Abstract
ATP synthase, the assembly which makes ATP in mitochondria, chloroplasts and bacteria, uses transmembrane proton gradients generated by respiration or photosynthesis to drive the phosphorylation of ADP. Its membrane domain is joined by a slender stalk to a peripheral catalytic domain, F1-ATPase. This domain is made of five subunits with stoichiometries of 3 alpha: 3 beta: 1 gamma: 1 delta: 1 epsilon, and in bovine mitochondria has a molecular mass of 371,000. We have determined the 3-dimensional structure of bovine mitochondrial F1-ATPase to 6.5 A resolution by X-ray crystallography. It is an approximately spherical globule 110 A in diameter, on a 40 A stem which contains two alpha-helices in a coiled-coil. This stem is presumed to be part of the stalk that connects F1 with the membrane domain in the intact ATP synthase. A pit next to the stem penetrates approximately 35 A into the F1 particle. The stem and the pit are two examples of the many asymmetric features of the structure. The central element in the asymmetry is the longer of the two alpha-helices in the stem, which extends for 90 A through the centre of the assembly and emerges on top into a dimple 15 A deep. Features with threefold and sixfold symmetry, presumed to be parts of homologous alpha and beta subunits, are arranged around the central rod and pit, but the overall structure is asymmetric. The central helix provides a possible mechanism for transmission of conformational changes induced by the proton gradient from the stalk to the catalytic sites of the enzyme.
Full text
PDF
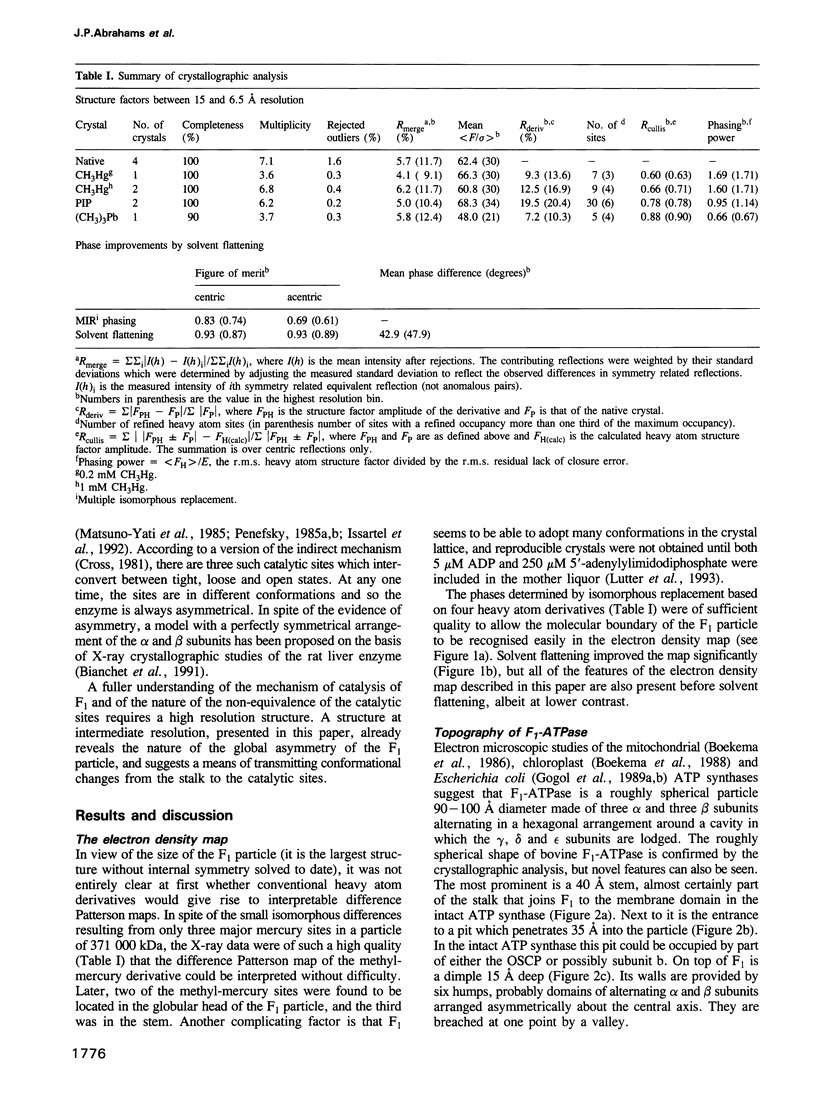

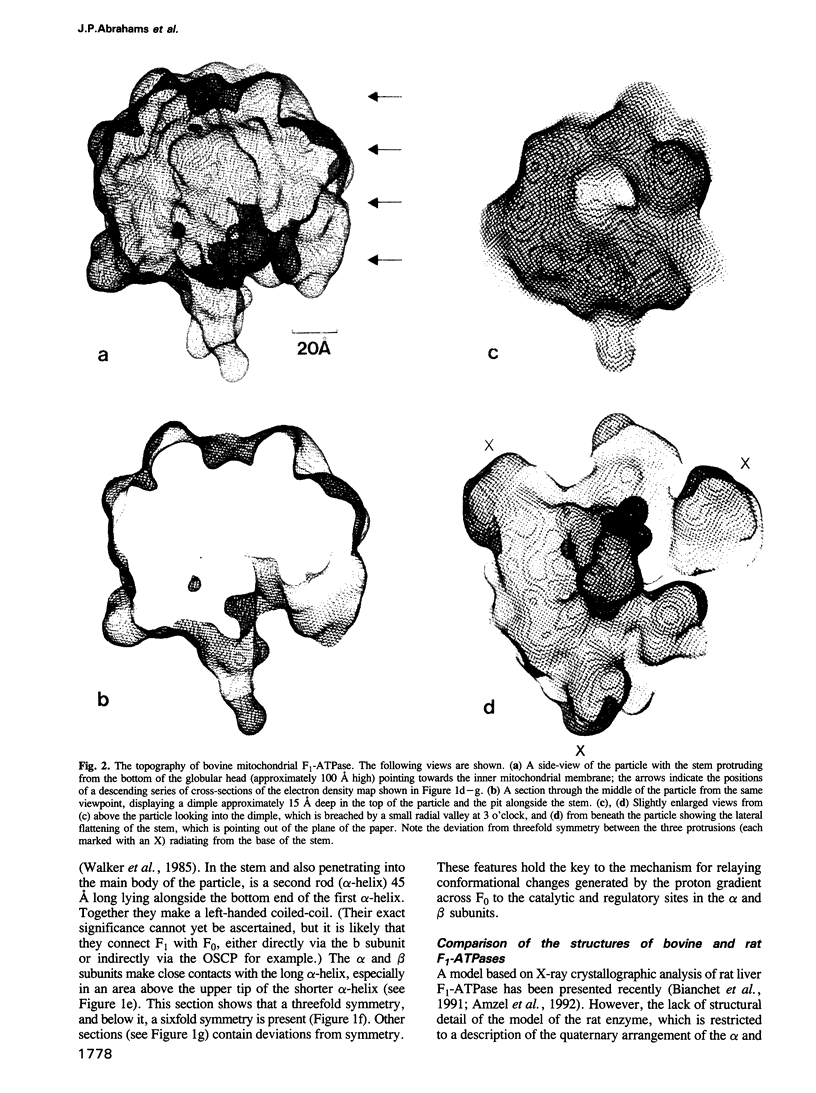


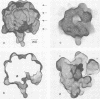
Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Amzel L. M., Bianchet M. A., Pedersen P. L. Quaternary structure of ATP synthases: symmetry and asymmetry in the F1 moiety. J Bioenerg Biomembr. 1992 Oct;24(5):429–433. doi: 10.1007/BF00762358. [DOI] [PubMed] [Google Scholar]
- Berden J. A., Hartog A. F., Edel C. M. Hydrolysis of ATP by F1 can be described only on the basis of a dual-site mechanism. Biochim Biophys Acta. 1991 Mar 29;1057(2):151–156. doi: 10.1016/s0005-2728(05)80099-4. [DOI] [PubMed] [Google Scholar]
- Bianchet M., Ysern X., Hullihen J., Pedersen P. L., Amzel L. M. Mitochondrial ATP synthase. Quaternary structure of the F1 moiety at 3.6 A determined by x-ray diffraction analysis. J Biol Chem. 1991 Nov 5;266(31):21197–21201. [PubMed] [Google Scholar]
