Abstract
Understanding the Maxam-Gilbert and Sanger sequencing as the first generation, in recent years there has been an explosion of newly-developed sequencing strategies, which are usually referred to as next generation sequencing (NGS) techniques. NGS techniques have high-throughputs and produce thousands or even millions of sequences at the same time. These sequences allow for the accurate identification of microbial taxa, including uncultivable organisms and those present in small numbers. In specific applications, NGS provides a complete inventory of all microbial operons and genes present or being expressed under different study conditions. NGS techniques are revolutionizing the field of microbial ecology and have recently been used to examine several food ecosystems. After a short introduction to the most common NGS systems and platforms, this review addresses how NGS techniques have been employed in the study of food microbiota and food fermentations, and discusses their limits and perspectives. The most important findings are reviewed, including those made in the study of the microbiota of milk, fermented dairy products, and plant-, meat- and fish-derived fermented foods. The knowledge that can be gained on microbial diversity, population structure and population dynamics via the use of these technologies could be vital in improving the monitoring and manipulation of foods and fermented food products. They should also improve their safety.
Keywords: Next generation sequencing, NGS techniques, Pyrosequencing, Illumina, Food microbiology, Molecular microbial ecology, Food ecology.
1. GENERAL INTRODUCTION
Foods harbor complex microbial communities composed of viruses, bacteria and fungi. Some of these microorganisms are of technological importance and invest foods with desirable sensorial (organoleptic and rheological) characteristics [1]. However, undesirable microorganisms may also be present; these can reduce the quality of foods (spoilage microorganisms) or even negatively affect their safety (pathogens) [2].
The traditional way of determining the composition of food-associated microbiotas has been through culturing methods. These relay on the isolation and cultivation of microorganisms before their identification and typing. However, it has been repeatedly shown that culturing is unreliable for the complete microbial characterization of many ecosystems, including those of foods [3]. The selective isolation of microorganisms may require unknown growth factors and/or growth conditions present in natural habitats that are not reproduced by laboratory media. Foods may also have a low pH, a reduced aw, or have to be kept under harsh storageconditions, etc., which might leave microbes in a physiologically viable but not cultivable state. Moreover, microbes present in low numbers can be outcompeted by numerically more abundant species, impeding their detection in culture. Such limitations lead these techniques to underestimate microbial diversity, and sometimes even the failure to detect the majority microbial groups [4]. Over recent decades, a great number of culture-independent, molecular methods have been developed that help overcome these problems; most of which have been used extensively in food systems (for reviews see [5, 6]). These techniques allow the identification and, in some cases, quantification of food-associated microbial groups, and provide sensitive and rapid methods for determining the composition and diversity of complex microbial communities.
Culture-independent techniques are mostly based on the analysis of microbial nucleic acid sequences (DNA and/or RNA). Most rely on the amplification of these nucleic acids by the polymerase chain reaction (PCR) technique. The majority of amplification techniques target the rRNA genes (rDNA). Comparing the sequences obtained to one another, and to those held in databases, allows phylogenetic relationships between microbes to be established. Culture-independent, PCR-based techniques include denaturing gradient gel electrophoresis (DGGE), temporal temperature gradient gel electrophoresis (TTGE), single stranded conformation polymorphism (SSCP), real-time quantitative PCR (qPCR), the construction and analysis of 16S rRNA gene libraries, terminal restriction fragment length polymorphism (TRFLP), and a few others [7-16]. Of these, DGGE (qualitative/semiquantitative analysis) and qPCR (quantitative analysis) have been widely used to microbiologically characterize food environments and to analyze the course of food fermentations [reviewed in 2, 6]. Changes in microbial populations furnish useful information regarding the dynamics of food fermentation, the monitoring of the growth of starter and adjunct cultures, and the fate of pathogen and spoiling microorganisms. A number of new microbial types with no cultured relatives –and occasionally of potential technological interest– have also been detected using these techniques.
NGS techniques have promoted the emergence of new, high-throughput technologies, such as genomics, metagenomics, transcriptomics and metatranscriptomics, etc. As compared to previous culture-independent methods, the number of nucleic acid sequences analyzed by NGS techniques is exceedingly higher, allowing a deeper description of the microbial constituents of the ecosystems. These technologies can be used in two substantially different ways: sequencing the total microbial nucleic acids (shotgun sequencing) and gene-specific sequencing (targeted sequencing). For the latter, segments of highly conserved DNA or cDNA sequences are first amplified by PCR using universal or group-specific primers. Shotgun sequencing returns information far beyond that regarding phylogenetic composition, providing insights into the number and potential function of genes within the community [17, 18]. Both shotgun and targeted techniques have already been used to study the microbiology of a series of foods and food fermentations, and pertinent reviews have recently been compiled [18-21]. However, research in this area is so active that findings must be continually reviewed, and the current and potential applications of these constantly updated.
2. NGS PLATFORMS
NGS platforms involve many different technologies [22] all of which generate large, genome-scale datasets. However, they differ substantially in terms of their engineering, sequencing chemistry, output (length of reads, number of sequences), accuracy and cost. Current commercial platforms include the 454 (Roche), Illumina (Illumina), SOLiD and Ion Torrent (Life Technologies), and PacBio (Pacific Biosciences) systems. Comparisons of their advantages and disadvantages have recently been published [23, 24]. Many other ‘third-generation’ techniques, such as DNA nanoball sequencing, heliscope single molecule sequencing, nanopore DNA sequencing, tunneling current DNA sequencing, sequencing with mass spectrometry, and microscopy-based techniques, are currently under development [25].
2.1. The Roche 454 Pyrosequencer
The NGS era started with the release of the first array-based 454 pyrosequencing system by Roche in 2004 [26]. In this system, double stranded DNA fragments are ligated to specific adaptors, diluted, and immobilized on tiny beads (one molecule per bead). The attached DNA is copied millions of times by PCR, with one of the primers biotinylated, in an oil-water emulsion. The beads are then housed in separate picotiter-size wells, in which the non-biotinylated strand is denatured and washed away. The plate array consists of approximately one million wells, and independent sequencing reactions occur within each. After the addition of the sequencing primer, the pyrosequencing reaction proceeds via DNA synthesis using single stranded DNA molecules as templates [27]. The sequencer runs the A, T, C and G nucleotide building blocks – one type at a time - over the wells. When a nucleotide is incorporated, pyrophosphate is released and converted into ATP, which fuels a coupled luciferin-luciferease reaction producing light (the intensity proportional to the number of incorporated nucleotides). Light is recorded by a camera, and the unincorporated nucleotides degraded enzymatically. At present, 454 pyrosequencers offer reads longer than 1000 bp, with a high sequence output (1 megabase (Mb) in the case of the 454 GS FLX+ system). Difficulties in sequencing homopolymer stretches and the rather high cost per Mb are the main weaknesses of the 454 platform.
2.2. The Illumina Platform
The technology used by this platform was first released by Solexa in 2006, a company later purchased by Illumina. In this method, primers with specific adaptors complementary to those on the DNA are attached to a slide. Individual DNA molecules are locally amplified by PCR so that clonal DNA clusters remain attached to the primers on the slide forming bridge-like structures. Sequencing proceeds by synthesis, using primers complementary to adaptors and four types of reversibly-blocked terminator nucleotides (RT-nucleotides). Non-incorporated RT-nucleotides are washed away. The dye, along with the terminal 3’ blocker, is then chemically removed from the DNA, allowing for incorporation and detection, and for the next sequencing cycle to begin [28]. Unlike in pyrosequencing, DNA chains are extended one nucleotide at a time. The method only uses DNA polymerase as opposed to multiple, expensive enzymes required by other techniques. Additionally, image acquisition can be delayed from nucleotide incorporation, allowing for very large arrays of DNA clusters to be captured by sequential images taken by a single camera. Currently, the Illumina platform generates typical reads of 150-300 bp that can be increased to 300-600 bp via “paired-end” sequencing (i.e., sequencing both ends of the same DNA cluster). The reduced price for the high output of sequences per run (up to 3000 Mb for the HiSeq) is the greatest advantage of this technology.
2.3. The SOLiD System
The first SOLiD (Sequencing by Oligonucleotide Ligation and Detection) sequencer was released in 2007 by Life Technologies. True to its name, and unlike any other technology, the SOLiD technique carries out sequencing by ligation instead of synthesis [29]. DNA fragments are ligated to a universal adapter, and then attached to a magnetic bead. After emulsion PCR, the resulting amplicons attached to the beads are bound covalently to a glass slide. A set of four fluorescently-labeled di-nucleotide probes compete for ligation to the sequencing primer (complementary to the adaptor). Following a series of cycles of probe ligation, dye cleavage and detection, the extension product is removed and the template reset with a primer complementary to the n-1 position for a second round of ligation. Sequencing by ligation is very accurate (99.99%) since ligases make fewer mistakes than polymerases. However, the maximum-length reads obtained with SOLiD measure just 75 bp, insufficient for most metagenomic studies. This also makes the assembly of the sequences a very challenging task. SOLiD has uses in whole genome resequencing, targeted sequencing, and transcriptome research.
2.4. Ion Torrent
The Ion Torrent Company (now acquired by Life Technologies) released its first sequencer based on semiconductor technology in 2010. As in other methods, sequencing is performed via the synthesis of a complementary strand, but detection is based on the hydrogen ions released during DNA polymerization; the number of releases is proportional to the number of incorporated nucleotides [30]. This sequencing technology differs from others in that no modified nucleotides or optics are used. After library construction and emulsion PCR, microwells containing single template pools of the DNA to be sequenced are flooded in turn with single nucleotide species. The Ion Torrent sequencer can generate up to 4 Mb in 7 h at a moderate cost per Mb. The length of the reads has risen in recent years up to 400 bp. This technology is less accurate (98%) than others, but is useful in small studies, such as the sequencing of microbial genomes and targeted metagenomics.
2.5. PacBio SMRT
The PacBio platform, marketed in 2010 by Pacific Biosciences, uses single molecule real-time (SMRT) sequencing technology. As in other methods, sequencing is based on synthesis, using nucleotides labeled with distinct fluorescent dyes, but sequencing proceeds without the need for prior PCR amplification. The single stranded DNA molecules to be sequenced are deposited in zeptoliter wells, where a single polymerase molecule is immobilized [31]. The wells are constructed in such a way that only the fluorescence occurring at the bottom of the well is detected. The fluorescent label is detached from the nucleotide during its incorporation into the DNA strand. Theoretically, large DNA molecules (up to 30 kb) can be sequenced by this technology. The latest protocols have improved the average read length to over 7 kb. However, the low output obtained with this platform (0.05 Mb per run) is a major shortcoming, rendering it unsuitable for large sequencing projects. Nevertheless, its high accuracy can be exploited for sequencing difficult DNA regions and for genome scaffolding in metagenomic studies.
3. EXAMINING THE MICROBIAL ECOLOGY OF FOODS BY NGS
Attempts to characterize whole community profiles of traditional fermented foods were first performed using rDNA amplification, cloning, sequencing and sequence analysis [12, 32-34]. These methods are now being replaced by high-throughput NGS techniques. Targeted techniques provide a snapshot of the diversity and phylogeny of the different elements making up microbial populations. The term phylobiome has been introduced recently to refer to the phylogenetic information gathered using this approach [35]. In addition, shotgun techniques inform on the genetics and functional capabilities of the microbial constituents of food ecosystems. NGS is not only contributing towards the study of food microbial ecology but also to the genome sequencing of food-borne microorganisms. Food microbial genomics has already established pivotal connections between phenotype, genotype and environmental adaptations. However, this topic falls beyond the subject of this review.
3.1. Workflow, Pitfalls and Care
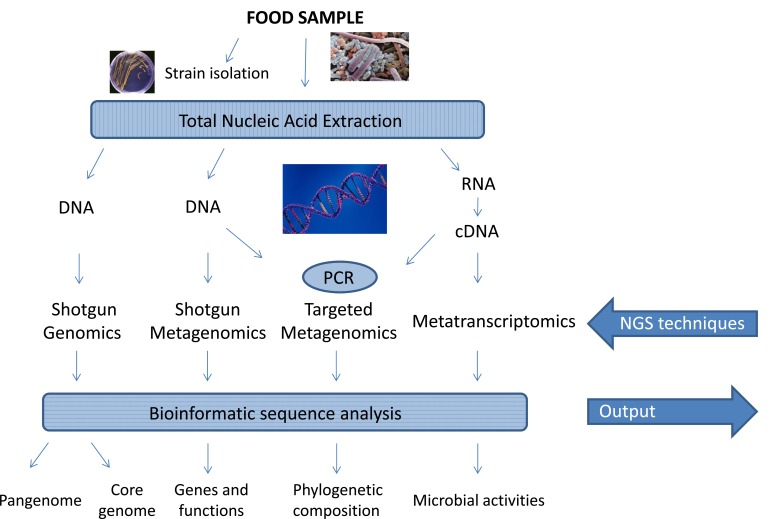
The NGS analysis of nucleic acids from food samples requires a series of steps, each of which must be optimized: nucleic acid extraction and purification, library construction, sequencing and analysis of the raw sequences using computer software, and the searching of different databases (Fig. 1). These steps are discussed below. NGS techniques have further witnessed the appearance and spreading of a myriad of molecular microbiological terms (Table 1), to which food microbiologists were unfamiliar until recently.
