one should have in mind the role of each parameter on the entire evolutionary process. For example, the population size of a deme can increase with the population growth rate, the carrying capacity, and the number of immigrants. The number of emigrants from a deme depends on the migration rate and the population size. Thus, one can understand for example that a scenario with low carrying capacities (e.g., as a consequence of a climate change) and low migration rates may lead a population to extinction [e.g., 49, 86].
After the forward simulation, one could be interested in the evolutionary analysis of a particular sample. Here, some methods allow for the recovery of the history of a sample from the history of the entire population by using the coalescent (Fig. 1B) and then simulate genetic data for only such a sample [80, 85]. As expected, the main advantage of this procedure is the low computational cost for simulating genetic data. By contrast, other methods can simulate genetic data during the forward simulation leading to higher computational costs but allowing additional capabilities such as the ability to follow multi-locus genotypes within individuals or the analysis of all individuals of a deme [74].
SPATIALLY EXPLICIT EVOLUTIONARY FRAMEWORKS APPLIED IN HUMAN EVOLUTION
The implementation of spatially explicit models in available evolutionary frameworks is a recent development and is becoming increasingly more common with time. To date several spatially explicit computer simulators exist and implement different capabilities. (Table 2) shows a list of current spatially explicit simulators. These simulators can be classified as individual-based or deme-based population modeling (see Table 2). In theory, individual-based simulations can be more realistic than deme-based simulators but in practice, a similar performance was observed from both approaches [e.g., 74, 87].
Table 2.
The main publicly available evolutionary frameworks based on 2D spatially explicit models that can be applied to simulate human evolution. “Method” includes forward and coalescent approaches. “Category” indicates if the simulator is deme or individual-modeling oriented. “Scenario” indicates the implementation of the following evolutionary scenarios: de-mographics (D), population history and migration models (Pm), recombination or gene flow (R) and molecular adaptation or selection (S). “Genetic Marker” indicates the kind of genetic data that can be simulated, the implemented substitution models of evolution are described within a parenthesis. “Other capabilities” includes other interesting evolutionary fea-tures implemented in the simulator that may help generate more realistic simulations.
| Program | Method | Category | Scenario | Genetic Marker | Other Capabilities | Reference |
|---|---|---|---|---|---|---|
| Splatche/ Splatche2 |
Forward/coalescent | Deme | D, Pm, R | DNA (JC, K2P)1, SNP, STR (SMM)2 and RFLP | Long-distance dispersal Anisotropic migration Two populations and admixture |
[80, 85] |
| KernelPop | Forward | Individual | D, Pm | STR (IAM, SMM)2, DNA (JC)1 | Long-distance dispersal | [150] |
| IBDsim | Forward/coalescent | Individual | D, Pm | STR (IAM, KAM, GSM, SMM)2 | - | [151] |
| CDPop | Forward | Individual | D, Pm, S3 | STR (KAM)2 | Sex-biased migration and mating Variable dispersal distance |
[130] |
| EcoGenetics | Forward | Individual | D, Pm | STR (KAM, SMM)2 | Sex-biased migration and mating | Unpublished. See http://www2.unil.ch/ biomapper/ecogenetics/ |
1. JC and K2P refer to the Jukes and Cantor [152] and Kimura two parameters [153] DNA substitution models, respectively.
2. Microsatellite (STR) models: IAM, SMM, KAM and GSM refer to the infinite alleles model [154], the stepwise model [155], K-allele model [156] and generalized stepwise model [e.g., 157], respectively.
3. CDPop can simulate natural selection by considering local selective pressures [further details in 131, 132].
Unfortunately, these simulators only implement a few substitution models and, for example, this limitation could generate unrealistic simulation of genome-wide data [e.g., 88-91]. Note that an incorrect substitution model (a model which does not fit well with the real data) may lead to “incorrect” simulations and derived estimations [e.g., 57, 92]. Other demanded capabilities can be the variation of demographic parameters with time (e.g., variable long-distance dispersal (LDD) rate and growth rate with time), covarion models of evolution [93], genomic rearrangement [94] and longitudinal sampling [95].
To our knowledge, only the spatially explicit available simulators SPLATCHE [85] and its new version SPLATCHE2 [80] have been applied in human evolution. This is probably due to their practical graphical user interface (GUI), the implementation of realistic evolutionary processes, and the simulation of genetic material evolution under a variety of genetic markers (see Table 1). Unfortunately, other spatially explicit simulators that have been applied to study human evolution are not publicly available. For example, in 1986, Rendine et al. [96] developed a simple spatially-explicit tool to simulate a European Paleolithic and
Neolithic expansion with admixture (discussed in the following section). Recently, Rasteiro et al. [74] implemented a simulator similar to SPLATCHE which was individual-based and allowed for the consideration of sex-biased migration. Liu et al. [97] also developed a 1D spatially explicit simulator that was applied to simulate the world-wide human settlement. In the next section we describe several interesting applications of these simulators in human evolution.
HUMAN EVOLUTIONARY STUDIES BASED ON SPATIALLY EXPLICIT SIMULATIONS
The application of spatially explicit simulations in human evolution is becoming more popular with the passage of time. These realistic computer simulations are mainly applied in human evolution for comparing alternative models (i.e., geographic origins or different climate changes) and for estimating evolutionary parameters (i.e., rate of interbreeding in population admixtures). Below we describe some applications of spatially explicit simulations that can be of general interest.
Geographic origin of early modern humans. As indicated in the Introduction, a geographic origin of early modern humans in Central or South Africa is commonly assumed [e.g., 6, 7] but there are other studies that suggest a North Africa origin [e.g., 9, 10] or even multiregional origins [11, 12]. These scenarios were evaluated by Ray et al. [98] through SPLATCHE simulations of the Old World human settlement. They performed simulations of a range expansion from 25 evenly distributed geographic origins. They also considered scenarios with a unique origin and multiregional origins (nine models based on different combinations of population sizes and migration rates between continents). Concerning the evolutionary parameters, they assumed an onset of the expansions of 120 kya, a generation time of 30 years according to [99], a growth rate of 0.3 [100], a migration rate of 0.05 (the number of emigrants is 5% of the population size), and a realistic carrying capacity for each deme (environmental heterogeneity). For each scenario, they simulated 10,000 samples of STR data for a total of 22 populations. Real samples from these 22 populations were collected from Rosenberg et al. [101] and this real data was applied to evaluate the different scenarios through statistical correlations. First results suggested a unique North African origin. Nevertheless, the consideration of ascertainment bias in the simulations suggested a unique East African origin. Liu et al. [97] also found this result by using worldwide spatially explicit computer simulations.