- Boekema E. J., Berden J. A., van Heel M. G. Structure of mitochondrial F1-ATPase studied by electron microscopy and image processing. Biochim Biophys Acta. 1986 Oct 8;851(3):353–360. doi: 10.1016/0005-2728(86)90071-x. [DOI] [PubMed] [Google Scholar]
- Boyer P. D. A perspective of the binding change mechanism for ATP synthesis. FASEB J. 1989 Aug;3(10):2164–2178. doi: 10.1096/fasebj.3.10.2526771. [DOI] [PubMed] [Google Scholar]
- Cross R. L. The mechanism and regulation of ATP synthesis by F1-ATPases. Annu Rev Biochem. 1981;50:681–714. doi: 10.1146/annurev.bi.50.070181.003341. [DOI] [PubMed] [Google Scholar]
- Dimroth P. Mechanisms of sodium transport in bacteria. Philos Trans R Soc Lond B Biol Sci. 1990 Jan 30;326(1236):465–477. doi: 10.1098/rstb.1990.0025. [DOI] [PubMed] [Google Scholar]
- Dunn S. D. The polar domain of the b subunit of Escherichia coli F1F0-ATPase forms an elongated dimer that interacts with the F1 sector. J Biol Chem. 1992 Apr 15;267(11):7630–7636. [PubMed] [Google Scholar]
- FERNANDEZ-MORAN H. Cell-membrane ultrastructure. Low-temperature electron microsopy and x-ray diffraction studies of lipoprotein components in lamellar systems. Circulation. 1962 Nov;26:1039–1065. doi: 10.1161/01.cir.26.5.1039. [DOI] [PubMed] [Google Scholar]
- Gogol E. P., Aggeler R., Sagermann M., Capaldi R. A. Cryoelectron microscopy of Escherichia coli F1 adenosinetriphosphatase decorated with monoclonal antibodies to individual subunits of the complex. Biochemistry. 1989 May 30;28(11):4717–4724. doi: 10.1021/bi00437a031. [DOI] [PubMed] [Google Scholar]
- Gogol E. P., Lücken U., Bork T., Capaldi R. A. Molecular architecture of Escherichia coli F1 adenosinetriphosphatase. Biochemistry. 1989 May 30;28(11):4709–4716. doi: 10.1021/bi00437a030. [DOI] [PubMed] [Google Scholar]
- Issartel J. P., Dupuis A., Garin J., Lunardi J., Michel L., Vignais P. V. The ATP synthase (F0-F1) complex in oxidative phosphorylation. Experientia. 1992 Apr 15;48(4):351–362. doi: 10.1007/BF01923429. [DOI] [PubMed] [Google Scholar]
- Jones T. A., Zou J. Y., Cowan S. W., Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr A. 1991 Mar 1;47(Pt 2):110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- Kagawa Y., Racker E. Partial resolution of the enzymes catalyzing oxidative phosphorylation. IX. Reconstruction of oligomycin-sensitive adenosine triphosphatase. J Biol Chem. 1966 May 25;241(10):2467–2474. [PubMed] [Google Scholar]