Fig. (1).
Next-Generation Sequencing (NGS) applications in Food Microbiology and general flow chart.
Table 1.
Usual terms utilized in most NGS techniques.
| Term | Definition |
|---|---|
| α-diversity | Measures the diversity associated with a single sample (e.g. OTU number, Shannon Index, rarefaction curve, etc.) |
| Assembly | Partial reconstruction of genes or genomes by aligning and merging short sequencing reads |
| Barcodes | Short nucleotides sequences merged with primers and/or adaptors allowing simultaneous sequencing of DNA from multiple samples and further separation in silico |
| β-diversity | Measures the diversity among samples (e.g. heatmaps, venn diagrams, similarity trees, PCAs, ordinations etc.) |
| Binning | Separation of all fragments originating from a common taxon or OTU |
| Contig | Set of small overlapping DNA segments representing a consensus region of DNA |
| Coverage | Means how deep was the sequencing effort in sampling a given community; number of times a nucleotide is read during sequencing |
| Denoising | Quality processing applied to 16S rDNA reads, correcting the “noise” (errors) artificially generated during sequencing |
| Diversity estimators | e.i. Shannon – Estimate the diversity, taking into account the number of species and how even they are distributed |
| OTU | Operational taxonomic unit. A cluster of sequences within a given similarity cut-off (e.g. 3%, which is usually utilized to define bacterial species by 16S rDNA) |
| Sequencing trimming | Processing of sequencing reads, which includes the removal of primers and barcodes, deletion of a given sequence region and elimination of low quality and very short reads |
| Phylogenetic assignment | Assignment of each sequence or OTU to its known closest relative organism |
| Rarefaction curve | Curve representing the richness of the sample according to the number of sequences. The shape of the curve reflects the sample diversity |
| Richness estimators | OTU, Ace, Chao1 - Estimate the number of species (or genus, orders etc.) in a given sample by different methods |
3.1.1. Nucleic Acid Extraction and Purification
In microbial studies, the handling and storage of food samples is critical. Rapid processing is always recommendable, and all preparations and conditions that might alter the original proportion of microbial cells should be avoided. Food systems are almost always heterogeneous. Thus, strategies must be carefully designed to ensure that samples are representative of the different conditions found in the food, and that sufficient homogenization is obtained. Incomplete cell lysis, and therefore inadequate DNA extraction, can also provide unrealistic views of a food microbial community. This can be avoided by performing supplementary lysis steps to improve extraction efficiency, e.g., via the combination of enzymatic and mechanical treatments [36]. Further, some food components, such as lipids and proteins, may inhibit PCR amplification. These compounds must be removed during nucleic acid extraction [37]. Extracting the same sample multiple times and making a final pool can further minimize the bias associated with single extractions.
Quality control and nucleic acid quantification are vital in any sample preparation method. Accurate quantification is preferentially achieved via the use of fluorometers [36, 38]. The amount and quality of DNA required from a food sample for total DNA sequencing (shotgun metagenomics) is usually higher than that needed for gene-specific sequencing (targeted metagenomics).
In foods, it is of great importance to distinguish between viable, active and inactive cells. Dead cells may contribute to the DNA pool which will be further processed. Though persistence of RNA molecules in dead cells from foods has yet to be addressed adequately, the use of NGS techniques with cDNA, produced by reverse transcription from RNA, focuses on active microbial populations, thus avoiding these problems [39, 40]. In metatranscriptomic studies, interest focuses on the non-ribosomal fraction, which includes mRNA, micro-RNA, tRNA and other non-coding RNAs [36]. However, the majority of total RNA corresponds to rRNA, which may account for >90% of the reads obtained [41, 42]. Indeed, in RNA-based studies, sample preparation remains an active area of investigation; the recovery and enrichment of high quality mRNA, its short half-life, and the selective removal of rRNA, are all challenges that must be adequately met [35, 43, 44].
Alternatively, complex communities can be ‘enriched’ in viable cells via the use of propidium monoazide (PMA) or ethidium monoazide (EMA) prior to DNA extraction. These compounds enter cells with damaged membranes, intercalate into the double strand of DNA and form covalent linkages upon exposure to intense light, thus preventing subsequent PCR amplification [45]. The effectiveness of these azides has been tested in the microbial analysis of water samples [45].
3.1.2. Molecular Target
The target sequence should be long enough to contain nucleotide heterogeneity that distinguishes between organisms [46]. Hypervariable regions within the 16S rRNA gene are commonly chosen for the assessment of taxonomic bacterial and archaeal diversity [47-52]. Primers, however, should be designed in well-conserved, universal regions to cover most (if not all) of the microorganisms present in the sample [53-55]. This is a difficult task, but online resources devoted to primer design (such as Primer Prospector [56]) are already available. Primer design on 16S rRNA genes should also avoid homology between prokaryotic and eukaryotic sequences, or between prokaryotic and eukaryotic organelle sequences. Design is also important because the taxonomic information provided by different regions of the 16S rRNA gene varies widely. Moreover, targeting specific microbial groups may require the amplification of specific hypervariable regions [46]. For instance, Bokulich et al. [48] showed the V4 domain to be more appropriate than the V5 domain for wine ecology studies since it provided greater taxonomic resolution for lactic acid bacteria (LAB) communities. Furthermore, some closely related species cannot be truly differentiated by 16S rDNA sequences [57, 58]. The preferential amplification of sequences from certain microbial types (due to different gene lengths and/or GC contents) is always a danger [46, 58]. A further problem associated with the amplification of ribosomal genes is the operon copy number, which varies widely across taxa, distorting quantitative diversity estimates [59], as well as the intra-species 16S rRNA gene heterogeneity found in natural populations [60]. Single-copy target genes, such as rpoB recA, radA, rpoA, gyrB and others have therefore been proposed as alternatives [57, 59-62]. However, the absence of well-represented databases for comparison is an important limiting factor in their widespread use [18]. Finally, the use of state-of-the-art, high-fidelity polymerases is recommended to reduce error rates. This can also help prevent base-calling errors and chimera formation, common problems associated with PCR [63].
Several molecular target genes have already been used to track fungal communities. The most commonly used have been the 18S, 28S and the intergenic regions of rDNA [64]. Well established primers and databases are described in the literature [65]. However, they cannot be used to determine with confidence the phylogeny of fungi at the species level [64, 66]. Therefore, when this level of discrimination is required, primers targeting the ITS regions are more appropriate [66, 67]. However, these sorts of primer do not align with guarantee across genetically distant taxa [68].
3.1.3. Library Construction
In the context of NGS, the library is the entire number of fragments to be sequenced. The total number of sequences obtained is known as the coverage, and besides specificity is key variable that must be considered for the characterization of any ecosystem [46]. Library construction includes the joining of platform-specific adaptors and sample-specific identifiers to double stranded DNA fragments. A key difference in library construction for total nucleic acid sequencing (shotgun metagenomics) and gene-specific sequencing (targeted metagenomics) is the latter’s need for the prior PCR-amplification of conserved DNA or cDNA sequences. For targeted metagenomics, platform-specific adaptors are designed as part of the PCR primers, resulting in template molecules ready to be sequenced. The multiplexing of several samples is possible if short sample-specific DNA sequences known as barcodes, tags or multiplex-identifiers (MIDs) are used [69]. These are also designed as part of the PCR primers. Normalized mixtures of the amplicons should be made prior to sequencing, thus ensuring that each amplicon is adequately represented in the sequencing reaction [36, 70]. In shotgun sequencing, DNA amplification is not necessary (except when only tiny amounts of DNA are available) [71]; amplification of total environmental DNA is obtained via the use of Phi29 DNA polymerase [72]. Since no amplification is required, shotgun metagenomics avoids the inherent bias of PCR-based techniques. The preparation of the shotgun library commonly starts by fragmenting the DNA using mechanical force. An attractive alternative is digestion by restriction enzymes [36]. In either case, platform-specific adaptors are then ligated to the resulting DNA fragments. Barcodes can also be included as part of the adaptors, thus allowing multiplexing in shotgun sequencing projects.
3.1.4. Data Analysis
High- quality read length and abundance are key factors to achieve accurate taxonomy assignment and diversity assessment. In targeted metagenomics, reads are generally clustered into operational taxonomic units (OTUs) (Fig. 2) based on the similarities of the reads themselves (similarity-based approach), or by taxonomic assignment (composition approach method), i.e., classifying the reads at different taxonomic levels (phylum, class, order, family, or genus) based on the similarities of the sequences with those in a database. Both approaches are currently used and produce similar results [73]. Taxonomic assignment has the advantage of providing information about the relationship of the reads with known microbial groups; this helps in ecological and/or functional interpretations. Additionally, it allows for comparisons between different studies, even when sequences come from different regions of the marker gene (rDNA, for example). The ribosomal database (RDP) [74], Silva [75] and Greengenes projects [76] provide updated databases for the small (16S and 18S) and large (23S and 28S) subunits of rDNA sequences. Besides, the UNITE database focuses mainly on ITS sequences from eukaryotes [77]. In the absence of a consensus definition of genetic species, as a rule of thumb, rDNA sequences sharing a 95% of identity are considered to belong to the same genus, and sequences sharing 97% or higher identity are considered to belong to the same species. These percentages are considered for the full rRNA gene molecule; therefore, lower resolution could actually be achieved by analyzing shorter reads. When working with other phylogenetic markers (like rpoB, gyrG, recA, etc.) and functional genes (such as those coding for antibiotic resistance, biodegradation activities, etc.) others databases such as the FunGene (http://fungene.cme.msu.edu) should be used. Finally, web tools, such as RAST [78] and MG-RAST [79] provide databases designed to help in the interpretation of genomic and metagenomic data, respectively. All these databases have user-friendly web-based pipelines and are good options for users with little bioinformatic training. However, all analyses are pre-formatted, do not allow for customization, and provide poor quality controls. Moreover, a vast majority of microbial types from some ecosystems have never been characterized or classified into taxonomic groups. Thus, there is a lack of representative organisms at several taxonomic levels that limits any taxonomic assignment approach [80].
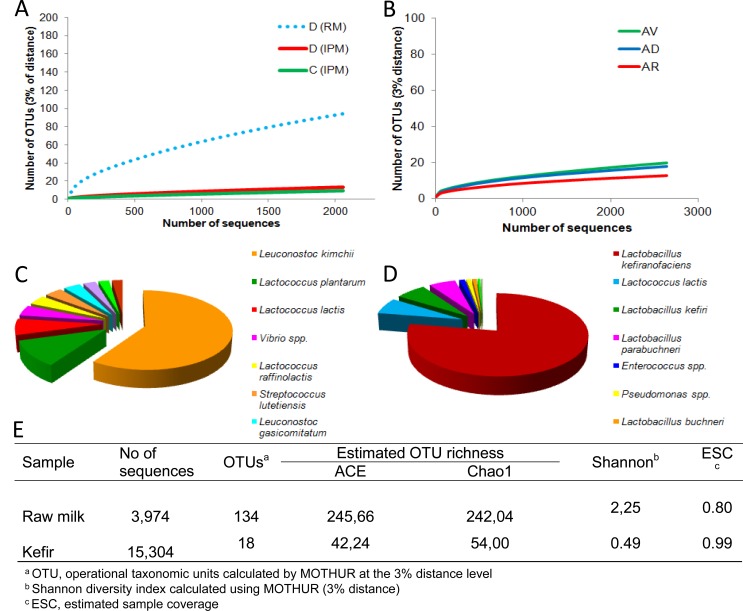
Fig. (2).