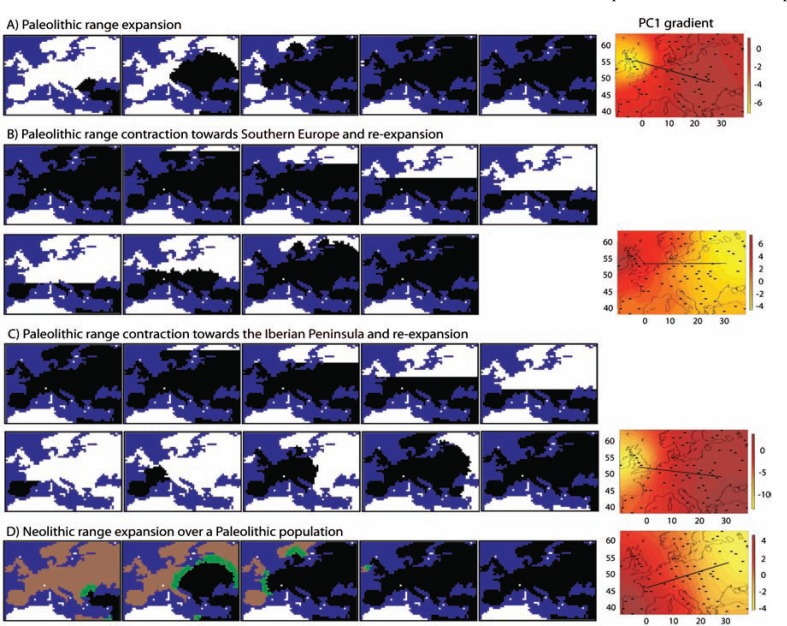
Human genetic diversity gradients in Europe. Evidence for admixture between Paleolithic and Neolithic humans and past range contractions followed by re-expansions. The Paleolithic European colonization was dated between approximately 45 and 40 kya [102] and, as noted in the Introduction, it was initially studied by Cavalli-Sforza et al. [20] by applying PCA on spatially distributed allele frequencies. The resulting PC gradients presented a southeast (SE)-northwest (NW) axis [103, 104], and were interpreted to be a consequence of DD of Neolithic farmers that replaced Paleolithic hunter-gatherer populations with some admixture [21-23]. This interpretation was largely discussed. In 2005, Currat and Excoffier [105] performed spatially explicit simulations of the colonization of Europe by pure Neolithic populations and by Paleolithic and Neolithic populations under different levels of admixture. They found that both Paleolithic and Neolithic populations resulted in SE-NW genetic diversity gradients as a consequence of allele surfing in the wave of the expansion, but not as a consequence of DD as suggested by Cavalli-Sforza et al. In 2010, François et al. [106] repeated these simulations with updates in the evolutionary scenarios. They computed PC gradients from the simulated data. Counterintuitively, the resulting PC1 gradient presented a SW-NE orientation that is orthogonal to the range expansion (Fig. 2D, right). This PC1 gradient was explained as a consequence of allele surfing based on geographic sectors along the Neolithic wave of the expansion [107]. In 2013, Arenas et al. [53] performed extensive simulations of more sophisticated evolutionary scenarios including range contractions (as a consequence of the LGM) towards Southern Europe (Fig. 2B, left) or towards the Iberian Peninsula (Fig. 2C, left) [see 49, 108]. These simulations included a refugial isolation period that was followed by a re-expansion. Pure Paleolithic expansion (Fig. 2A, left) and different admixture levels were also evaluated. This study showed that pure Paleolithic populations lead to a SE-NW PC1 gradient (Fig. 2A, right; similar to the gradients obtained by Cavalli-Sforza et al.) caused by a homogenization of molecular diversity [53]. By contrast, pure Neolithic populations resulted in a SW-NE PC1 gradient (Fig. 2D, right; similar to the gradients obtained by François et al.). In addition, PC1 gradients varied between those orientations as a function of the amount of admixture [53]. On the other hand, range contraction scenarios generated PC1 gradients orthogonal to the axis of the range re-expansion (Fig. 2B and 2C, right), especially for scenarios with higher Paleolithic contribution (note that the LGM occurs during the Paleolithic period). Overall, both the location of the refugia and the level of admixture influence PC gradients. The gradients by Cavalli-Sforza et al. are reproducible under a large Paleolithic contribution (Fig. 2A, right) or under a range contraction towards the Iberian Peninsula (Fig. 2C, right).
Admixture between modern humans and Neanderthals. The admixture between modern humans and preexisting humans was studied by Currat and Excoffier [109]. They simulated mitochondrial DNA (mtDNA) data by spatially explicit simulations under a scenario where early modern humans colonize Europe with different amounts of admixture with Neanderthals. In case of admixture, massive introgression genes from Neanderthals to modern humans might have taken place during the invasion [110]. However, the authors found that the maximum possible contribution of Neanderthals into modern humans was smaller than 0.1%, suggesting almost complete sterility between modern humans and Neanderthals. More recently, these authors published a more sophisticated study on admixture in Eurasians [111]. In particular, they performed extensive spatially explicit simulations with variable amounts of admixture and under a variety of evolutionary scenarios based on different levels of migration rates, carrying capacities and growth rates. They then computed the maximum likelihood for each model and selected the best model through the Akaike information criterion (AIC) [112]. The results indicated a very low rate of interbreeding (smaller than 2%), suggesting an important barrier to gene flow between both species.
Sex-biased migration during the Neolithic transition. An important feature in human evolution is the different demographic histories for males and females [113, 114]. For example, Hamilton et al. [87] performed spatially explicit simulations of human populations from northern Thailand [115] to show that “the number of male immigrants is much smaller (8 times) in patrilocal populations than in matrilocal populations”, and by contrast, “females move 2.5 times more in patrilocal populations than in matrilocal populations” [87]. Recently, Rasteiro et al. [74] studied the role of post-marital residence (PMR) and admixture between Paleolithic and Neolithic populations. They developed an individual-based spatially explicit simulator (not publicly available) that implements sex-biased migration. They then simulated scenarios where Neolithic populations colonize Europe under different amounts of admixture with Paleolithic populations, and under different patterns of PMR. To study the role of PMR, they simulated both mtDNA and Y-chromosome (NRY) data. Simulated datasets were evaluated with real data [38, 116] to select which model best fit the real information. The results indicated that patrilocality in farmers explained the genetic diversity better than matrilocality or bilocality. In addition, they observed that the genetic diversity of farmers can also be influenced by Paleolithic PMR rules.
Fig. (2).
Illustrative examples of spatially explicit simulation of modern human colonization of Europe and principal component analysis derived from the simulated genetic diversity. Left: Snapshots of SPLATCHE2 to simulate an example of a: (A) Paleolithic range expansion over Europe; (B) Paleolithic range contraction towards Southern Europe and posterior re-expansion; (C) Paleolithic range contraction towards the Iberian Peninsula and posterior re-expansion; (D) Neolithic range expansion over Europe where the brown area is colonized by Paleolithic populations, the black area is colonized by Neolithic populations and the green region indicates a zone of cohabitation; at the end of this simulation Paleolithic populations are totally replaced by Neolithic populations. Settings (demographic parameter values) that we have applied to perform these simulations follow Arenas et al. [53]. The simulated population range expansions always start from the Middle East. Snapshots are taken each 50 generations. Right: Illustrative example of PC1 gradients for each above-described scenario. The black lines represent the PC1 gradient orientation, namely NW-SE for (A), W-E for (B), NW-SE for (C) and SW-NE for (D).
THE FUTURE OF SPATIALLY EXPLICIT COMPUTER SIMULATIONS IN HUMAN EVOLUTION
Spatially explicit models are fundamental to mimic the evolutionary history of terrestrial species because the consideration of spatio-temporal phenomena like range expansions, range contractions, range shifts, LDD, and habitat fragmentation, can influence genetic diversity [e.g., 49, 76, 77, 117]. Different evolutionary frameworks have been developed for the simulation of molecular data under spatially explicit models. However, these simulators implement very simple substitution models of evolution, and it is known that an assumed model, that is more simple than the true model, may lead to incorrect results [e.g., 57, 92]. Indeed, some of these simulators ignore recombination, which can bias evolutionary inferences [e.g., 118-121]. As a consequence, there is a need for more realistic computer simulators that implement complex substitution models of evolution, not only at the nucleotide level, but also at the codon [e.g., 43, 69], protein [e.g., 122-124] and genome-wide levels [88, 90]. Indeed, recombination (as well as other processes of exchange of genetic material) may generate evolutionary networks [67, 125] that should be considered to properly describe the history of human populations [see 18, 126].
As expected, most of the studies mentioned in the previous section could be improved with the consideration of additional evolutionary processes. For example, some available spatially explicit simulators implement LDD. However, this feature has not been applied yet in studies of human evolution. This fact could be explained by the complexity in the definition of the LDD, which includes priors for the LDD rate, dispersal distance, and direction of the dispersal events [see 76, 127] that should be studied from real observations [76]. Furthermore, other complex migration forms like anisotropic migration [49, 53] and sex-biased dispersal [e.g., 74, 87] can also influence genetic diversity and should always be considered. One could also expect that some of these evolutionary processes could vary with time. For example, population growth rates and dispersal distances could increase with time due to acculturation [105]. In addition, the topographic map and its resources could also change with time [e.g., 29]. As noted above, the LGM period could lead to past range contractions towards refugia areas [49, 53] and could allow the colonization of the Americas through the Bering Strait [28, 29]. Natural selection is another evolutionary force that should be considered to simulate human evolution [see 128, 129]. In spatially explicit simulations, to date only CDPop [130] implements natural selection [131, 132] and unfortunately, it was not applied to humans yet. All together, to obtain accurate and realistic results it is important to consider complex evolutionary models and model updates according with the simulated evolutionary time. Of course, more complex models do not necessarily lead to more realistic simulations, but if the complexity comes from real features and observations, such complex models should be taken into account.