- Knowles A. F., Guillory R. J., Racker E. Partial resolution of the enzymes catalyzing oxidative phosphorylation. XXIV. A factor required for the binding of mitochondrial adenosine triphosphatase to the inner mitochondrial membrane. J Biol Chem. 1971 Apr 25;246(8):2672–2679. [PubMed] [Google Scholar]
- Lien S., Racker E. Partial resolution of the enzymes catalyzing photophosphorylation. 8. Properties of silicotungstate-treated subchloroplast particles. J Biol Chem. 1971 Jul 10;246(13):4298–4307. [PubMed] [Google Scholar]
- Lutter R., Abrahams J. P., van Raaij M. J., Todd R. J., Lundqvist T., Buchanan S. K., Leslie A. G., Walker J. E. Crystallization of F1-ATPase from bovine heart mitochondria. J Mol Biol. 1993 Feb 5;229(3):787–790. doi: 10.1006/jmbi.1993.1081. [DOI] [PubMed] [Google Scholar]
- Matsuno-Yagi A., Yagi T., Hatefi Y. Studies on the mechanism of oxidative phosphorylation: effects of specific F0 modifiers on ligand-induced conformation changes of F1. Proc Natl Acad Sci U S A. 1985 Nov;82(22):7550–7554. doi: 10.1073/pnas.82.22.7550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell P. A chemiosmotic molecular mechanism for proton-translocating adenosine triphosphatases. FEBS Lett. 1974 Jul 15;43(2):189–194. doi: 10.1016/0014-5793(74)80997-x. [DOI] [PubMed] [Google Scholar]
- PULLMAN M. E., MONROY G. C. A NATURALLY OCCURRING INHIBITOR OF MITOCHONDRIAL ADENOSINE TRIPHOSPHATASE. J Biol Chem. 1963 Nov;238:3762–3769. [PubMed] [Google Scholar]
- PULLMAN M. E., PENEFSKY H. S., DATTA A., RACKER E. Partial resolution of the enzymes catalyzing oxidative phosphorylation. I. Purification and properties of soluble dinitrophenol-stimulated adenosine triphosphatase. J Biol Chem. 1960 Nov;235:3322–3329. [PubMed] [Google Scholar]
- Penefsky H. S. Mechanism of inhibition of mitochondrial adenosine triphosphatase by dicyclohexylcarbodiimide and oligomycin: relationship to ATP synthesis. Proc Natl Acad Sci U S A. 1985 Mar;82(6):1589–1593. doi: 10.1073/pnas.82.6.1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penefsky H. S. Reaction mechanism of the membrane-bound ATPase of submitochondrial particles from beef heart. J Biol Chem. 1985 Nov 5;260(25):13728–13734. [PubMed] [Google Scholar]
- Schnebli H. P., Vatter A. E., Abrams A. Membrane adenosine triphosphatase from Streptococcus faecalis. Molecular weight, subunit structure, and amino acid composition. J Biol Chem. 1970 Mar 10;245(5):1122–1127. [PubMed] [Google Scholar]
- Senior A. E. ATP synthesis by oxidative phosphorylation. Physiol Rev. 1988 Jan;68(1):177–231. doi: 10.1152/physrev.1988.68.1.177. [DOI] [PubMed] [Google Scholar]