Rarefaction curves of raw milk (dotted blue line) and pasteurized milk samples incubated at 42ºC for 24 h (red and green lines (Panel A). Rarefaction curves of several samples of Brazilian kefir grains (Panel B). Schematic representation of the microbiota from raw milk (Panel C) and a kefir (Panel D) samples. Estimated OTU richness, sample coverage and diversity indexes are indicated on the table (Panel E) (data from Leite et al., 2012 and Delgado et al., 2013).
An alternative and very common way to analyze sequencing information is that based on the genetic similarity between reads. In this way, the diversity of the sample can be measured as the number of different OTUs; this is represented by the clusters constructed at a given level of sequence dissimilarity (normally at 1-3% for bacteria). This approach is advantageous when dealing with complex communities in which most of the microorganisms cannot be classified [80]. However, it provides no information about “who is there”, thus limiting the interpretation of the data. Some of the more recent bioinformatic tools that integrate the taxonomic-dependent and -independent approaches, offer several means of customizing analyses, and provide high quality data, which can easily be presented in comprehensive tables and didactic graphs (Fig. 2). The most popular software programs are Mothur [81] and QIMME [82], both of which are freely distributed and come with manuals and data analysis examples. These programs are periodically updated, improving analytical options as new concepts arise. Initial bioinformatic concerns were restricted to the verification of primer and barcode sequences, fragment length, and sequence quality. However, other control steps are now required, including noise removal [83], the detection of chimeric sequences, procedures for preclustering, and the exclusion of singletons [84-86]. Control steps facilitate downstream analysis and interpretation of the data, but they are usually platform-specific and need to be independently addressed [87].
Another promising method of NGS data analysis relies on the link between community phylogeny and functionality. Based on a large number of genome and metagenomic datasets available at the IMG database (https://img.jgi. doe.gov/cgi-bin/w/main.cgi), the PICRUSt software is able to infer the number of genes shared by different microbial taxa. This program provides information on the phenotypic relationships within a microbial community based solely on its phylogenetic composition [88].
3.2. Composition, Structure and Dynamics of Food-associated Microbial Populations
The pyrosequencing of tagged 16S rRNA gene amplicons of DNA and cDNA was first used to analyze the bacterial community structure and dynamics of ten samples of “ben-saalga” pear millet slurry (a traditional African fermented product) [89] (Table 2). This allowed 80.7% of the sequences to be attributed to a family and 70% to a genus, but did not permit identification of sequences at the species level. The amplification of large numbers of Archaea (4.8%) and chloroplast-related Eukarya (43%) sequences was rather surprising since bacteria-specific primers were used. Further, the number of sequences per sample varied widely (from 641 to 13,380), highlighting the under-representation of the diversity of some samples. With the exception of a few proteobacteria, almost all the bacterial sequences were attributed to cultivable bacteria. As expected, the sequences attributed to LAB genera were the most common (ben-saalga is made via lactic acid fermentation). Further, the results obtained from the analysis of cDNA revealed the same microbiome pattern, suggesting that the assignments made through the DNA analysis corresponded to living bacteria. Considerable diversity was found at the beginning of fermentation, whereas at the end only representatives of the Lactobacillaceae (Pediococcus and Lactobacillus), Leuconostocaceae and Enterococcaceae were found. Nevertheless, differences in community dynamics were seen between samples fermented spontaneously, those inoculated by backslopping (using a portion of a previous batch), and those inoculated with pure LAB cultures. Despite unspecific amplification and inaccurate identification, the authors concluded this NGS method to be very promising for the preliminary and/or rapid microbial characterization of food samples.
Table 2.
Summary of NGS projects analyzing food-associated microbiotas, including working conditions and major findings.
| Food sample/source | Sequencing project | Amplicon length (bp) | Sequencing platform | Taxonomic resolution | Database useda | Major findings | Reference |
|---|---|---|---|---|---|---|---|
| Material of plant origin | |||||||
| Table olives fermentation | V1-V3 16S rDNA and cDNA | >250 | 454FLX | Genus/ species |
NCIB nr | Agreement for the DNA and RNA data; high level of halophilic bacteria at the beginning of fermentation; Lactobacillus spp. at the end | [40] |
| Wine made from botrytized grapes | Separate V4 and V5 16S rDNA | >150 | Illumina | Family/ genus |
RDP | Similar community structure with V4 and V5 amplicons; Acetobacteriaceae and Proteobacteria dominant organisms | [48] |
| Ray and wheat sourdough fermentation | V1-V3 16S rDNA and cDNA | >300 | 454FLX | Genus/ species |
RDP | Grain-associated bacteria do not progress in dough, except for E. coli; Weissella, Lactobacillus and Leuconostoc dominate the fermentation | [51] |
| Fermentation of African pearl millets | V3 16S rDNA | >180 | 454 FLX | Genus | RDP | Weissella, Pediococcus and Lactobacillus were the only genera after 24 h of fermentation; high intersample variability | [89] |
| “Cheonggukjang” fermentation | V1-V2 16S rDNA | >300 | 454 FLX | Genus | SILVA | Bacillus spp. dominates the fermentation, but the actual majority species was different in different samples | [91] |
| Korean soybean pastes | V1-V2 16S rDNA | >300 | 454 FLX | Genus | SILVA | High diversity in different brands; Bacillus spp. dominant in a majority of products; LAB species occasionally dominant | [92] |
| “Kochujang” fermentation | V1-V2 16S rDNA | >300 | 454 FLX | Genus | SILVA | Bacillus spp. dominates the fermentation, but the actual majority species was different in different samples | [93] |
| Ten kinds of “kimchi” | V1-V3 16S rDNA | >300 | 454 FLX | Genus/ species |
ExTaxon database | Bacteria diversity and richness varied highly and depended on the type of “kimchi” | [94] |
| Fermented shrimp, kimchi and sauerkraut | V1-V3 16S rDNA, total DNA | >300 na |
454 FLX | Genus | RDP | Viral and hosts communities; discrepancy on phage hosts via homology comparison and rDAN sequencing | [94] |
| Fermentation of “kimchi” | Total DNA | na | 454 FLX | Species | RDP and MG-RAST | Leuconostoc, Lactobacillus and Weissella dominant organisms; Leuc. mesenteroides and Lb. sakei genomes highly represented; phage-related sequences | [95] |
| Winery-associated microbiota before, during and after harvest | V4 16S and ITS1 from rDNA of bacteria and fungi | >150 | Illumina | Family/ genus |
Greengenes and UNITE | Saccharomyces colonized winery surfaces; microbial communities were dependent on the production context at each site, shaped by technological practices, processing stage and season | [106] |
| Cocoa bean fermentation | Total DNA | na | 454 FLX | Species | RDP and NCBI nr | Complex fermentation including bacteria (Acetobacter pasteurianus and Lb. fermentum) and yeasts Hansenulla uvarum, Hansenulla opuntiae and Saccharomyces cerevisieae) | [111] |
| Fermented sushi (“narezushi”) | V1-V2 16S rDNA | >300 | 454 FLX | Genus | RDP | Species of Lactobacillus and Pediococcus always present; occasional presence of genera of LAB and other bacterial groups | [113] |
| Fermented rice brand mash (“nukadoko”) | V6-V8 16S rDNA | >400 | 454 FLX | Genus/ species |
RDP | Rice-associated bacteria are replace by Lb. namurensis, Lb. acetotolerans, and some other Lactobacillus during fermentation | [114] |
| American coolship ale beer fermentation | V4 16S rDNA | >150 | Illumina | Family/ genus |
Greengenes | Initial Enterobacteriaceae are overgrowth by LAB species though fermentation; Pediococcus spp. becomes dominant after few weeks | [115] |
| Korean rice beer fermentation | V1-V3 16S rDNA and ITS1 from bacteria and fungi | >350 | 454 FLX | Genus/ species |
Genus | Proteobacteria are replaced by LAB species through fermentation; amylolytic yeasts drive saccharification; alcohol-producing S. cerevisiae) dominate at the end of the fermentation | [116] |
| Fermentation of medieval sushi (“kaburazushi”) | V1-V2 16S rDNA | >300 | 454 FLX | Genus | NCIB nr | Bacillus and Staphylococcus are replaced by Lactobacillus through fermentation; Lb. sakei constitutes 80% of the microbiota. | [117] |
| Milk and dairy products | |||||||
| Raw milk cheeses | V3-V4 16S rDNA and cDNA | nr | 454 FLX | Genus | RDP | Fate of starters and inoculated pathogens in cheese. Listeria innocua and Stap. aureus do not progress in cheese but E. coli does | [39] |
| Brazilian kefir grains | V3 16S rDNA | >300 | 454 FLX | Genus/ species |
RDP | Dominant species Lb. kefiranofaciens, Lb. kefiri, Lb. parakefiri; Acetobacter spp. in only one sample | [49] |
| Pasteurized, cultured raw milk | V3 16S rDNA | >300 | 454 FLX | Genus/ species |
RDP | S. thermophilus dominant organism; Lb. delbrueckii, Lb. helveticus, and Enterococcus spp. subdominant components | [50] |
| Healthy and culture-negative mastitic milk | V1-V2 16S rDNA | >200 | 454 FLX | Genus | SILVA RDP |
Pseudomonas, Ralstonia, Psychrobacter reads higher in milk from healthy udders; uncultured bacteria as causative agents in some mastitis | [71] |
| Healthy and mastitic milk | V1-V2 16S rDNA | 250-500 | 454 FLX | Genus | RDP | DNA sequences from recognized pathogens, from pathogens not associated with mastitis and from bacteria not known to be pathogens | [96] |
| Irish kefir grain and associated kefir beverage | V4 16S rDNA | nr | 454 FLX | Genus/ species |
RPD | Lactobacillus spp., as majority organisms including Lb. kefiranofaciens, Lb. kefiri, Lb. parabuchneri, and Lb. helveticus. | [97] |
| Danish raw milk cheeses made with starter cultures | V3-V4 16S rDNA | >200 na |
454 FLX | Genus | RDP | Streptococcus, Lactococcus and Lactobacillus constitute the largest groups; minor discrepancies between pyrosequencing and DGGE | [98] |
| Oscypeck traditional PDO Polish cheese | V5-V6 16S rDNA | >150 | 454 FLX | Genus | SILVA | Lactococcus spp. as dominant organism; presence of Bifidobacterium spp. and Enhydrobacter spp. Reads | [99] |
| Milk, whey starters and Mozzarella cheese | V1-V3 16S rDNA | >200 | 454 FLX | Genus/ species |
RDP |
Acinetobacter spp. and Pseudomonas spp. dominant in milk; S. thermophilus, Lb. delbrueckii and Lb. helveticus in cheese. |
[102] |
| Latin-style cheeses | V1-V3 16S rDNA | >200 | 454 FLX | Genus/ species |
RDP | High bacterial diversity in different brands; presence of high numbers of Exiguobacteirum in one of the brands | [101] |
| Artisanal Irish cheeses and associated cheese rinds | V4 16S rDNA | >150 | 454 FLX | Genus | NCBI nr | Detection for the first time in cheese of Arthrobacter, Brachybacterium, Faecalibacterium, Helcococcus and Prevotella | [100] |
| Artisan American cheeses | V4 16S and ITS1 from rDNA and cDNA of bacteria and fungi | >150 | Illumina | Family/ genus |
Greengenes and UNITE | Debaryomyces and Lactococcus as dominant fungi and bacteria, respectively; similar microbial communities occupy the same type surfaces; facility-specific microorganisms | [103] |
| Subclinical mastitic milk | Total DNA | na | 454 FLX | Species | SEED | Presence of E. coli, Pseudomonas mendocina, Staphylococcus aureus and Klebsiella pneumoniae and their phages | [107] |
| Industrial starter for Gouda cheese | Genomes, total DNA | na | 454 FLX, Illumina | Strain | NCBI nr | Metabolic complementation of starter components, Lc. Lactis and Leuc. mesenteroides; starter stability by “kill-the-winner”-phage dynamics | [109] |
| Meat and fish products | |||||||
| “Jeotgal” (fermented fish and seafood products) | V3 16S rRNA, archaea, bacteria | >100 | 454 FLX | Family | Greengenes | High microbial diversity, Halorobum, Alalkalicoccus, dominant archaeal genera; Weissella and Lactobacillus major LAB genera | [47] |
| Packaging of beef meat | V1-V3 16S rDNA | 500 | 454 FLX | Genus/ species |
RDP | Inhibition of Enterobacteriaceae by modified atmospheres; nisin-active vacuum packaging inhibited Brochrotys thermosphacta | [105] |
| Marinated and unmarinated broiler meat | V1-V3 16S rDNA, total DNA | >250 na |
454 FLX | Family/ genus |
SILVA NCBI nr |
Carnobacteriaceae, Clostridiaceae, Entorococcaceae, Enterobacteriaceae, and Vibrionaceae more common in unmarinated meat samples | [110] |
RDP, Ribosomal Database Project; NCIB nr, National Center for Biotechnology Information non-redundant nucleotide database; MG-RAST, Meta Genome Rapid Annotation server based on Subsystem Technology; SILVA (quality-controlled database of aligned ribosomal RNA gene sequences); Greengenes (a chimera-checked 16S rRNA gene database); EzTaxon (a web-based tool for identification of prokaryotes based on 16S rDNA sequences).
nr, not reported.
na, not applicable.