On the other hand, robust inferences of human evolution will probably require the use of genome data. In this concern, next-generation sequencing (NGS) technologies now deliver fast and accurate genome sequences [133]. Note that complete and near-complete ancient human genomes are currently being obtained [e.g., 34, 134, 135]. However, the complexity of genome evolution [88, 89] may result in models and data where a likelihood function cannot be computed [136, 137]. As an alternative, analytical methods based on computer simulations are emerging since last years, in particular, the Bayesian model-choice [138] and the approximate Bayesian computation (ABC) approach [see for a review, 55, 139]. An additional goal of these methods is their ability to co-estimate evolutionary parameters. Since molecular evolution consists of the joint action of all the evolutionary processes together, ideally, one would want to estimate these parameters simultaneously to avoid potential biases [54, 56]. By contrast, these methods require extensive simulations and therefore, fast computer simulators are desired. For example, SPLATCHE2 has combined the forward and the coalescent methods to perform rapid simulations by multiple sampling of genetic data (coalescent) from a previously simulated entire population (forward-time). In addition, this program allows parallelization of the simulations on a cluster, which can alleviate computer times. Since last years, ABC is more frequently applied to the analysis of human evolution [e.g., 50, 87, 140-142]. Nevertheless, to our knowledge only the study by Hamilton et al. [87] applies an ABC method based on spatially explicit simulations. We believe the application of ABC in spatially explicit contexts will benefit future human evolutionary inferences.
Altogether, this review examines current methods and software applied to perform spatially explicit simulations of human evolution. We found that to date only a few simulators have been developed for this purpose and they still assume a number of evolutionary aspects (i.e. too simple substitution models of evolution and neutral evolution). Therefore, there is a continuous need for fast and more realistic spatially explicit simulators and we expect future advances in this concern. As a consequence, we also expect much more application of spatially explicit simulations in analysis of human evolution.
ACKNOWLEDGEMENTS
We want to thank the Editor of Current Genomics for the invitation to contribute with this review. We also want to thank Laurent Excoffier (as well as other members of the CMPG Lab), Mathias Currat, Nicolas Ray and Olivier François for fascinating discussions and studies during last years. We thank David A. Liberles, Russell A. Hermansen and Anke Konrad for helpful comments. We thank two reviewers for insightful comments and suggestions. M.A. was supported by the “Juan de la Cierva” fellowship JCI-2011-10452 (Spanish Government) and the EMBO fellowship “ASTF 367-2013”.
CONFLICT OF INTEREST
The author(s) confirm that this article content has no conflict of interest.
REFERENCES
- 1.Perez-Losada M, Posada D, Arenas M, Jobes DV, Sinangil F, Berman PW, Crandall KA. Ethnic differences in the adaptation rate of HIV gp120 from a vaccine trial. Retrovirology. 2009;6:67. doi: 10.1186/1742-4690-6-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wiencke JK. Impact of race/ethnicity on molecular pathways in human cancer. Nat. Rev. Cancer. 2004;4(1):79–84. doi: 10.1038/nrc1257. [DOI] [PubMed] [Google Scholar]
- 3.Weinberger PM, Merkley MA, Khichi SS, Lee JR, Psyrri A, Jackson LL, Dynan WS. Human papillomavirus-active head and neck cancer and ethnic health disparities. Laryngoscope. 2010;120(8):1531–1537. doi: 10.1002/lary.20984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Novembre J, Stephens M. Response to Cavalli-Sforza interview [Human Biology 82(3): 245-266 (June 2010,)] Hum. Biol. 2010;82(4):469–470. doi: 10.3378/027.082.0408. [DOI] [PubMed] [Google Scholar]
- 5.Relethford J.H. Genetic evidence and the modern human origins debate. Heredity (Edinb) 2008;100(6):555–563. doi: 10.1038/hdy.2008.14. [DOI] [PubMed] [Google Scholar]
- 6.White TD, Asfaw B, DeGusta D, Gilbert H, Richards GD, Suwa G, Howell FC. Pleistocene Homo sapiens from Middle Awash Ethiopia. Nature. 2003;423(6941):742–747. doi: 10.1038/nature01669. [DOI] [PubMed] [Google Scholar]
- 7.Stringer C. Human evolution: Out of Ethiopia. Nature. 2003;423(6941):692–693-695. doi: 10.1038/423692a. [DOI] [PubMed] [Google Scholar]
- 8.Rito T, Richards MB, Fernandes V, Alshamali F, Cerny V, Pereira L, Soares P. The first modern human dispersals across Africa. PLoS One. 2013;8(11):e80031. doi: 10.1371/journal.pone.0080031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Balter M. Was North Africa the launch pad for modern human migrations? Science 2011. 331(6013):20–23. doi: 10.1126/science.331.6013.20. [DOI] [PubMed] [Google Scholar]
- 10.Smith TM, Tafforeau P, Reid DJ, Grun R, Eggins S, Bou-takiout M, Hublin JJ. Earliest evidence of modern human life history in North African early Homo sapiens. Proc. Natl. Acad. Sci. U S A. 2007;104(15):6128–6133. doi: 10.1073/pnas.0700747104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wolpoff MH, Hawks J, Caspari R. Multiregional not multiple origins. AM. J. Phys. Anthrop. 2000;112(1):129–136. doi: 10.1002/(SICI)1096-8644(200005)112:1<129::AID-AJPA11>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 12.Wolpoff MH, editor. In The human revolution Biological perspectives in the origins of modern humans. Princeton NJ: Princeton University Press ; 1989. Multiregional evolution: The fossil alternative to Eden; pp. 62–108. [Google Scholar]
- 13.Stringer C. Palaeoanthropology.Coasting out of Africa. Nature. 2000;405(6782):24–25 27. doi: 10.1038/35011166. [DOI] [PubMed] [Google Scholar]
- 14.Lahr MM, Foley RA. Towards a theory of modern human origins.geography demography and diversity in recent human evolution. AM. J. Phys. Anthropol. 1998;(Suppl 27):137–176. doi: 10.1002/(sici)1096-8644(1998)107:27+<137::aid-ajpa6>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- 15.Krings M, Salem AE, Bauer K, Geisert H, Malek AK, Chaix L, Simon C, Welsby D, Di Rienzo A, Utermann G, Sajantila A, Paabo S, Stoneking M. mtDNA analysis of Nile River Valley populations: A genetic corridor or a barrier to migration? AM.J. Hum. Genet. 1999;64(4):1166–1176. doi: 10.1086/302314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stanyon R, Sazzini M, Luiselli D. Timing the first human migration into eastern Asia. J. Biol. 2009;8(2):18. doi: 10.1186/jbiol115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Walter RC, Buffler RT, Bruggemann JH, Guillaume MM, Berhe SM, Negassi B, Libsekal Y, Cheng H, Edwards RL, von Cosel R, Neraudeau D, Gagnon M. Early human occupation of the Red Sea coast of Eritrea during the last interglacial. Nature. 2000;405(6782):65–69. doi: 10.1038/35011048. [DOI] [PubMed] [Google Scholar]