- Tzagoloff A., Maclennan D. H., Byington K. H. Studies on the mitochondrial adenosine triphosphatase system. 3. Isolation from the oligomycin-sensitive adenosine triphosphatase complex of the factors which bind F-1 and determine oligomycin sensitivity of bound F-1. Biochemistry. 1968 Apr;7(4):1596–1602. doi: 10.1021/bi00844a049. [DOI] [PubMed] [Google Scholar]
- WILLIAMS R. J. Possible functions of chains of catalysts. J Theor Biol. 1961 Jan;1:1–17. doi: 10.1016/0022-5193(61)90023-6. [DOI] [PubMed] [Google Scholar]
- Walker J. E., Fearnley I. M., Gay N. J., Gibson B. W., Northrop F. D., Powell S. J., Runswick M. J., Saraste M., Tybulewicz V. L. Primary structure and subunit stoichiometry of F1-ATPase from bovine mitochondria. J Mol Biol. 1985 Aug 20;184(4):677–701. doi: 10.1016/0022-2836(85)90313-4. [DOI] [PubMed] [Google Scholar]
- Walker J. E., Fearnley I. M., Lutter R., Todd R. J., Runswick M. J. Structural aspects of proton-pumping ATPases. Philos Trans R Soc Lond B Biol Sci. 1990 Jan 30;326(1236):367–378. doi: 10.1098/rstb.1990.0018. [DOI] [PubMed] [Google Scholar]
- Walker J. E., Lutter R., Dupuis A., Runswick M. J. Identification of the subunits of F1F0-ATPase from bovine heart mitochondria. Biochemistry. 1991 Jun 4;30(22):5369–5378. doi: 10.1021/bi00236a007. [DOI] [PubMed] [Google Scholar]
- Walker J. E., Runswick M. J., Poulter L. ATP synthase from bovine mitochondria. The characterization and sequence analysis of two membrane-associated sub-units and of the corresponding cDNAs. J Mol Biol. 1987 Sep 5;197(1):89–100. doi: 10.1016/0022-2836(87)90611-5. [DOI] [PubMed] [Google Scholar]
- Walker J. E., Saraste M., Gay N. J. E. coli F1-ATPase interacts with a membrane protein component of a proton channel. Nature. 1982 Aug 26;298(5877):867–869. doi: 10.1038/298867a0. [DOI] [PubMed] [Google Scholar]
- Walker J. E., Saraste M., Runswick M. J., Gay N. J. Distantly related sequences in the alpha- and beta-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1982;1(8):945–951. doi: 10.1002/j.1460-2075.1982.tb01276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B. C. Resolution of phase ambiguity in macromolecular crystallography. Methods Enzymol. 1985;115:90–112. doi: 10.1016/0076-6879(85)15009-3. [DOI] [PubMed] [Google Scholar]
- Williams R. J. The history and the hypotheses concerning ATP-formation by energised protons. FEBS Lett. 1978 Jan 1;85(1):9–19. doi: 10.1016/0014-5793(78)81238-1. [DOI] [PubMed] [Google Scholar]