Next paragraphs summarize pioneering and representative works studying the microbiota of different foods and food fermentation by NGS approaches.
3.2.1. Microbial Fingerprinting of Plant-derived Fermentations
The bacterial community of “meju”, a Korean, soybean-derived, fermented product, was characterized in one of the first food microbial ecology studies involving NGS [90]. Meju is similar to the Japanese “natto” and “miso”, and to the Indonesian “tempeh”, and is the basis of many Asian products in which fermented soybean pastes are mixed with seasonings, vegetables and rice, e.g., “cheonggukjang” [91], “doenjang” [92], and “kochjang” [93]. The microbial populations of all these products have now been examined using NGS techniques. Bacillus spp. (B. subtilis, B. licheniformis, B. amyloliquefaciens, and others) seemed to be dominant throughout fermentation, followed by Enterococcus spp. (E. faecium, E. faecalis), Lactobacillus spp., and species of other LAB genera (Leuconostoc, Weissella, Tetragenococcus). However, the actual majority species varied widely between samples, and sometimes the number of LAB reads surpassed that of the bacilli [91-93]. Most of the bacterial species had been identified in these products by culture and culture-independent techniques. However, notable discrepancies have been noted between the results obtained by these methods and those provided by pyrosequencing [91].
Another Korean product which has been actively investigated by pyrosequencing is “kimchi” [94, 95], a fermented product that combines vegetables, fermented seafood products (“jeotgal”), seasonings, and a starchy-rich paste (rice, wheat). There are hundreds of kimchi varieties, depending on the region, local area, and the availability of seasonal ingredients. In ten types of kimchi, the succession of microbes was reported to follow a few general trends [94]. Together with some species of Proteobacteria (Pseudomonas, Enterobacter), pyrosequencing showed that Leuconostoc spp. and Weissella spp. dominate the early stages of fermentation, while Lactobacillus spp. are dominant at the end [94].
The archaeal and bacterial communities of seven types of jeotgal - as mentioned above, an ingredient of “kimchi”, but also used as a side dish - have also been analyzed by pyrosequencing [47]. Since the raw ingredients are mixed with large amounts of salt, several members of the family Halobacteriaceae (of the archaeal phylum Euryarchaeota), such as Halorubum and Halalkalicoccus, were commonly detected in most samples. Lactobacillus and Weissella reads in similar proportions made up the majority bacteria, although Gammaproteobacteria belonging to the genus Salinivibrio were predominant in one sample. The authors concluded pyrosequencing to have revealed new phylotypes of both bacteria and archaea that were not detected by DGGE.
Pyrosequencing of both rDNA- and cDNA-derived amplicons has been used to study the microbial ecology of Spanish-style and Greek-style fermented table olives [40]. In this work, the olive surfaces and brines (8-12% NaCl) were sampled independently. Greek-style fermentation involves natural, untreated olives that are directly brined after picking; Spanish-style fermentation includes a previous washing step of the fruits with a dilute NaOH (2-3%) solution. This reduces bitterness through the degradation of oleuropein and polyphenols, and softens the olive pericarp increasing its permeability. The bacterial compositions revealed throughout fermentation by DNA and cDNA analyses were very similar, suggesting that microorganisms were all alive and active. The initial stages of both the Greek and Spanish fermentations were characterized by large numbers of halophilic bacteria (Chromohalobacter and Halomonas accounting for 50-60% of the total reads), while at the end of the fermentation (90 days), Lactobacillus species surpassed all other microorganisms. The Spanish-style fermentation was associated with larger numbers of Enterobacteriaceae, such as Enterobacter, Citrobacter, Escherichia and Klebsiella. These differences may account for the distinctive safety and preservation properties of these two forms of fermented olives.
rDNA pyrosequencing has been used to examine the bacterial ecology of rye and wheat sourdough fermentations [51]. With the exception of the Enterobacteriaceae, all other flour-associated contaminants were shown to be completely inhibited during sourdough propagation. Rye sourdough fermentation was dominated by Weissella spp., the dominant bacteria in rye flour. In contrast, in wheat sourdoughs, Lactobacillus sakei, Leuconostoc spp., Weissella spp. and Lactococcus lactis, fluctuated as the majority species. The rye and wheat flour types initially showed distinctive microbiota, but over fermentation their complexity was rapidly simplified and a core microbiota composed mainly of LAB was seen in both sourdough fermentations.
3.2.2. Microbiology of Milk and Fermented Dairy Products
The pyrosequencing of rDNA amplicons has been used to examine the microbiome of cow milk, with particular interest shown in identifying the microbial types involved in mastitis [96]. In most samples, the mastitis-causing pathogens identified by culturing (Escherichia coli, Klebsiella spp., Trueperella pyogenes, Streptococcus uberis, Staphylcoccus aureus) were among the most common detected by pyrosequencing. However, rDNA sequences of bacterial pathogens not previously associated with mastitis, as well as of bacteria never described as pathogens, were occasionally detected.
The communities in kefir grains and kefir beverages of different origin have also been studied by the pyrosequencing of rDNA amplicons [49, 97]. As expected, the dominant microorganisms detected by culturing were also identified by pyrosequencing; these included Lactobacillus kefiranofaciens, Lactobacillus kefiri, Lactobacillus parakefiri, Lactobacillus buchneri, and others. Pyrosequencing also revealed minor bacterial constituents such as species of Acetobacter and Lactococcus [49, 97]. This technique further confirmed and that the microbiota of the beverage is distinct to that of the grains [97].
Pyrosequencing has also been used to study the diversity and dynamics of bacterial populations present during the manufacture and ripening of traditional cheeses, including Danish raw milk cheeses [98], the Polish cheese Oscypek [99], artisanal Irish cheeses [100], Latin-style cheeses [101], and water buffalo Mozzarella [102] (Table 2). Different LAB species were shown dominant in all cheeses and at all stages of production, the actual types depending on the cheese technology employed and/or the use (or not) of starter cultures. Several bacterial types never before reported in traditional and artisanal cheeses were identified by pyrosequencing, including Bifidobacterium spp. in Oscypek [99], Faecalibacterium, Prevotella, and Helcococcus in Irish cheeses [100], and Exiguobacterium in Latin-style cheeses [101]. The importance of these and other minority populations on the sensory profiles of these cheeses and/or their safety remains uncertain.
The characterization of the diversity of thermophilic bacteria in milk following pasteurization and culturing at 42ºC for 24 h has also been examined by the pyrosequencing of rDNA amplicons [50]. Compared to raw milk, in which mesophilic LAB species were the majority, reads of Streptococcus thermophilus accounted for about 99% of the sequences found. The 4% and 0.1% of Vibrio spp. reads returned by the raw and treated milk, respectively, was not explained [50]. Vibrionaceae read sequences have recently been associated with the new species Vibrio casei [103], which was originally isolated from a French, washed-rind cheese [104]. Small numbers of DNA sequences assigned to species never before isolated from milk or dairy products were also noticed, including Geobacillus toebii and Methylobacterium populi; these may have been environmental contaminants.
3.2.3. Composition and Succession of Meat-associated Microbiota
Community profiling via rDNA pyrosequencing has been used in a meat system to evaluate microbial diversity shifts during storage under different conditions [105]. As expected, the meat microbiota was significantly affected by the storage conditions. The growth of Enterobacteriaceae and Pseudomonas spp. was inhibited in modified atmosphere conditions, at least during the first three weeks of storage. Vacuum packaging in nisin-containing bags reduced the growth of Brochothrix thermosphacta, a major meat-spoiling organism. Pyrosequencing further revealed complex shifts in most microbial populations; this might have a negative influence on meat quality. The microbial data correlated well with the volatile and non-volatile microbial metabolites produced during storage and detected by gas chromatography (SPME-GC-MS) and proton nuclear magnetic resonance (1H NMR). Combination of NGS with the analysis of microbial metabolites (metabolomics) would further contribute to characterize beneficial and detrimental activities of the microorganisms in food ecosystems.
3.3. Spatial and Temporal Distribution of Microbial Populations
Bokulich et al. [106] used the Illumina platform technology to study the bacteria and fungi colonizing the surfaces of a pilot plant winery and its manufacturing tools before, during and after harvest over two seasons (Table 2). Under normal conditions, the winery surfaces harbored seasonally fluctuating populations of both types of microorganism. Community composition was also dependent on the production context of each sampling site, and was shaped by the technological practices used and the processing stage. Interestingly, S. cerevisiae and other beneficial fermentation-related yeast were detected on the surfaces prior to harvest, while spoilage-related microbes were undetected or detected only at low levels.
In a similar study involving the same technology, the manufacturing and ripening areas of two artisanal cheese factories were analyzed for bacterial and fungal populations [103]. The Illumina technology was used to track the microbiome dwelling in the cheemaking facility and study its influence on the cheese microbiota [103]. Milk fermentation-associated microbes dominated most of the surfaces, among which Debaryomyces and Lactococcus were the dominant eukaryotic and prokaryotic organisms, respectively. The establishment of these organisms on processing surfaces may play a pivotal role in their transfer between sequential fermentations. Interestingly, similar microbial communities occupied the same surface types, again suggesting the strong selection of microbes on the basis of production technology and stage [103]. Facility-specific microorganisms were also detected, which might influence the sensorial properties of the cheeses they produce.
3.4. Shotgun Metagenomics
Analysis of total DNA allows the simultaneous monitoring of bacterial populations and their metabolic potential. In addition, whole DNA sequencing enables a vast array of microbial genes to be identified and annotated, including novel genes and operons.
Pyrosequencing of total microbial DNA from the milk of healthy cows, and that from cows with clinical and subclinical mastitis, has recently been undertaken [71, 107] (Table 2), and significant differences in their microbial compositions reported. Kuen et al. [71] associated healthy samples with larger proportions of Pseudomonas, Psychrobacter, and Raldstonia reads, while in the mastitis samples reads of Brevundimonas, Burkholderia, Sphingomonas and Strenotrophomonas were found in greater abundance. Based on these results the authors proposed that some cases of mastitis may be produced by a mixture of microbes rather than a single pathogen. Similar results (although involving different microbial species) were obtained by Bhatt et al. [107]. In the latter study, large numbers of viral sequences, together with sequences of the hosts species S. aureus, E. coli and Enterobacter and Yersinia species, were found. The authors speculated that phages might be involved in providing the milk ‘natural resistance’ against pathogens [107].
The first NGS-based metagenomic study of total DNA from a traditional fermented food was performed on kimchi [95] (Table 2). Assignment of metagenomic sequences to functional gene categories revealed that kimchi fermentation was mostly accomplished by heterofermentative LAB species, as reported previously by targeted metagenomic studies [94]. Analysis of rDNA sequences among the pyrosequencing reads showed that Leuconostoc, Lactobacillus, Weissella and Enterococcus were dominant throughout fermentation. Surprisingly, when the metagenomic reads were mapped onto the database of complete genomes, the two most often recruited genomes were those of Leuconostoc mesenteroides and Lb. sakei. However, the metagenome sequences poorly matched the genomes of Leuconostoc kimchi, a species isolated from this product, and Weissella paramesenteroides, a species that the analysis of rDNA libraries had suggested was dominant throughout fermentation [33]. Similar results were obtained by assembling the pyrosequencing reads into contigs and comparing them against the GenBank non-redundant nucleotide database. Most contigs shared a nucleotide similarity of >97% to the Leuc. mesenteroides and Lb. sakei genomes. It is noteworthy that a large number of kimchi metagenomic contigs were classified as belonging to bacteriophage DNA, suggesting that a high proportion of bacterial cells either contained integrated prophages or were infected with them [108]. Viral metagenomics could help us understand the ecological role of viruses in the bacterial dynamics of food environments [108, 109].