- 18.Mele M, Javed A, Pybus M, Zalloua P, Haber M, Comas D, Netea MG, Balanovsky O, Balanovska E, Jin L, Yang Y, Pitchappan RM, Arunkumar G, Parida L, Calafell F, Bertranpetit J. Recombination gives a new insight in the effective population size and the history of the old world human populations. Mol. Biol. Evol. 2012;29(1):25–30. doi: 10.1093/molbev/msr213. [DOI] [PubMed] [Google Scholar]
- 19.Armitage SJ, Jasim SA, Marks AE, Parker AG, Usik VI, Uerpmann H P. The southern route "out of Africa" evidence for an early expansion of modern humans into Arabia. Science. 2011;331(6016):453–456. doi: 10.1126/science.1199113. [DOI] [PubMed] [Google Scholar]
- 20.Cavalli-Sforza LL, Edwards A W F, editors. In: Genetics today: Proceedings of the 11th International Congress of Genetics. New York: The Hague The Netherlands.: Pergamon:; 1963. Analysis of human evolution; pp. 923–993. [Google Scholar]
- 21.Ammerman AJ, Cavalli-Sforza L.L, editors. Princeton NJ.: Princeton University Press; 1984. The neolithic transition and the genetics of populations in Europe. [Google Scholar]
- 22.Diamond J, Bellwood P. Farmers and their languages: the first expansions. Science. 2003;300(5619):597–603. doi: 10.1126/science.1078208. [DOI] [PubMed] [Google Scholar]
- 23.Sokal RR, Oden NL, Wilson C. Genetic evidence for the spread of agriculture in Europe by demic diffusion. Nature. 1991;351(6322):143–145. doi: 10.1038/351143a0. [DOI] [PubMed] [Google Scholar]
- 24.Novembre J, Stephens M. Interpreting principal component analyses of spatial population genetic variation. Nat. Genet. 2008;40(5):646–649. doi: 10.1038/ng.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reich D, Price AL, Patterson N. Principal component analysis of genetic data. Nat. Genet. 2008;40(5):491–492. doi: 10.1038/ng0508-491. [DOI] [PubMed] [Google Scholar]
- 26.Reich D, Patterson N, Campbell D, Tandon A, Mazieres S, Ray N, Parra MV, Rojas W, Duque C, Mesa N, Garcia LF, Triana O, Blair S, Maestre A, Dib JC, Bravi CM, Bailliet G, Corach D, Hunemeier T, Bortolini MC, Salzano FM, Petzl-Erler ML, Acuna-Alonzo V, Aguilar-Salinas C, Canizales-Quinteros S, Tusie-Luna T, Riba L, Rodriguez-Cruz M, Lopez-Alarcon M, Coral-Vazquez R, Canto-Cetina T, Silva-Zolezzi I, Fernandez-Lopez JC, Contreras AV, Jimenez-Sanchez G, Gomez-Vazquez MJ, Molina J, Carracedo A, Salas A, Gallo C, Poletti G, Witonsky DB, Alkorta-Aranburu G, Sukernik RI, Osipova L, Fedorova SA, Vasquez R, Villena M, Moreau C, Barrantes R, Pauls D, Excoffier L, Bedoya G, Rothhammer F, Dugoujon JM, Larrouy G, Klitz W, Labuda D, Kidd J, Kidd K, Di Rienzo A, Freimer NB, Price AL, Ruiz-Linares A. Reconstructing Native American population history. Nature. 2012;488(7411):370–374. doi: 10.1038/nature11258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Neves WA, Prous A, Gonzalez-Jose R, Kipnis R, Powell J. Early Holocene human skeletal remains from Santana do Riacho Brazil: implications for the settlement of the New World. J. Hum. Evol. 2003;45(1):19–42. doi: 10.1016/s0047-2484(03)00081-2. [DOI] [PubMed] [Google Scholar]
- 28.Goebel T, Waters MR, O'Rourke DH. The late Pleistocene dispersal of modern humans in the Americas. Science. 2008;319(5869):1497–1502. doi: 10.1126/science.1153569. [DOI] [PubMed] [Google Scholar]
- 29.Ray N, Adams J M. A GIS-based Vegetation Map of the World at the Last Glacial Maximum (25000-15000 BP) Internet Archaeology. 2001: 11. [Google Scholar]
- 30.Hellenthal G, Auton A, Falush D. Inferring human colonization history using a copying model. PLoS Genet. 2008;4(5):e1000078. doi: 10.1371/journal.pgen.1000078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang S, Lewis CM, Jakobsson M, Ramachandran S, Ray N, Bedoya G, Rojas W, Parra MV, Molina JA, Gallo C, Mazzotti G, Poletti G, Hill K, Hurtado AM, Labuda D, Klitz W, Barrantes R, Bortolini MC, Salzano FM, Petzl-Erler ML, Tsuneto LT, Llop E, Rothhammer F, Excoffier L, Feldman MW, Rosenberg NA, Ruiz-Linares A. Genetic variation and population structure in native Americans. PLoS Genet. 2007;3(11):e185. doi: 10.1371/journal.pgen.0030185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hammer MF, Woerner AE, Mendez FL, Watkins JC, Wall J D. Genetic evidence for archaic admixture in Africa. Proc. Natl. Acad. Sci. U S A. 2011;108(37):15123–15128. doi: 10.1073/pnas.1109300108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wall JD, Lohmueller KE, Plagnol V. Detecting ancient admixture and estimating demographic parameters in multiple human populations. Mol. Biol. Evol. 2009;26(8):1823–1827. doi: 10.1093/molbev/msp096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Prufer K, Racimo F, Patterson N, Jay F, Sankararaman S, Sawyer S, Heinze A, Renaud G, Sudmant PH, de Filippo C, Li H, Mallick S, Dannemann M, Fu Q, Kircher M, Kuhlwilm M, Lachmann M, Meyer M, Ongyerth M, Siebauer M, Theunert C, Tandon A, Moorjani P, Pickrell J, Mullikin JC, Vohr SH, Green RE, Hellmann I, Johnson PL, Blanche H, Cann H, Kitzman JO, Shendure J, Eichler E.E, Lein ES, Bakken TE, Golovanova LV, Doronichev VB, Shunkov MV, Derevianko AP, Viola B, Slatkin M, Reich D, Kelso J, Paabo S. The complete genome sequence of a Neanderthal from the Altai Mountains. Nature. 2014;505(7481):43–49. doi: 10.1038/nature12886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Alves I, Sramkova Hanulova A, Foll M, Excoffier L. Genomic data reveal a complex making of humans. PLoS Genet. 2012;8(7):e1002837. doi: 10.1371/journal.pgen.1002837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chikhi L, Destro-Bisol G, Bertorelle G, Pascali V, Barbujani G. Clines of nuclear DNA markers suggest a largely neolithic ancestry of the European gene pool. Proc. Natl. Acad. Sci. U S A. 1998;95(15):9053–9058. doi: 10.1073/pnas.95.15.9053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Barbujani G, Chikhi L. Population genetics: DNAs from the European Neolithic. Heredity. 2006;97(2):84–85. doi: 10.1038/sj.hdy.6800852. [DOI] [PubMed] [Google Scholar]
- 38.Richards M, Macaulay V, Hickey E, Vega E, Sykes B, Guida V, Rengo C, Sellitto D, Cruciani F, Kivisild T, Villems R, Thomas M, Rychkov S, Rychkov O, Rychkov Y, Golge M, Dimitrov D, Hill E, Bradley D, Romano V, Cali F, Vona G, Demaine A, Papiha S, Triantaphyllidis C, Stefanescu G, Hatina J, Belledi M, Di Rienzo A, Novelletto A, Oppenheim A, Norby S, Al-Zaheri N, Santachiara-Benerecetti S, Scozari R, Torroni A, Bandelt HJ. Tracing European founder lineages in the Near Eastern mtDNA pool. AM. J. Hum. Genet. 2000;67(5):1251–1276. [PMC free article] [PubMed] [Google Scholar]