Pyrosequencing has also been used to compare the composition and diversity of microbial communities in marinated and unmarinated broiler fillets [110]. Bacterial diversity and succession was analyzed by the sequencing of rDNA amplicons and then later by shotgun metagenomics. Smaller populations of Carnobacterium, Vagococcus, Brochothrix, Clostridium, Enterobacteriaceae and Vibrio were found in marinated meat compared to the unmarinated fillets [110]. Vagococcus and Vibrio, the predominant communities in the unmarinated samples, had never before been associated with the shelf-life of meat. In addition, increased proportions of LAB species belonging to the Lactobacillaceae and Leuconostocaceae families were encountered in the marinated samples. According to the functional analysis of the metagenomes, over-represented genes in the unmarinated strips were shown to be specific to Gram-negative bacteria. The differences in the genes between the marinated and unmarinated samples were thought to be due to the different phylogenetic composition of the bacterial communities. The authors concluded that marinating extended both the self-life and sensorial properties of broiler meat by delaying the growth of spoilage microorganisms.
A further shotgun metagenomic DNA study involving pyrosequencing was conducted to identify the prokaryotic and eukaryotic microorganisms involved in cocoa bean fermentation [111]. To determine the best computational approach for estimating the microbial diversity, taxonomic profiling was performed using both similarity-based and composition-based methods involving a vast array of taxonomic profiling tools. Overall, greater community diversity was detected by pyrosequencing than by culturing and previously-used culture-independent methods. The members of the family Lactobacillaceae were found to be the most abundant, with Lactobacillus the dominant genus. Subdominant populations of γ-Proteobacteria belonging to the genus Escherichia were also recorded. As in other food fermentations, a moderate proportion (0.25% of the total) of viral reads was encountered. Most phage sequences were assigned to those of Lactobacillus and enterobacteria, supporting the idea that an interaction exists between bacterial hosts and viral communities [111]. Metagenomic analysis indicated the most abundant yeast to be Hanseniaspora uvarum, followed by Hanseniaspora opuntiae and S. cerevisiae - species that have been commonly associated with cocoa bean fermentation. Other fungal species were also detected, including some which had never before been associated with cocoa fermentation.
4. COMBINATION OF NGS TECHNIQUES
In a recent study, NGS techniques were used in genomic and metagenomic studies to characterize a complex, undefined cheese starter culture for Gouda cheese with a long history of industrial use [109]. Analysis of rDNA sequences revealed the dominance of Lc. lactis (99%) and a minor population of Leuc. mesenteroides (1%). Culturing and typing techniques identified five genetic lineages of Lc. lactis subsp. cremoris, two of Lc. lactis subsp. lactis biovar diacetylactis, and one of Leuc. mesenteroides. Genome sequencing of all eight lineages allowed the contribution of individual strains to metagenomic data to be tracked during starter propagation and cheese manufacture. Lineage contribution was found to be in good agreement with the results obtained by culturing. Genome analysis was further used to reconstruct in silico metabolic maps, with which the authors evaluated complementary metabolic reactions that might contribute towards species and lineage maintenance. The superimposition of metabolic maps suggested that (-aminobutyric acid excreted by Lc. lactis during the acid stress response might serve as a substrate for succinate formation by Leuc. mesenteroides [109]. The presence of 1.15% phage-related DNA sequences in the metagenomic data was also reported. Using strains of Lc. lactis as indicators, three lytic phages, two of the P335 type and one of the P936 type, were isolated from the starter mixture. Surprisingly, the Lc. lactis strains showed wide variation in phage sensitivity within and between lineages. The stability and dynamics of the microbial community during backslopping propagation of this starter, and throughout cheese manufacture, strongly suggest phage resistance/susceptibility to be a key factor in the prevention of the loss of lineages as environmental conditions change. As in other ecosystems [112], phage predation was proposed to ensure diversity by suppressing abundant strains (“kill the winner” theory) [109], and thus stabilizing overall community functionality.
5. CONCLUSION AND FUTURE PROSPECTS
As in other fields of science, NGS techniques will ultimately revolutionize food microbiology. Pathogen detection, microbial profiling, genetic data mining, genotype-phenotype linking, determining the fate of starters and pathogens over food manufacturing and ripening, and predicting product shelf life etc., will be the realm of these high-throughput technologies. In fact, the microbiology of most foods and food fermentations will shortly be revisited with the aid of one or more of these new techniques.
Thought valuable and promising as stated above, current NGS technologies sill suffer from shortcomings and limitations that must be overcome. Much progress needs to be made before reliable data of biological significance can be generated. Recent discoveries and ensuing work have revealed limitations and biases that were previously ignored. These and other drawbacks should be addressed in the new future to improve all NGS constrains that limit at present the interpretation of data in the complex microbial diversity and physiology of foods.
5.1. NGS in Food Microbiology
To date, much work has been devoted to testing these techniques in a variety of food and food fermentation ecosystems. They have been mostly used for descriptive purposes, e.g., in the discovery of novel, low abundance, taxonomic lineages. New genes and operons have already been associated with certain foods and food processes, and a vast panoply of new species has already been associated with some foods. Such knowledge will allow for the determination of critical microbial variables during food production and therefore help better control food quality and safety. Community structure and population dynamics could ultimately be used for maximizing and optimization of fermentation/maturation and/or preservation processes. However, the significance in terms of quality and safety of the huge diversity present in most food environments, and particularly in food fermentations, is not yet known. Nonetheless, NGS techniques could fuel subsequent knowledge on relationships between microbial diversity and sensorial and safety properties. Subdominant and minority populations, which have been shown to vary widely between producers and batches, could contribute to generate, specific key components of aroma and taste. In addition, the isolation of strains belonging to newly discovered taxa will ultimately allow their functional characterization and perhaps their future use as improved starters. Besides, the presence of unexpected microorganisms (contaminants) in products and/or processes can be related to quality (spoiling) and safety (pathogens) issues.
Further, the use of NGS in the testing of scientific hypotheses has now begun. Among others, challenges may include exploring the complex interactions taking place between species and strains in mixed fermentations, which are pivotal in the production of taste and aroma compounds, and development of metagenomic and pan-genomic metabolic models, which would allow systematic exploration of the capabilities of the complex microbial consortia in foods, ecosystems of a high practical relevance.
5.2. Improving Existing and Implementing New Technologies
Comparison of the results from different works, products and/or processes is difficult as nucleic acid extraction and purification methods employed are usually different, as are the amplification primer pairs in targeted metagenomics, the analysis of data through different software programs, and so on. Harmonization of the methodology in a near future would allow obtaining deeper insights and stronger conclusions on the microbial diversity in and community structure of foods and food-related environments, as well as the gene content and functionality.
Refining the existing sequencing technologies should allow the analysis of longer reads, thus permitting genetic identification at lower taxonomic level (which has more biological meaning). Increasing the length of reads to cover the whole of the 16S rRNA gene would undoubtedly improve identification of microbes. Indeed, identification at the species level continues to be crucial in order to obtain biologically-meaning data. It is not relevant whether there is Listeria spp. on a food, but whether this is Listeria monocytogenes and, more importantly, if it is alive or dead. Thus, both sequence length and sequencing accuracy become critical. Sequencing technologies currently under development will further increase phylogenetic accuracy. Metagenomic and metatranscriptomic approaches could then be used with confidence to assign sequences to specific species and/or strains and to track their growth in foods.
The short half-life of mRNAs actually allows a better view of the microbial activities, metabolic processes and trophic chains occurring within the food at any particular time during manufacture. Metatranscriptomic approaches have been used successfully to examine complex microbial ecosystems such as the rumen and soil, and have been suggested promising for studying food fermentations. Though enrichment of messenger RNA from bacteria is still a challenge, metatranscriptomics could provide a snapshot of the transcriptional profiles at the time of sampling that correspond to discrete populations within a complex microbial community. Application of this technique could be employed to understand the diversity of the microbial communities of foods and food fermentations and their potential activity and regulatory mechanisms on the food matrices.
Presumably sequencing cost reductions in the near future will fuel the development of more shotgun projects to the detriment of targeted sequencing, because beyond the phylogenetic data additional information on genes and gene functionality of the components of the microbial communities can be obtained.
5.3. Well-managed Databases and User-friendly Software Programs
Reads obtained by NGS techniques need to be converted into more user-friendly information that allows comparisons between samples. There is still a great room for improvement in both databases and software programs to manage the massive data obtained through NGS projects. Enlarged and well-kept databases would also help in accurately determining the phylogenetic position of sequence data. As compared to those of bacteria, of particular shortage is the absence in databases of comprehensive fungal gene sequences. In spite of their universal spread and active role in most food fermentations, populations of fungi (yeasts, moulds) have received until recently little attention if at all. New software programs linking phylogeny with function should further enable the direct assessment of community functionality, in an effort to relate microorganisms to the sensorial properties of foods.
5.4. Combination of “Omic” Technologies
Finally, combination of NGS with some other high throughput “omic” techniques (metabolomics, proteomics, glycomics, etc.) and system biology approaches. Correlation of data from all these techniques would help link microbial diversity in and evolution on foods with their beneficial and/or detrimental activities contributing to the overall quality of food products. Particularly, metabolomics is a critical omic, as, whether desirable or detrimental, microbial development results in an accumulation of microbial metabolites. The presence and amount of such molecules could aid linking microbes with sensorial attributes.
ACKNOWLEDGEMENTS
All authors contribute to review the literature, writing the successive drafts, and revise and approve the final version of the submitted manuscript.
Financial support for working at the authors’ laboratories was provided by projects from the Spanish Ministry of Economy and Competitiveness (Ref. AGL2011-24300 and RM2011-00005-00-00) to B.M., and the Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq), Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (Capes) and Fundação de Amparo à Pesquisa do Estado do Rio de Janeiro (FAPERJ) to C.T.C.C.R., A.M.O.L., and R.S.P.
CONFLICT OF INTEREST
The author(s) confirm that this article content has no conflict of interest.
REFERENCES
- 1.Smit G, Smit BA, Engels W J M. Flavour formation by LAB and biochemical flavour profiling of cheese products. FEMS Microbiol. Rev. 2005;29:591–610. doi: 10.1016/j.femsre.2005.04.002. [DOI] [PubMed] [Google Scholar]
- 2.Quigley L, O'Sullivan O, Beresford TP, Ross RP, Fitzgerald GF, Cotter P D. Molecular approaches to analyzing the microbial composition of raw milk and raw milk cheeses. Int. J. Food Microbiol. 2011;150:81–94. doi: 10.1016/j.ijfoodmicro.2011.08.001. [DOI] [PubMed] [Google Scholar]
- 3.Amann RI, Ludwingg W, Schleifer K-F. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol. Rev. 1995;59:143–169. doi: 10.1128/mr.59.1.143-169.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ercolini D, Moschetti G, Blaiotta G, Coppola S. The potential of a polyphasic PCR-DGGE approach in evaluating microbial diversity of natural whey cultures for water-buffalo Mozzarella cheese production: bias of culture-dependent and culture-independent analyzes. Syst. Appl. Microbiol. 2001;24:610–617. doi: 10.1078/0723-2020-00076. [DOI] [PubMed] [Google Scholar]
- 5.Giraffa G, Neviani E. DNA-based culture-independent strategies for evaluating microbial communities in food-associated ecosystems. Int. J. Food Microbiol. 2001;67:19–34. doi: 10.1016/s0168-1605(01)00445-7. [DOI] [PubMed] [Google Scholar]
- 6.Jany JL, Barbier G. Culture-independent methods for identifying microbial communities in cheese. Food Microbiol. 2008;25:839–848. doi: 10.1016/j.fm.2008.06.003. [DOI] [PubMed] [Google Scholar]
- 7.Ampe F, ben Omar N, Moizan C, Wacher C, Guyot J P. Polyphasic study of the spatial distribution of microorganisms in Mexican pozol a fermented maize dough demonstrates the need for cultivation-independent methods to investigate traditional fermentations. Appl. Environ. Microbiol. 1999;65:5464–5473. doi: 10.1128/aem.65.12.5464-5473.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cocolin L, Manzano M, Cantoni C, Comi G. Denaturing gradient gel electrophoresis analysis of the 16S rRNA gene V1 region to monitor dynamic changes in the bacterial population during fermentation of Italian sausages. Appl. Environ. Microbiol. 2001;67:5113–5121. doi: 10.1128/AEM.67.11.5113-5121.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Randazzo CL, Torriani S, Akkermans A L D, de Vos WM, Vaughan E E. Diversity dynamics and activity of bacterial communities during production of an artisanal Sicilian cheese as evaluated by 16S rRNA analysis. Appl. Environ. Microbiol. 2002;68:1882–1892. doi: 10.1128/AEM.68.4.1882-1892.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cocolin L, Innocente N, Biasutti M, Comi G. The late blowing in cheese: a new molecular approach based on PCR and DGGE to study the microbial ecology of the alteration process. Int. J. Food Microbiol. 2004;90:83–91. doi: 10.1016/s0168-1605(03)00296-4. [DOI] [PubMed] [Google Scholar]
- 11.Ercolini D, Mauriello G, Blaiotta G, Moschetti G, Coppola S. PCR-DGGE fingerprints of microbial succession during a manufacture of traditional water buffalo mozzarella cheese. J. Appl. Microbiol. 2004;96:263–270. doi: 10.1046/j.1365-2672.2003.02146.x. [DOI] [PubMed] [Google Scholar]
- 12.Delbès C, Ali-Mandjee L, Montel M C. 2007.Monitoring bacterial communities in raw milk and cheese by culture-dependent and -independent 16S rRNA gene-based analysis. Appl. Environ. Microbiol. 2007;73:1882–1891. doi: 10.1128/AEM.01716-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Alegría A, Álvarez-Martín P, Sacristán N, Fernández E, Delgado S, Mayo B. Diversity and evolution of majority microbial populations during manufacturing and ripening of Casín a Spanish traditional starter-free cheese made of raw cow's milk. Int. J. Food Microbiol. 2009;136:44–51. doi: 10.1016/j.ijfoodmicro.2009.09.023. [DOI] [PubMed] [Google Scholar]
- 14.Carraro L, Maifreni M, Bartolomeoli I, Martino ME, Novelli E, Frigo F, Marino M, Cardazzo B. Comparison of culture-dependent and -independent methods for bacterial community monitoring during Montasio cheese manufacturing. Res. Microbiol. 2011;162:231–239. doi: 10.1016/j.resmic.2011.01.002. [DOI] [PubMed] [Google Scholar]
- 15.Bokulich N A, Mills D A. Differentiation of mixed lactic acid bacteria communities in beverage fermentations using targeted terminal restriction fragment length polymorphism. Food Microbiol. 2012;31:126–132. doi: 10.1016/j.fm.2012.02.007. [DOI] [PubMed] [Google Scholar]
- 16.Gkatzionis K, Yunita D, Linforth R S, Dickinson M, Dodd C E. Diversity and activities of yeasts from different parts of a Stilton cheese. Int. J. Food Microbiol. 2014;177:109–116. doi: 10.1016/j.ijfoodmicro.2014.02.016. [DOI] [PubMed] [Google Scholar]
- 17.Wilmes P, Simmons SL, Denef VJ, Banfield J F. The dynamic genetic repertoire of microbial communities. FEMS Microbiol. Rev. 2009;33:109–132. doi: 10.1111/j.1574-6976.2008.00144.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Solieri L, Dakal TC, Giudici P. Next-generation sequencing and its potential impact on food microbial genomics. Ann. Microbiol. 2013;63:21–37. [Google Scholar]
- 19.Liu G E. Recent applications of DNA sequencing technologies in food nutrition and agriculture. Recent Patents Food Nutr. Agricult. 2011;3:187–191. doi: 10.2174/2212798411103030187. [DOI] [PubMed] [Google Scholar]
- 20.Bokulich NA, Mills D A. Next-generation approaches to the microbial ecology of food fermentations. BMB Reports. 2012;45:377–389. doi: 10.5483/bmbrep.2012.45.7.148. [DOI] [PubMed] [Google Scholar]
- 21.Ercolini D. High-throughput sequencing and metagenomics: moving forward in the culture-independent analysis of food microbial ecology. Appl. Environ. Microbiol. 2013;79:3148–3155. doi: 10.1128/AEM.00256-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Glenn T C. Field Guide to Next Generation DNA Sequencers. Mol. Ecol. Resources. 2011;11:759–769. doi: 10.1111/j.1755-0998.2011.03024.x. [DOI] [PubMed] [Google Scholar]
- 23.Liu L, Li Y, Li S, Hu N, He Y, Pong R, Lin D, Lu L, Law M. Comparison of Next-Generation Sequencing Systems. J. Biomed. Biotechnol. 2012:251364. doi: 10.1155/2012/251364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Quail MA, Smith M, Coupland P, Otto TD, Harris SR, Connor TR, Bertoni A, Swerdlow HP, Gu Y. A tale of three next generation sequencing platforms: comparison of Ion torrent pacific biosciences and illumina MiSeq sequencers. BMC Genomics. 2012;13:341. doi: 10.1186/1471-2164-13-341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schadt EE, Turner S, Kasarskis A. A window into third-generation sequencing. Human Mol. Gen. 2010;19:R227–240. doi: 10.1093/hmg/ddq416. [DOI] [PubMed] [Google Scholar]
- 26.Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, Bemben LA, Berka J, Braverman MS, Chen YJ, Chen Z, Dewell SB, Du L, Fierro JM, Gomes XV, Godwin BC, He W, Helgesen S, Ho CH, Irzyk GP, Jando SC, Alenquer ML, Jarvie TP, Jirage KB, Kim JB, Knight JR, Lanza JR, Leamon JH, Lefkowitz SM, Lei M, Li J, Lohman KL, Lu H, Makhijani VB, McDade KE, McKenna MP, Myers EW, Nickerson E, Nobile JR, Plant R, Puc BP, Ronan MT, Roth GT, Sarkis GJ, Simons JF, Simpson JW, Srinivasan M, Tartaro KR, Tomasz A, Vogt KA, Volkmer GA, Wang SH, Wang Y, Weiner MP, Yu P, Begley RF, Rothberg J M. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005; 437:376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dressman D, Yan H, Traverso G, Kinzler KW, Vogelstein B. Transforming single DNAmolecules into fluorescent magnetic particles for detection and enumeration of genetic variations. Proc. Ntl. Acad. Sci. USA. 2003;100:8817–8822. doi: 10.1073/pnas.1133470100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bentley DR, Balasubramanian S, Swerdlow HP, Smith GP, Milton J, Brown CG, Hall KP, Evers DJ, Barnes CL, Bignell HR, Boutell JM, Bryant J, Carter RJ, Keira Cheetham R, Cox AJ, Ellis DJ, Flatbush MR, Gormley NA, Humphray SJ, Irving LJ, Karbelashvili MS, Kirk SM, Li H, Liu X, Maisinger KS, Murray LJ, Obradovic B, Ost T, Parkinson ML, Pratt MR, Rasolonjatovo IM, Reed MT, Rigatti R, Rodighiero C, Ross MT, Sabot A, Sankar SV, Scally A, Schroth GP, Smith ME, Smith VP, Spiridou A, Torrance PE, Tzonev SS, Vermaas EH, Walter K, Wu X, Zhang L, Alam MD, Anastasi C, Aniebo IC, Bailey DM, Bancarz IR, Banerjee S, Barbour SG, Baybayan PA, Benoit VA, Benson KF, Bevis C, Black PJ, Boodhun A, Brennan JS, Bridgham J A, Brown RC, Brown AA, Buermann DH, Bundu AA, Burrows JC, Carter NP, Castillo N, Chiara E C M, Chang S, Neil Cooley R, Crake NR, Dada OO, Diakoumakos KD, Dominguez-Fernandez B, Earnshaw DJ, Egbujor UC, Elmore DW, Etchin SS, Ewan MR, Fedurco M, Fraser LJ, Fuentes Fajardo KV, Scott Furey W, George D, Gietzen KJ, Goddard CP, Golda GS, Granieri PA, Green DE, Gustafson DL, Hansen NF, Harnish K, Haudenschild CD, Heyer NI, Hims MM, Ho JT, Horgan AM, Hoschler K, Hurwitz S, Ivanov DV, Johnson MQ, James T, Huw Jones TA, Kang GD, Kerelska TH, Kersey AD, Khrebtukova I, Kindwall AP, Kingsbury Z, Kokko-Gonzales PI, Kumar A, Laurent MA, Lawley CT, Lee SE, Lee X, Liao AK, Loch JA, Lok M, Luo S, Mammen RM, Martin JW, McCauley PG, McNitt P, Mehta P, Moon KW, Mullens JW, Newington T, Ning Z, Ling Ng B, Novo SM, O'Neill MJ, Osborne MA, Osnowski A, Ostadan O, Paraschos LL, Pickering L, Pike AC, Chris Pinkard D, Pliskin DP, Podhasky J, Quijano VJ, Raczy C, Rae VH, Rawlings SR, Chiva Rodriguez A, Roe PM, Rogers J, Rogert Bacigalupo MC, Romanov N, Romieu A, Roth RK, Rourke NJ, Ruediger ST, Rusman E, Sanches-Kuiper RM, Schenker MR, Seoane JM, Shaw RJ, Shiver MK, Short SW, Sizto NL, Sluis JP, Smith MA, Ernest Sohna Sohna J, Spence EJ, Stevens K, Sutton N, Szajkowski L, Tregidgo CL, Turcatti G, Vandevondele S, Verhovsky Y, Virk SM, Wakelin S, Walcott GC, Wang J, Worsley GJ, Yan J, Yau L, Zuerlein M, Mullikin JC, Hurles ME, McCooke NJ, West JS, Oaks FL, Lundberg PL, Klenerman D, Durbin R, Smith A J. Accurate whole human genome sequencing using reversible terminator chemistry. Nature. 2008;456:53–59. doi: 10.1038/nature07517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Valouev A, Ichikawa J, Tonthat T, Stuart J, Ranade S, Peckham H, Zeng K, Malek JA, Costa G, MaKernan K, Sidow A, Fire A, Johnson S M. A high-resolution nucleosome position map of, C.elegans reveals a lack of universal sequence-dictated positioning. Genome Res. 2008;18:1051–1063. doi: 10.1101/gr.076463.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rothberg JM, Hinz W, Rearick TM, Schultz J, Mileski W, Davey M, Leamon JH, Johnson K, Milgrew MJ, Edwards M, Hoon J, Simons JF, Marran D, Myers JW, Davidson JF, Branting A, Nobile JR, Puc BP, Light D, Clark TA, Huber M, Branciforte JT, Stoner IB, Cawley SE, Lyons M, Fu Y, Homer N, Sedova M, Miao X, Reed B, Sabina J, Feierstein E, Schorn M, Alanjary M, Dimalanta E, Dressman D, Kasinskas R, Sokolsky T, Fidanza JA, Namsaraev E, McKernan KJ, Williams A, Roth GT, Bustillo J. An integrated semiconductor device enabling non-optical genome sequencing. Nature. 2011;475:348–352. doi: 10.1038/nature10242. [DOI] [PubMed] [Google Scholar]
- 31.Eid J, Fehr A, Gray J, Luong K, Lyle J, Otto G, Peluso P, Rank D, Baybayan P, Bettman B, Bibillo A, Bjornson K, Chaudhuri B, Christians F, Cicero R, Clark S, Dalal R, Dewinter A, Dixon J, Foquet M, Gaertner A, Hardenbol P, Heiner C, Hester K, Holden D, Kearns G, Kong X, Kuse R, Lacroix Y, Lin S, Lundquist P, Ma C, Marks P, Maxham M, Murphy D, Park I, Pham T, Phillips M, Roy J, Sebra R, Shen G, Sorenson J, Tomaney A, Travers K, Trulson M, Vieceli J, Wegener J, Wu D, Yang A, Zaccarin D, Zhao P, Zhong F, Korlach J, Turner S. Real-time DNA sequencing from single polymerase molecules. Science. 2009;323:133–138. doi: 10.1126/science.1162986. [DOI] [PubMed] [Google Scholar]