- 39.Belle EM, Landry PA, Barbujani G. Origins and evolution of the Europeans' genome: evidence from multiple microsatellite loci. Proc. Biol. Sci. 2006;273(1594):1595–1602. doi: 10.1098/rspb.2006.3494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Balaresque P, Bowden GR, Adams SM, Leung HY, King TE, Rosser ZH, Goodwin J, Moisan JP, Richard C, Millward A, Demaine AG, Barbujani G, Previdere C, Wilson IJ, Tyler-Smith C, Jobling MA. A predominantly neolithic origin for European paternal lineages. PLoS Biol. 2010;8(1):e1000285. doi: 10.1371/journal.pbio.1000285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Arenas M. Simulation of Molecular Data under Diverse Evolutionary Scenarios. PLoS Comput. Biol. 2012;8(5):e1002495. doi: 10.1371/journal.pcbi.1002495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Arenas M. Computer programs and methodologies for the simulation of DNA sequence data with recombination. Front. Genet. 2013;4(9) doi: 10.3389/fgene.2013.00009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Arenas M, Posada D, editors. In: Codon Evolution. Oxford: Oxford University Press:; 2012. Simulation of coding sequence evolution; pp. 126–132. [Google Scholar]
- 44.Hoban S, Bertorelle G, Gaggiotti O E. Computer simulations: tools for population and evolutionary genetics. Nat. Rev. Genet. 2012;13(2):110–122. doi: 10.1038/nrg3130. [DOI] [PubMed] [Google Scholar]
- 45.Epperson BK, McRae BH, Scribner K, Cushman SA, Rosenberg MS, Fortin MJ, James PM, Murphy M, Manel S, Legendre P, Dale MR. Utility of computer simulations in landscape genetics. Mol. Ecol. 2010;19(17):3549–3564. doi: 10.1111/j.1365-294X.2010.04678.x. [DOI] [PubMed] [Google Scholar]
- 46.Peck SL. Simulation as experiment: a philosophical reassessment for biological modeling. Trends Ecol. Evol. 2004;19(10):530–534. doi: 10.1016/j.tree.2004.07.019. [DOI] [PubMed] [Google Scholar]
- 47.Arenas M, Valiente G, Posada D. Characterization of reticulate networks based on the coalescent with recombination. Mol. Biol. Evol. 2008;25(12):2517–2520. doi: 10.1093/molbev/msn219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pierron D, Chang I, Arachiche A, Heiske M, Thomas O, Borlin M, Pennarun E, Murail P, Thoraval D, Rocher C, Letellier T. Mutation rate switch inside Eurasian mitochondrial haplogroups: impact of selection and consequences for dating settlement in Europe. PLoS One. 2011;6(6):e21543. doi: 10.1371/journal.pone.0021543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Arenas M, Ray N, Currat M, Excoffier L. Consequences of Range Contractions and Range Shifts on Molecular Diversity. Mol. Biol. Evol. 2012;29(1):207–218. doi: 10.1093/molbev/msr187. [DOI] [PubMed] [Google Scholar]
- 50.Fagundes NJ, Ray N, Beaumont M, Neuenschwander S, Salzano FM, Bonatto SL, Excoffier L. Statistical evaluation of alternative models of human evolution. Proc. Natl. Acad. Sci. U S A. 2007;104(45):17614–17619. doi: 10.1073/pnas.0708280104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Westesson O, Holmes I. Accurate detection of recombinant breakpoints in whole-genome alignments. PLoS Comput. Biol. 2009;5(3):e1000318. doi: 10.1371/journal.pcbi.1000318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Marttinen P, Hanage WP, Croucher NJ, Connor TR, Harris SR, Bentley SD, Corander J. Detection of recombination events in bacterial genomes from large population samples. Nucleic. Acids Res. 2012;40(1):e6. doi: 10.1093/nar/gkr928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Arenas M, Francois O, Currat M, Ray N, Excoffier L. Influence of admixture and paleolithic range contractions on current European diversity gradients. Mol. Biol. Evol. 2013;30(1):57–61. doi: 10.1093/molbev/mss203. [DOI] [PubMed] [Google Scholar]
- 54.Wilson DJ, Gabriel E, Leatherbarrow AJ, Cheesbrough J, Gee S, Bolton E, Fox A, Hart CA, Diggle PJ, Fearnhead P. Rapid evolution and the importance of recombination to the gastroenteric pathogen Campylobacter jejuni. Mol. Biol. Evol. 2009;26(2):385–397. doi: 10.1093/molbev/msn264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Beaumont MA. Approximate Bayesian Computation in Evolution and Ecology. Annu. Rev. Ecol. Evol. Syst. 2010;41:379–405. [Google Scholar]
- 56.Lopes JS, Arenas M, Posada D, Beaumont MA. Coestimation of Recombination Substitution and Molecular Adaptation rates by approximate Bayesian computation. Heredity. 2014;112(3):255–264. doi: 10.1038/hdy.2013.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lemmon AR, Moriarty EC. The importance of proper model assumption in bayesian phylogenetics. Syst. Biol. 2004;53(2):265–277. doi: 10.1080/10635150490423520. [DOI] [PubMed] [Google Scholar]
- 58.Ray N, Excoffier L. Inferring past demography using spatially explicit population genetic models. Hum. Biol. 2009;81(2-3):141–157. doi: 10.3378/027.081.0303. [DOI] [PubMed] [Google Scholar]
- 59.Dunning JB, Stewart DJ, Danielson BJ, Noon BR, Root TL, Lamberson RH, Stevens EE. Spatially explicit population models: current forms and future uses. Ecol. Appl. 1995;5(1):3–11. [Google Scholar]
- 60.Kingman J F C. The coalescent. Stochastic Processes and their Applications. 1982;13:235–248. [Google Scholar]
- 61.Nordborg M, editor. In: Handbook of Statistical Genetics . Third Edition. Chichester UK: John Wiley & Sons Ltd:; 2007. Coalescent theory; pp. 843–877. [Google Scholar]
- 62.Wakeley J, editor. Greenwood Village Colorado 2008: Roberts and Company Publishers.; Coalescent Theory: An Introduction. [Google Scholar]
- 63.Slatkin M. Simulating genealogies of selected alleles in a population of variable size. Genet. Res. 2001;78(1):49–57. doi: 10.1017/s0016672301005183. [DOI] [PubMed] [Google Scholar]
- 64.Hudson R R. Island models and the coalescent process. Mol. Ecol. 1998;7(4):413–418. [Google Scholar]
- 65.Arenas M, Posada D. Recodon coalescent simulation of coding DNA sequences with recombination migration and demography. BMC Bioinformatics. 2007;8(458) doi: 10.1186/1471-2105-8-458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hudson RR. Properties of a neutral allele model with intragenic recombination. Theor. Popul. Biol. 1983;23(2):183–201. doi: 10.1016/0040-5809(83)90013-8. [DOI] [PubMed] [Google Scholar]
- 67.Arenas M. The Importance and Application of the Ancestral Recombination Graph. Front. Genet. 2013;4(206) doi: 10.3389/fgene.2013.00206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hudson RR, Kaplan N L. The coalescent process in models with selection and recombination. Genetics. 1988;120(3):831–840. doi: 10.1093/genetics/120.3.831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Arenas M, Posada D. Coalescent simulation of intracodon recombination. Genetics. 2010;184(2):429–437. doi: 10.1534/genetics.109.109736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Laval G, Patin E, Barreiro LB, Quintana-Murci L. Formulating a historical and demographic model of recent human evolution based on resequencing data from noncoding regions. PLoS One. 2010;5(4):e10284. doi: 10.1371/journal.pone.0010284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Carvajal-Rodriguez A. Simulation of genes and genomes forward in time. Curr. Genomics. 2010;11(1):58–61. doi: 10.2174/138920210790218007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Carvajal-Rodriguez A. Simulation of genomes: a review. Curr. Genomics. 2008;9(3):155–159. doi: 10.2174/138920208784340759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Peng B, Amos CI, Kimmel M. Forward-time simulations of human populations with complex diseases. PLoS Genet. 2007;3(3):e47. doi: 10.1371/journal.pgen.0030047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Rasteiro R, Bouttier PA, Sousa VC, Chikhi L. Investigating sex-biased migration during the Neolithic transition in Europe using an explicit spatial simulation framework. Proc. Biol. Sci. 2012;279(1737):2409–2416. doi: 10.1098/rspb.2011.2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Calafell F, Grigorenko EL, Chikanian AA, Kidd KK. Haplotype evolution and linkage disequilibrium.A simulation study. . Hum. Hered. 2001;51(1-2):85–96. doi: 10.1159/000022963. [DOI] [PubMed] [Google Scholar]