- 32.Escalante A, Wacher C, Farrés A. Lactic acid bacterial diversity in the traditional Mexican fermented dough pozol as determined by 16S rDNA sequence analysis. Int. J. Food Microbiol. 2001;64:21–31. doi: 10.1016/s0168-1605(00)00428-1. [DOI] [PubMed] [Google Scholar]
- 33.Kim M, Chun J. Bacterial community structure in kimchi a Korean fermented vegetable food as revealed by 16S rRNA gene analysis. Int. J. Food Microbiol. 2005;103:91–96. doi: 10.1016/j.ijfoodmicro.2004.11.030. [DOI] [PubMed] [Google Scholar]
- 34.Ninane V, Mukandayambaje R, Berben G. Identification of lactic acid bacteria within the consortium of a kefir grain by sequencing 16S rDNA variable regions. J. AOAC Int. 2007;90:1111–1117. [PubMed] [Google Scholar]
- 35.van Hijum S A F.T, Vaughan EE, Vogel RF. Application of state-of-art sequencing technologies to indigenous food fermentations. Curr. Opin. Biotechnol. 2013;24:178–186. doi: 10.1016/j.copbio.2012.08.004. [DOI] [PubMed] [Google Scholar]
- 36.Linnarsson S. Recent advances in DNA sequencing methods - general principles of sample preparation. Exp. Cell Res. 2010;316:1339–1343. doi: 10.1016/j.yexcr.2010.02.036. [DOI] [PubMed] [Google Scholar]
- 37.Bonaiti C, Parayre S, Irlinger F. Novel extraction strategy of ribosomal RNA and genomic DNA from cheese for PCR-based investigations. Int. J. Food Microbiol. 2006;107:171–179. doi: 10.1016/j.ijfoodmicro.2005.08.028. [DOI] [PubMed] [Google Scholar]
- 38.Quail MA, Kozarewa I, Smith F, Scally A, Stephens PJ, Durbin R, Swerdlow H, Turner D J. A large genome center's improvements to the Illumina sequencing system. Nature Methods. 2008;5:1005–1010. doi: 10.1038/nmeth.1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Masoud W, Vogensen FK, Lillevang S, Al-Soud WA, Sørensen SJ, Jakobsen M. The fate of indigenous microbiota starter cultures Escherichia coli Listeria innocua and Staphylococcus aureus in Danish raw milk and cheeses determined by pyrosequencing and quantitative real-time (qRT)-PCR. Int. J. Food Microbiol. 2012;153:192–202. doi: 10.1016/j.ijfoodmicro.2011.11.014. [DOI] [PubMed] [Google Scholar]
- 40.Cocolin L, Alessandria V, Botta C, Gorra R, de Filippis F, Ercolini D, Rantsiou K. NaOH-Debittering induces changes in bacterial ecology during table olives fermentation. PLoS One. 2013;8:e69074. doi: 10.1371/journal.pone.0069074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schmieder R, Lim YW, Edwards R. Identification and removal of ribosomal RNA sequences from metatranscriptomes. Bioinformatics. 2012;28:433–435. doi: 10.1093/bioinformatics/btr669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Blazewicz SJ, Barnard RL, Daly RA, Firestone M K. Evaluating rRNA as an indicator of microbial activity in environmental communities: limitations and uses. ISME J. 2013;7:2061–2068. doi: 10.1038/ismej.2013.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mader U, Nicolas P, Richard H, Bessieres P, Aymerich S. Comprehensive identification and quantification of microbial transcriptomes by genome-wide unbiased methods. Curr. Opin. Biotechnol. 2011;22:32–41. doi: 10.1016/j.copbio.2010.10.003. [DOI] [PubMed] [Google Scholar]
- 44.Simon C, Daniel R. Metagenomic analyses past and future trends. Appl. Environ. Microbiol. 2011;77:1153–1161. doi: 10.1128/AEM.02345-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nocker A, Cheung C-Y, Camper A K. Comparison of propidium monoazide with ethidium monoazide for differentiation of live, VS.dead bacteria by selective removal of DNA from dead cells. J. Microbiol. Methods. 2006;67:310–320. doi: 10.1016/j.mimet.2006.04.015. [DOI] [PubMed] [Google Scholar]
- 46.Soergel DA, Dey N, Knight R, Brenner S E. Selection of primers for optimal taxonomic classification of environmental 16S rRNA gene sequences. ISME J. 2012;6:1440–1444. doi: 10.1038/ismej.2011.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Roh SW, Kim K-H, Nam Y-D, Chang H-W, Park E-J, Bae J-W. Investigation of archaeal and bacterial diversity in fermented seafood using barcoded pyrosequencing. The ISME J. 2010;4:1–16. doi: 10.1038/ismej.2009.83. [DOI] [PubMed] [Google Scholar]
- 48.Bokulich NA, Joseph CM, Allen G, Benson AK, Mills D A. Next-generation sequencing reveals significant bacterial diversity of botrytized wine. PLoS One. 2012;7:e36357. doi: 10.1371/journal.pone.0036357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Leite AM, Mayo B, Rachid CT, Peixoto RS, Silva JT, Paschoalin VM, Delgado S. Assessment of the microbial diversity of Brazilian kefir grains by PCR-DGGE and pyrosequencing analysis. Food Microbiol. 2012;31:215–221. doi: 10.1016/j.fm.2012.03.011. [DOI] [PubMed] [Google Scholar]
- 50.Delgado S, Rachid C T CC, Fernández E, Rychlik T, Alegría A, Peixoto RS, Mayo B. Diversity of thermophilic bacteria in raw pasteurized and selectively-cultured milk as assessed by culturing PCR-DGGE and pyrosequencing. Food Microbiol. 2013;36:103–111. doi: 10.1016/j.fm.2013.04.015. [DOI] [PubMed] [Google Scholar]
- 51.Ercolini D, Pontonino E, de Filippis F, Minervani F, La Storia A, Gobbetti M, Di Cagnio R. Microbial ecology dynamics during rye and wheat sourdough preparation. Appl. Environ. Microbiol. 2013;79:7827–7836. doi: 10.1128/AEM.02955-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lee SH, Jung JY, Jeon C O. Effects of temperature on microbial succession and metabolite change during saeu-jeot fermentation. Food Microbiol. 2014;38:16–25. doi: 10.1016/j.fm.2013.08.004. [DOI] [PubMed] [Google Scholar]
- 53.Liu Z, Lozupone C, Hamady M, Bushman FD, Knight R. Short pyrosequencing reads suffice for accurate microbial community analysis. Nucleic Acids Res. 2007;35:e120. doi: 10.1093/nar/gkm541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Petrosino JF, Highlander S, Luna RA, Gibbs RA, Versalovic J. Metagenomic pyrosequencing and microbial identification. Clin. Chem. 2009;55:856–866. doi: 10.1373/clinchem.2008.107565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rajendhran J, Gunasekaran P. Microbial phylogeny and diversity: small subunit ribosomal RNA sequence analysis and beyond. Microbiol Res. 2011;166:99–110. doi: 10.1016/j.micres.2010.02.003. [DOI] [PubMed] [Google Scholar]
- 56.Walters W A, Caporaso J G, Lauber C L, Berg-Lyons D, Fierer N, Knight R. PrimerProspector de novo design and taxonomic analysis of PCR primers. Bioinformatics. 2011;27:1159–1161. doi: 10.1093/bioinformatics/btr087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Renouf V, Claisse O, Miot-Sertier C, Lonvaud-Funel A. Lactic acid bacteria evolution during winemaking: use of rpoB gene as a target for PCR-DGGE analysis. Food Microbiol. 2006;23:136–145. doi: 10.1016/j.fm.2005.01.019. [DOI] [PubMed] [Google Scholar]
- 58.Ansorge W J. Next-generation DNA sequencing techniques. Nature Biotechnol. 2009;25:195–203. doi: 10.1016/j.nbt.2008.12.009. [DOI] [PubMed] [Google Scholar]
- 59.Kembel SW, Wu M, Eisen J, Green J L. Incorporating 16S gene copy number information improves estimates of microbial diversity and abundance. PLoS Computational Biol. 2012;8:e1002743. doi: 10.1371/journal.pcbi.1002743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.V?trovský T, Baldrian P. The variability of the 16S rRNA gene in bacterial genomes and its consequences for bacterial community analyses. PloS One. 2013;8:e57923. doi: 10.1371/journal.pone.0057923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Volokhov DV, Simonyan V, Davidson MK, Chizhikov V E. RNA polymerase beta subunit (rpoB) gene and the 16S-23S rRNA intergenic transcribed spacer region (ITS) as complementary molecular markers in addition to the 16S rRNA gene for phylogenetic analysis and identification of the species of the family Mycoplasmataceae. Mol. Phylogenet. Evol. 2012;62:515–528. doi: 10.1016/j.ympev.2011.11.002. [DOI] [PubMed] [Google Scholar]
- 62.Sibley CD, Peirano G, Church D L. Molecular methods for pathogen and microbial community detection and characterization: current and potential application in diagnostic microbiology. Infect. Genet. Evol. 2012;12:505–521. doi: 10.1016/j.meegid.2012.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Acinas SG, Sarma-rupavtarm R, Klepac-ceraj V, Polz M F. Reducing the effects of PCR amplification and sequencing artifacts on 16S rRNA-based studies. Appl. Environ. Microbiol. 2005;71:8966–8969. doi: 10.1128/AEM.71.12.8966-8969.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Anderson IC, Campbell CD, Prosser J I. Potential bias of fungal 18S rDNA and internal transcribed spacer polymerase chain reaction primers for estimating fungal biodiversity in soil. Environ. Microbiol. 2003;5:36–47. doi: 10.1046/j.1462-2920.2003.00383.x. [DOI] [PubMed] [Google Scholar]
- 65.Bokulich NA, Mills D A. Improved selection of internal transcribed spacer-specific primers enables quantitative ultra-high-throughput profiling of fungal communities. Appl. Environ. Microbiol. 2103;79:2519–2526. doi: 10.1128/AEM.03870-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Groenewald M, Robert V, Smith M T. The value of the D1/D2 and internal transcribed spacers (ITS) domains for the identification of yeast species belonging to the genus Yamadazyma. Persoonia. 2011;26:40–46. doi: 10.3767/003158511X559610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Nilsson RH, Ryberg M, Abarenkov K, Sjokvist E, Kristiansson E. The ITS region as a target for characterization of fungal communities using emerging sequencing technologies. FEMS Microbiol. Lett. 2009;296:97–101. doi: 10.1111/j.1574-6968.2009.01618.x. [DOI] [PubMed] [Google Scholar]
- 68.Kruger D, Kapturska D, Fischer C, Daniel R, Wubet T. Diversity measures in environmental sequences are highly dependent on alignment quality-data from ITS and new LSU primers targeting basidiomycetes. PLoS One. 2012;7:e32139. doi: 10.1371/journal.pone.0032139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Binladen J, Gilbert MT, Bollback JP, Panitz F, Bendixen C, Nielsen R, Willerslev E. The use of coded PCR primers enables high-throughput sequencing of multiple homolog amplification products by 454 parallel sequencing. PLoS One. 2007;2:e197. doi: 10.1371/journal.pone.0000197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mamanova L, Coffey AJ, Scott CE, Kozarewa I, Turner EH, Kumar A, Howard E, Shendure J, Turner D J. Target-enrichment strategies for next-generation sequencing. Nature Methods. 2010;7:111–118. doi: 10.1038/nmeth.1419. [DOI] [PubMed] [Google Scholar]
- 71.Kuehn JS, Gorden PJ, Munro D, Rong R, Dong Q, Plummer PJ, Wang C, Phillips G J. Bacterial community profiling of milk samples as a means to understand culture-netative bovine clinical mastitis. PLoS One. 2013;8:e61959. doi: 10.1371/journal.pone.0061959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Silander K, Saarela J. Whole genome amplification with Phi29 DNA polymerase to enable genetic or genomic analysis of samples of low DNA yield. Methods Mol. Biol. 2008;439:1–18. doi: 10.1007/978-1-59745-188-8_1. [DOI] [PubMed] [Google Scholar]
- 73.Sul WJ, Cole JR, Jesus E D C, Wang Q, Farris RJ, Fish JA, Tiedje J M. Bacterial community comparisons by taxonomy-supervised analysis independent of sequence alignment and clustering. Proc. Natl. Acad. Sci. USA. 2011;108:14637–14642. doi: 10.1073/pnas.1111435108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Cole JR, Wang Q, Cardenas E, Fish J, Chai B, Farris RJ, Kulam-Syed-Mohideen AS, McGarrell DM, Marsh T, Garrity GM, Tiedje J M. The Ribosomal Database Project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res. 2009;37:D141–D145. doi: 10.1093/nar/gkn879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Pelies J, Glöckner F O. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 2013;41:590–596. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, Huber T, Dalevi D, Hu P, Andersen G L. Greengenes a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 2006;72:5069–5072. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Abarenkov K, Nilsson R H, Larsson K-H, Alexander I J, Eberhardt U, Erland S, Høiland K, Kjøller R, Larsson E, Pennanen T, Sen R, Taylor A F S, Tedersool L, Ursin B M, Vrålstad7 T, Liimatainen K, Peintner U, Kõljalg U. The UNITE database for molecular identification of fungi - recent updates and future perspectives. New Phytologist. 2010;186:281–285. doi: 10.1111/j.1469-8137.2009.03160.x. [DOI] [PubMed] [Google Scholar]