- 76.Ray N, Excoffier L. A first step towards inferring levels of long-distance dispersal during past expansions. Molecular Ecology Resources. 2010;10(5):902–914. doi: 10.1111/j.1755-0998.2010.02881.x. [DOI] [PubMed] [Google Scholar]
- 77.Mona S, Ray N, Arenas M, Excoffier L. Genetic consequences of habitat fragmentation during a range expansion. Heredity. 2014;112(3):291–299. doi: 10.1038/hdy.2013.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Padhukasahasram B, Marjoram P, Wall JD, Bustamante CD, Nordborg M. Exploring population genetic models with recombination using efficient forward-time simulations. Genetics. 2008;178(4):2417–2427. doi: 10.1534/genetics.107.085332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ewing G, Hermisson J. MSMS: a coalescent simulation program including recombination demographic structure and selection at a single locus. Bioinformatics. 2010;26(16):2064–2065. doi: 10.1093/bioinformatics/btq322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ray N, Currat M, Foll M, Excoffier L. SPLATCHE2: a spatially explicit simulation framework for complex demography genetic admixture and recombination. Bioinformatics. 2010;26(23):2993–2994. doi: 10.1093/bioinformatics/btq579. [DOI] [PubMed] [Google Scholar]
- 81.Excoffier L, Foll M, Petit R J. Genetic consequences of range expansions. Annu. Rev. Ecol. Evol. Syst. 2009;40:481–501. [Google Scholar]
- 82.Arenas M, Mona S, Trochet A, Sramkova Hanulova A, Currat M, Ray N, Chikhi L, Rasteiro R, Schmeller DS, Excoffier L, editors. In Scaling in ecology and conservation; Pensoft Publishers. In press.: The scaling of genetic diversity in a changing and fragmented world. [Google Scholar]
- 83.Bivand R. Using the R statistical data analysis language on GRASS 5.0 GIS database files. Computers & Geosciences. 2000;26:1043–1052. [Google Scholar]
- 84.Kimura M, Weiss G H. The Stepping Stone Model of Population Structure and the Decrease of Genetic Correlation with Distance. Genetics. 1964;49(4):561–576. doi: 10.1093/genetics/49.4.561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Currat M, Ray N, Excoffier L. SPLATCHE: a program to simulate genetic diversity taking into account environmental heterogeneity. Mol. Ecol. Notes. 2004;4(1):139–142. [Google Scholar]
- 86.Atkins KE, Travis J M. Local adaptation and the evolution of species' ranges under climate change. J. Theor. Biol. 2010;266(3):449–457. doi: 10.1016/j.jtbi.2010.07.014. [DOI] [PubMed] [Google Scholar]
- 87.Hamilton G, Stoneking M, Excoffier L. Molecular analysis reveals tighter social regulation of immigration in patrilocal populations than in matrilocal populations. Proc. Natl. Acad. Sci. U S A. 2005;102(21):7476–7480. doi: 10.1073/pnas.0409253102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Arbiza L, Patricio M, Dopazo H, Posada D. Genome-wide heterogeneity of nucleotide substitution model fit. Genome Biol. Evol. 2011;3:896–908. doi: 10.1093/gbe/evr080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Arenas M, Posada D. Simulation of Genome-wide Evolution under Heterogeneous Substitution models and Complex Multispecies Coalescent Histories. Mol. Biol. Evol. 2014;31(5):1295–1301. doi: 10.1093/molbev/msu078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Liberles DA, Teufel AI, Liu L, Stadler T. On the need for mechanistic models in computational genomics and metagenomics. Genome Biol. Evol. 2013;5(10):2008–2018. doi: 10.1093/gbe/evt151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Hughes AL, Friedman R, Glenn N L. The Future of Data Analysis in Evolutionary Genomics. Curr. Genomics. 2006;7(4):227–234. [Google Scholar]
- 92.Zhang J. Performance of likelihood ratio tests of evolutionary hypotheses under inadequate substitution models. Mol. Biol. Evol. 1999;16(6):868–875. doi: 10.1093/oxfordjournals.molbev.a026171. [DOI] [PubMed] [Google Scholar]
- 93.Penny D, McComish BJ, Charleston MA, Hendy M D. Mathematical elegance with biochemical realism: the covarion model of molecular evolution. J. Mol. Evol. 2001;53(6):711–723. doi: 10.1007/s002390010258. [DOI] [PubMed] [Google Scholar]
- 94.Katsura Y, Iwase M, Satta Y. Evolution of Genomic Structures on Mammalian Sex Chromosomes. Curr. Genomics. 2012;13(2):115–123. doi: 10.2174/138920212799860625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Drummond AJ, Nicholls GK, Rodrigo AG, Solomon W. Estimating mutation parameters population history and genealogy simultaneously from temporally spaced sequence data. Genetics. 2002;161(3):1307–1320. doi: 10.1093/genetics/161.3.1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Rendine S, Piazza A, Cavalli-Sforza L L. Simulation and Separation by Principal Components of Multiple Demic Expansions in Europe. AM. Nat. 1986;128:681–706. [Google Scholar]
- 97.Liu H, Prugnolle F, Manica A, Balloux F. A geographically explicit genetic model of worldwide human-settlement history. AM. J. Hum. Genet. 2006;79(2):230–237. doi: 10.1086/505436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Ray N, Currat M, Berthier P, Excoffier L. Recovering the geographic origin of early modern humans by realistic and spatially explicit simulations. Genome Res. 2005;15(8):1161–1167. doi: 10.1101/gr.3708505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Tremblay M, Vezina H. New estimates of intergenerational time intervals for the calculation of age and origins of mutations. AM. J. Hum. Genet. 2000;66(2):651–658. doi: 10.1086/302770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Cavalli-Sforza LL, Menozzi P, Piazza A, editors. Princeton New Jersey 1994.: The history and geography of human genesPrinceton University Press: . [Google Scholar]
- 101.Rosenberg NA, Pritchard JK, Weber JL, Cann HM, Kidd KK, Zhivotovsky LA, Feldman M W. Genetic structure of human populations. Science. 2002;298(5602):2381–2385. doi: 10.1126/science.1078311. [DOI] [PubMed] [Google Scholar]
- 102.Higham T, Compton T, Stringer C, Jacobi R, Shapiro B, Trinkaus E, Chandler B, Groning F, Collins C, Hillson S, O'Higgins P, FitzGerald C, Fagan M. The earliest evidence for anatomically modern humans in northwestern Europe. Nature. 2011;479(7374):521–524. doi: 10.1038/nature10484. [DOI] [PubMed] [Google Scholar]
- 103.Menozzi P, Piazza A, Cavalli-Sforza L. Synthetic maps of human gene frequencies in Europeans. Science. 1978;201(4358):786–792. doi: 10.1126/science.356262. [DOI] [PubMed] [Google Scholar]
- 104.Piazza A, Rendine S, Minch E, Menozzi P, Mountain J, Cavalli-Sforza L L. Genetics and the origin of European languages. Proc. Natl. Acad. Sci. U S A. 1995;92(13):5836–5840. doi: 10.1073/pnas.92.13.5836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Currat M, Excoffier L. The effect of the Neolithic expansion on European molecular diversity. Proc. Biol. Sci. 2005;272(1564):679–688. doi: 10.1098/rspb.2004.2999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.François O, Currat M, Ray N, Han E, Excoffier L, Novembre J. Principal component analysis under population genetic models of range expansion and admixture. Mol. Biol. Evol. 2010;27(6):1257–1268. doi: 10.1093/molbev/msq010. [DOI] [PubMed] [Google Scholar]
- 107.Hallatschek O, Hersen P, Ramanathan S, Nelson D R. Genetic drift at expanding frontiers promotes gene segregation. Proc. Natl. Acad. Sci. U S A. 2007;104(50):19926–19930. doi: 10.1073/pnas.0710150104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Straus L G. Southwestern Europe at the Last Glacial Maximum. Curr. Anthropol. 1991;32:189–199. [Google Scholar]
- 109.Currat M, Excoffier L. Modern Humans Did Not Admix with Neanderthals during Their Range Expansion into Europe. PLoS Biol. 2004;2(12):e421. doi: 10.1371/journal.pbio.0020421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Currat M, Ruedi M, Petit RJ, Excoffier L. The hidden side of invasions: massive introgression by local genes. Evolution. 2008;62(8):1908–1920. doi: 10.1111/j.1558-5646.2008.00413.x. [DOI] [PubMed] [Google Scholar]
- 111.Currat M, Excoffier L. Strong reproductive isolation between humans and Neanderthals inferred from observed patterns of introgression. Proc. Natl. Acad. Sci. U S A. 2011;108(37):15129–15134. doi: 10.1073/pnas.1107450108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Akaike H. A new look at the statistical model identification. IEEE Trans. Aut. Control. 1974;19(6):716–723. [Google Scholar]
- 113.Bentley RA, Layton RH, Tehrani J. Kinship marriage and the genetics of past human dispersals. Hum. Biol. 2009;81(2-3):159–179. doi: 10.3378/027.081.0304. [DOI] [PubMed] [Google Scholar]
- 114.Cavalli-Sforza LL, Minch E. Paleolithic and Neolithic lineages in the European mitochondrial gene pool. AM. J. Hum. Genet. 1997;61(1):247–254. doi: 10.1016/S0002-9297(07)64303-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Oota H, Settheetham-Ishida W, Tiwawech D, Ishida T, Stoneking M. Human mtDNA and Y-chromosome variation is correlated with matrilocal versus patrilocal residence. Nat. Genet. 2001;29(1):20–21. doi: 10.1038/ng711. [DOI] [PubMed] [Google Scholar]
- 116.Rosser ZH, Zerjal T, Hurles ME, Adojaan M, Alavantic D, Amorim A, Amos W, Armenteros M, Arroyo E, Barbujani G, Beckman G, Beckman L, Bertranpetit J, Bosch E, Bradley DG, Brede G, Cooper G, Corte-Real HB, de Knijff P, Decorte R, Dubrova YE, Evgrafov O, Gilissen A, Glisic S, Golge M, Hill E.W, Jeziorowska A., Kalaydjieva L., Kayser M., Kivisild T., Kravchenko S.A., Krumina A., Kucinskas V., Lavinha J., Livshits L.A., Malaspina P., Maria S., McElreavey K., Meitinger T.A., Mikelsaar A.V., Mitchell R.J., Nafa K., Nicholson J., Norby S., Pandya A., Parik J., Patsalis P.C., Pereira L., Peterlin B., Pielberg G., Prata M.J., Previdere C., Roewer L., Rootsi S., Rubinsztein D.C., Saillard J., Santos FR, Stefanescu G, Sykes BC, Tolun A, Villems R, Tyler-Smith C, Jobling MA. Y-chromosomal diversity in Europe is clinal and influenced primarily by geography rather than by language. AM. J. Hum. Genet. 2000;67(6):1526–1543. doi: 10.1086/316890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Wegmann D, Currat M, Excoffier L. Molecular diversity after a range expansion in heterogeneous environments. Genetics. 2006;174(4):2009–2020. doi: 10.1534/genetics.106.062851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Schierup MH, Hein J. Recombination and the molecular clock. Mol. Biol. Evol. 2000;17(10):1578–1579. doi: 10.1093/oxfordjournals.molbev.a026256. [DOI] [PubMed] [Google Scholar]
- 119.Arenas M, Posada D. The effect of recombination on the reconstruction of ancestral sequences. Genetics. 2010;184(4):1133–1139. doi: 10.1534/genetics.109.113423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Arenas M, Posada D. Computational Design of Centralized HIV-1 Genes. Curr. HIV Res. 2010;8(8):613–621. doi: 10.2174/157016210794088263. [DOI] [PubMed] [Google Scholar]
- 121.Perez-Losada M, Jobes DV, Sinangil F, Crandall KA, Arenas M, Posada D, Berman P W. Phylodynamics of HIV-1 from a phase III AIDS vaccine trial in Bangkok Thailand. PLoS One. 2011;6(3):e16902. doi: 10.1371/journal.pone.0016902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Arenas M, Dos Santos HG, Posada D, Bastolla U. Protein evolution along phylogenetic histories under structurally constrained substitution models. Bioinformatics. 2013;29(23):3020–3028. doi: 10.1093/bioinformatics/btt530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Arenas M, Villaverde MC, Sussman F. Prediction and analysis of binding affinities for chemically diverse HIV-1 PR inhibitors by the modified SAFE_p approach. J. Comput. Chem. 2009;30(8):1229–1240. doi: 10.1002/jcc.21147. [DOI] [PubMed] [Google Scholar]
- 124.Grahnen JA, Liberles D A. CASS: Protein sequence simulation with explicit genotype-phenotype mapping. Trends in Evolutionary Biology. 2012;4:1. [Google Scholar]
- 125.Arenas M, Patricio M, Posada D, Valiente G. Characterization of phylogenetic networks with NetTest. BMC Bioinformatics. 2010;11(1):268. doi: 10.1186/1471-2105-11-268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Javed A, Mele M, Pybus M, Zalloua P, Haber M, Comas D, Netea MG, Balanovsky O, Balanovska E, Jin L, Yang Y, Arunkumar G, Pitchappan R, Bertranpetit J, Calafell F, Parida L. Recombination networks as genetic markers in a human variation study of the Old World. Hum. Genet. 2012;131(4):601–613. doi: 10.1007/s00439-011-1104-8. [DOI] [PubMed] [Google Scholar]
- 127.Novembre J, Galvani AP, Slatkin M. The geographic spread of the CCR5 Delta32 HIV-resistance allele. PLoS Biol. 2005;3(11):e339. doi: 10.1371/journal.pbio.0030339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Bamshad M, Wooding S P. Signatures of natural selection in the human genome. Nat. Rev. Genet. 2003;4(2):99–111. doi: 10.1038/nrg999. [DOI] [PubMed] [Google Scholar]
- 129.Sabeti PC, Schaffner SF, Fry B, Lohmueller J, Varilly P, Shamovsky O, Palma A, Mikkelsen TS, Altshuler D, Lander E S. Positive natural selection in the human lineage. Science. 2006;312(5780):1614–1620. doi: 10.1126/science.1124309. [DOI] [PubMed] [Google Scholar]
- 130.Landguth EL, Cushman S A. cdpop: A spatially explicit cost distance population genetics program. Mol. Ecol. Resour. 2010;10(1):156–161. doi: 10.1111/j.1755-0998.2009.02719.x. [DOI] [PubMed] [Google Scholar]
- 131.Landguth EL, Cushman SA, Johnson N A. Simulating natural selection in landscape genetics. Mol. Ecol. Resour. 2012;12(2):363–368. doi: 10.1111/j.1755-0998.2011.03075.x. [DOI] [PubMed] [Google Scholar]
- 132.Jones MR, Forester BR, Teufel AI, Adams RV, Anstett DN, Goodrich BA, Landguth EL, Joost S, Manel S. Integrating landscape genomics and spatially explicit approaches to detect loci under selection in clinal populations. Evolution. 2013;67(12):3455–3468. doi: 10.1111/evo.12237. [DOI] [PubMed] [Google Scholar]