- 78.Aziz R, Bartels D, Best A, DeJongh M, Disz T, Edwards R, Formsma K, Gerdes S, Glass E, Kubal M, Meyer F, Olsen G, Olson R, Osterman A, Overbeek R, McNeil L, Paarmann D, Paczian T, Parrello B, Pusch G, Reich C, Stevens R, Vassieva O, Vonstein V, Wilke A, Zagnitko O. The RAST Server Rapid Annotations using Subsystems Technology. BMC genomics. 2008;9:75. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Meyer F, Paarmann D, D'Souza M, Olson R, Glass EM, Kubal M, Paczian T, Rodriguez A, Stevens R, Wilke A, Wilkening J, Edwards R A. The metagenomics RAST server-a public resource for the automatic phylogenetic and functional analysis of metagenomes. BMC bioinformatics. 2008;9:386. doi: 10.1186/1471-2105-9-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Rachid C T CC, Santos AL, Piccolo MC, Balieiro FC, Coutinho H L C, Peixoto RS, Tiedje JM, Rosado A S. Effect of sugarcane burning or green harvest methods on the Brazilian Cerrado soil bacterial community structure. PloS One. 2013;8:e59342. doi: 10.1371/journal.pone.0059342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, Sahl JW, Stres B, Thallinger GG, van Horn DJ, Weber C F. Introducing mothur Open source platform-independent community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 2009;75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman F.D, Costello EK, Fierer N, Pena AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. QIIME allows analysis of high-throughput community sequencing data. Na-ture Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Quince C, Lanzen A, Davenport RJ, Turnbaugh P J. Removing noise from pyrosequenced amplicons. BMC Bioinformatics. 2011;12:38. doi: 10.1186/1471-2105-12-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Quince C, Lanzén A, Curtis TP, Davenport RJ, Hall N, Head IM, Read LF, Sloan W T. Accurate determination of microbial diversity from 454 pyrosequencing data. Nature Methods. 2009;6:639–641. doi: 10.1038/nmeth.1361. [DOI] [PubMed] [Google Scholar]
- 85.Reeder J, Knight R. Rapidly denoising pyrosequencing amplicon reads by exploiting rank-abundance distributions. Nature Methods. 2010;7:668–669. doi: 10.1038/nmeth0910-668b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Sogin M L, Morrison H, Mclellan S, Welch DM, Huse S. The rare biosphere? sorting out fact from fiction. Genome Biol. 2010;11:SI19. [Google Scholar]
- 87.Bokulich N A, Subramanian S, Faith J J, Gevers D, Gordon J I, Knight R, Mills D A, Caporaso J G. Quality-filtering vastly improves diversity estimates from Illumina amplicon sequencing. Nature Methods. 2013;10:57–59. doi: 10.1038/nmeth.2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Langille MG, Zaneveld J, Caporaso JG, McDonald D, Knights D, Reyes JA, Clemente JC, Burkepile DE, Vega Thurber RL, Knight R, Beiko RG, and Huttenhower C. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nature Biotechnol. 2013;31:814–821. doi: 10.1038/nbt.2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Humblot C, and Guyot J P. Pyrosequencing of tagged 16S rRNA gene amplicons for rapid deciphering of the microbiomes of fermented foods such as pearl millet slurries. Appl. Environ. Microbiol. 2009;75:4354–4361. doi: 10.1128/AEM.00451-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kim YS, Kim MC, Kwon SW, Kim SJ, Park IC, Ka JO, Weon H Y. Analyses of bacterial communities in meju a Korean traditional fermented soybean bricks by cultivation-based and pyrosequencing methods. J. Microbiol. 2011;49:340–348. doi: 10.1007/s12275-011-0302-3. [DOI] [PubMed] [Google Scholar]
- 91.Nam Y-D, Yi S-H, Lim S-I. Bacterial diversity of cheonggukjang a traditional Korean fermented food analyzed by barcoded pyrosequencing. Food Control. 2012;28:135–142. [Google Scholar]
- 92.Nam Y-D, Lee S-Y, Lim S-I. Microbial community analysis of Korean soybean pastes by next-generation sequencing. Int. J. Food Microbiol. 2012;155:36–42. doi: 10.1016/j.ijfoodmicro.2012.01.013. [DOI] [PubMed] [Google Scholar]
- 93.Nam Y-D, Park S-I, Lim S-I. Microbial composition of the Korean traditional food "kochujang" analyzed by a massive sequencing technique. J. Food Sci. 2012;77:250–256. doi: 10.1111/j.1750-3841.2012.02656.x. [DOI] [PubMed] [Google Scholar]
- 94.Park E-J, Chun J, Cha C-J, Park W-S, Jeon CO, Bae J-W. Bacterial community analysis during fermentation of ten representative kinds of "kimchi" with barcoded pyrosequencing. Food Microbiol. 2012;30:197–204. doi: 10.1016/j.fm.2011.10.011. [DOI] [PubMed] [Google Scholar]
- 95.Jung JY, Lee SH, Kim JM, Park MS, Bae J.-I Hahn, Y. Madsen EL, Jeon C O. Metagenomic analysis of "kimchi" a traditional Korean fermented food. Appl. Environ. Microbiol. 2011;77:2264–2274. doi: 10.1128/AEM.02157-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Oikonomou G, Machado VS, Santisteban C, Schukken YH, Bicalho R C. Microbial diversity of bovine mastitis milk as described by pyrosequencing of metagenomic 16S rDNA. PLoS One. 2012;7:e47671. doi: 10.1371/journal.pone.0047671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Dobson A, O'sullivan O, Cotter PD, Ross P, Hill C. High-throughput sequence-based analysis of the bacterial composition of kefir and an associated kefir grain. FEMS Microbiol. Lett. 2011;320:56–62. doi: 10.1111/j.1574-6968.2011.02290.x. [DOI] [PubMed] [Google Scholar]
- 98.Masoud W, Takamiya M, Vogensen FK, Lillevang S, Al-Soud WA, Sørensen SJ, Jakobsen M. Characterization of bacterial populations in Danish raw milk cheeses made with different starter cultures by denaturing gradient gel electrophoresis and pyrosequencing. J. airy J. 2010; 21:142–148. [Google Scholar]
- 99.Alegría A, Szczesny P, Mayo B, Bardowski J, Kowalczyk M. Biodiversity in Oscypek a traditional Polish cheese determined by culture-dependent and -independent approaches. Appl. Environ. Microbiol. 2012;78:1890–1898. doi: 10.1128/AEM.06081-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Quigley L, O'Sullivan O, Beresford TP, Ross RP, Fitzgerald GF, Cotter P D. High-throughput sequencing for detection of subpopulations of bacteria not previously associated with artisanal cheeses. Appl. Environ. Microbiol. 2012;78:5717–5723. doi: 10.1128/AEM.00918-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Lusk TS, Ottensen AR, White JR, Allard MW, Brown EW, Kase J E. Characterization of microflora in Latin-style cheeses by next-generation sequencing technology. BCM Microbiol. 2012;12:254. doi: 10.1186/1471-2180-12-254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Ercolini D, De Filippis F, La Storia A, Iacono M. "Remake" by high-throughput sequencing of the microbiota involved in the production of water buffalo Mozzarella cheese. Appl. Environ. Microbiol. 2012;78:8142–8145. doi: 10.1128/AEM.02218-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Bokulich NA, Mills D A. Facility-specific "house"microbiome drives microbial landscapes of artisan cheesemaking plants. Appl. Environ. Microbiol. 2013;79:5214–5223. doi: 10.1128/AEM.00934-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Blicher A, Neuhaus K, Scherer S, Vibrio casei SP. nov.isolated from the surface of two French red smear soft cheeses. Int. J. Syst. Evol. Microbiol. 2010;60:1745–1749. doi: 10.1099/ijs.0.016493-0. [DOI] [PubMed] [Google Scholar]
- 105.Ercolini D, Ferrocino I, Nasi A, Ndagijimana M, Vernocchi P, La Storia A, Laghi L, Mauriello G, Guerzoni ME, and Villani F. Monitoring of microbial metabolites and bacterial diversity in beef stored under different packaging conditions. Appl. Environ. Microbiol. 2011;77:7372–7381. doi: 10.1128/AEM.05521-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Bokulich NA, Ohta M, Richardson PM, Mills D A. Monitoring seasonal changes in winery-resident microbiota. PLoS One. 2013;8:e66437. doi: 10.1371/journal.pone.0066437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Bhatt VD, Ahir VB, Koringa PG, Jakhesara SJ, Rank DN, Nauriyal DS, Kunjadia AP, Joshi C G. Milk microbiome signatures of subclinical mastitis-affected cattle analysed by shotgun sequencing. J. Appl. Microbiol. 2012;112:639–650. doi: 10.1111/j.1365-2672.2012.05244.x. [DOI] [PubMed] [Google Scholar]
- 108.Park E-J, Kim K-H, Abell G C J, Kim M-S, Roh SW, Bae J-W. Metagenomic analysis of the viral communities in fermented foods. Appl. Environ. Microbiol. 2011;77:1284–1291. doi: 10.1128/AEM.01859-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Erkus O, de Jager V C L, Spus M, van Alen-Boerritger IJ, van Rijswijck I M H, Hazelwood L, Janssen P W M, van Hijum S A FT, Kleerebezem M, Smid E J. Multifactorial diversity sustains microbial community stability. The ISME J. 2013;7:2126–2136. doi: 10.1038/ismej.2013.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Nieminen TT, Koskinen K, Laine P, Hultman J, Säde E, Paloranta A, Johansson P, Björkroth J, Auvinen P. Comparison of microbial communities in marinated and unmarinated broiler meat by metagenomics. Int. J. Food Microbiol. 2012;157:142–149. doi: 10.1016/j.ijfoodmicro.2012.04.016. [DOI] [PubMed] [Google Scholar]
- 111.Illeghems K, De Vuyst L, Papalexandratou Z, Weckx S. Phylogenetic analysis of a spontaneous cocoa bean fermentation metagenome reveals new insights into its bacterial and fungal community diversity. PLoS One. 2012;7:e38040. doi: 10.1371/journal.pone.0038040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Rodríguez-Valera F, Martín-Cuadrado A B, Rodríguez-Brito B, Pasic L, Thingstad T F, Rohwer F, Mira A. Explaining microbial population genomics through phage predation. Nature Rev. Microbiol. 2009;7:828–836. doi: 10.1038/nrmicro2235. [DOI] [PubMed] [Google Scholar]
- 113.Koyanagi T, Kiyohara M, Matsui H, Yamamoto K, Kondo T, Katayama T, Kumagai H. Pyrosequencing survey of the microbial diversity of narezushi an archetype of modern Japanese sushi. Lett. Appl. Microbiol. 2011;53:635–640. doi: 10.1111/j.1472-765X.2011.03155.x. [DOI] [PubMed] [Google Scholar]
- 114.Sakamoto N, Tanaka S, Sonomoto K, Nakayama J. 16S rRNA pyrosequencing-based investigation of the bacterial community in nukadoko a pickling bed of fermented rice bran. Int. J. Food Microbiol. 2011;144:352–359. doi: 10.1016/j.ijfoodmicro.2010.10.017. [DOI] [PubMed] [Google Scholar]
- 115.Bokulich NA, Bamforth CW, Mills D A. Brewhouse-resident microbiota are responsible for multi-stage fermentation of American coolship ale. PLoS One. 2012;7:e35507. doi: 10.1371/journal.pone.0035507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Jung MJ, Nam YD, Roh SW, Bae J W. Unexpected convergence of fungal and bacterial communities during fermentation of traditional Korean alcoholic beverages inoculated with various natural starters. Food Microbiol. 2012;30:112–123. doi: 10.1016/j.fm.2011.09.008. [DOI] [PubMed] [Google Scholar]
- 117.Koyanagi T, Nakagawa A, Kiyohara M, Matsui H, Yamamoto K, Barla F, Take H, Katsuyama Y, Tsuji A, Shijimaya M, Nakamura S, Minami H, Enomoto T, Katayama T, Kumagai H. Pyrosequencing analysis of microbiota in kaburazushi a traditional medieval sushi in Japan. Biosci. Biotechnol. Biochem. 2013;77:2125–2130. doi: 10.1271/bbb.130550. [DOI] [PubMed] [Google Scholar]