- 133.Metzker M L. Sequencing technologies - the next generation. Nat. Rev. Genet. 2010;11(1):31–46. doi: 10.1038/nrg2626. [DOI] [PubMed] [Google Scholar]
- 134.Rasmussen M, Li Y, Lindgreen S, Pedersen JS, Albrechtsen A, Moltke I, Metspalu M, Metspalu E, Kivisild T, Gupta R, Bertalan M, Nielsen K, Gilbert MT, Wang Y, Raghavan M, Campos PF, Kamp HM, Wilson AS, Gledhill A, Tridico S, Bunce M, Lorenzen E.D, Binladen J., Guo X., Zhao J., Zhang X., Zhang H., Li Z., Chen M., Orlando L., Kristiansen K., Bak M., Tommerup N., Bendixen C., Pierre T.L., Gronnow B., Meldgaard M., Andreasen C., Fedorova S.A., Osipova L.P., Higham T.F., Ramsey C.B., Hansen T.V., Nielsen FC, Crawford MH, Brunak S, Sicheritz-Ponten T, Villems R, Nielsen R, Krogh A, Wang J, Willerslev E. Ancient human genome sequence of an extinct Palaeo-Eskimo. Nature. 2010;463(7282):757–762. doi: 10.1038/nature08835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Pedersen JS, Valen E, Vargas Velazquez AM, Parker BJ, Rasmussen M, Lindgreen S, Lilje B, Tobin DJ, Kelly TK, Vang S, Andersson R, Jones PA, Hoover CA, Tikhonov A, Prokhortchouk E, Rubin EM, Sandelin A, Gilbert MT, Krogh A, Willerslev E, Orlando L. Genome-wide nucleosome map and cytosine methylation levels of an ancient human genome. Genome Res. 2014;24(3):454–466. doi: 10.1101/gr.163592.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Beaumont MA, Rannala B. The Bayesian revolution in genetics. Nat Rev Genet. 2004;5(4):251–261. doi: 10.1038/nrg1318. [DOI] [PubMed] [Google Scholar]
- 137.Li S, Jakobsson M. Estimating demographic parameters from large-scale population genomic data using Approximate Bayesian Computation. BMC Genet. 2012;13:22. doi: 10.1186/1471-2156-13-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Sohn KA, Ghahramani Z, Xing EP. Robust estimation of local genetic ancestry in admixed populations using a nonparametric Bayesian approach. Genetics. 2012;191(4):1295–1308. doi: 10.1534/genetics.112.140228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Csillery K, Blum M G B, Gaggiotti OE, Francois O. Approximate Bayesian Computation (ABC) in practice. Trends Ecol. Evol. 2010;25(7):410–418. doi: 10.1016/j.tree.2010.04.001. [DOI] [PubMed] [Google Scholar]
- 140.Lorenzen ED, Nogues-Bravo D, Orlando L, Weinstock J, Binladen J, Marske KA, Ugan A, Borregaard MK, Gilbert MT, Nielsen R, Ho SY, Goebel T, Graf KE, Byers D, Stenderup JT, Rasmussen M, Campos PF, Leonard JA, Koepfli KP, Froese D, Zazula G, Stafford T W , Jr , Aaris-Sorensen K, Batra P, Haywood AM, Singarayer JS, Valdes PJ, Boeskorov G, Burns JA, Davydov SP, Haile J, Jenkins DL, Kosintsev P, Kuznetsova T, Lai X, Martin LD, McDonald HG, Mol D, Meldgaard M, Munch K, Stephan E, Sablin M, Sommer RS, Sipko T, Scott E, Suchard MA, Tikhonov A, Willerslev R, Wayne RK, Cooper A, Hofreiter M, Sher A, Shapiro B, Rahbek C, Willerslev E. Species-specific responses of Late Quaternary megafauna to climate and humans. Nature. 2011;479(7373):359–364. doi: 10.1038/nature10574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Tsutaya T, Yoneda M. Quantitative reconstruction of weaning ages in archaeological human populations using bone collagen nitrogen isotope ratios and approximate Bayesian computation. PLoS One. 2013;8(8):e72327. doi: 10.1371/journal.pone.0072327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Veeramah KR, Wegmann D, Woerner A, Mendez FL, Watkins JC, Destro-Bisol G, Soodyall H, Louie L, Hammer M F. An early divergence of KhoeSan ancestors from those of other modern humans is supported by an ABC-based analysis of autosomal resequencing data. Mol. Biol. Evol. 2012;29(2):617–630. doi: 10.1093/molbev/msr212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Hallatschek O, Nelson D R. Gene surfing in expanding populations. Theor. Popul. Biol. 2008;73(1):158–170. doi: 10.1016/j.tpb.2007.08.008. [DOI] [PubMed] [Google Scholar]
- 144.Excoffier L, Ray N. Surfing during population expansions promotes genetic revolutions and structuration. Trends Ecol. Evol. 2008;23(7):347–351. doi: 10.1016/j.tree.2008.04.004. [DOI] [PubMed] [Google Scholar]
- 145.Klopfstein S, Currat M, Excoffier L. The fate of mutations surfing on the wave of a range expansion. Mol. Biol. Evol. 2006;23(3):482–490. doi: 10.1093/molbev/msj057. [DOI] [PubMed] [Google Scholar]
- 146.Hewitt G M. Some genetic consequences of ice ages and their role in divergence and speciation. Biol. J. Linn. Soc. 1996;58(3):247–276. [Google Scholar]
- 147.Lister A M. The impact of Quaternary Ice Ages on mammalian evolution. Philos. Tans.R. Soc. Lond. B Biol. Sci. . 2004;359(1442):221–241. doi: 10.1098/rstb.2003.1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Jakob SS, Ihlow A, Blattner F R. Combined ecological niche modelling and molecular phylogeography revealed the evolutionary history of Hordeum marinum (Poaceae)--niche differentiation loss of genetic diversity and speciation in Mediterranean Quaternary refugia. Mol. Ecol. 2007;16(8):1713–1727. doi: 10.1111/j.1365-294X.2007.03228.x. [DOI] [PubMed] [Google Scholar]
- 149.Miller MP, Bellinger MR, Forsman ED, Haig S M. Effects of historical climate change habitat connectivity and vicariance on genetic structure and diversity across the range of the red tree vole (Phenacomys longicaudus) in the Pacific Northwestern United States. Mol. Ecol. 2006;15(1):145–159. doi: 10.1111/j.1365-294X.2005.02765.x. [DOI] [PubMed] [Google Scholar]
- 150.Strand AE, Niehaus J M. KERNELPOP a spatially explicit population genetic simulation engine. Mol. Ecol. Notes. 2007;7:969–973. [Google Scholar]
- 151.Leblois R, Estoup A, Rousset F. IBDSim: a computer program to simulate genotypic data under isolation by distance. Mol. Ecol. Resour. 2009;9(1):107–109. doi: 10.1111/j.1755-0998.2008.02417.x. [DOI] [PubMed] [Google Scholar]
- 152.Jukes TH, Cantor C R, editors. Mammalian Protein Metabolism; New York NY.: Academic Press: ; 1969. Evolution of protein molecules; pp. 21–132. [Google Scholar]
- 153.Kimura M. A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 1980;16(2):111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- 154.Kimura M, Crow JF. The Number of Alleles That Can Be Maintained in a Finite Population. Genetics. 1964;49(4):725–738. doi: 10.1093/genetics/49.4.725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Ota T, Kimura M. A model of mutation appropriate to estimate the number of electrophoretically detectable alleles in a finite population. Genet. Res. 1973;22(2):201–204. doi: 10.1017/s0016672300012994. [DOI] [PubMed] [Google Scholar]
- 156.Crow JF, Kimura M, editors. New York: : Harper & Row 1970.; An Introduction to Population Genetics Theory. [Google Scholar]
- 157.Estoup A, Jarne P, Cornuet J M. Homoplasy and mutation model at microsatellite loci and their consequences for population genetics analysis. Mol. Ecol. 2002;11(9):1591–1604. doi: 10.1046/j.1365-294x.2002.01576.x. [DOI] [PubMed] [Google Scholar]