Abstract
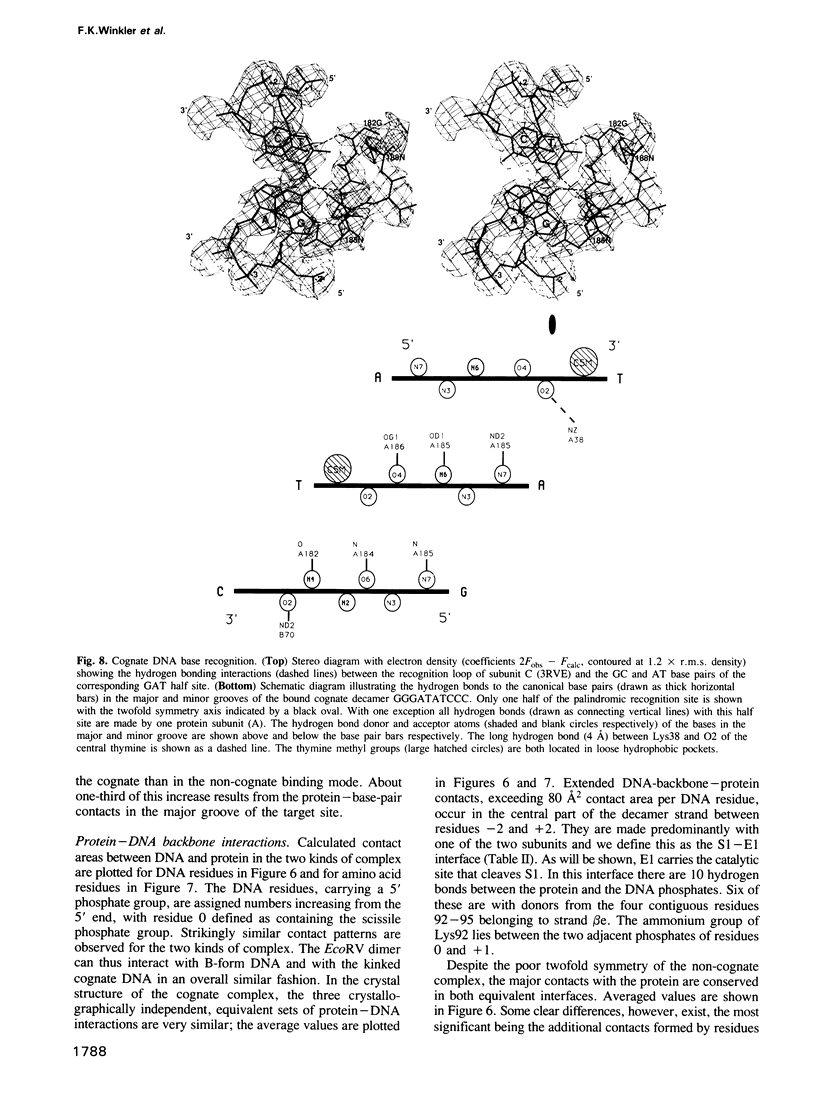
The crystal structure of EcoRV endonuclease has been determined at 2.5 A resolution and that of its complexes with the cognate DNA decamer GGGATATCCC (recognition sequence underlined) and the non-cognate DNA octamer CGAGCTCG at 3.0 A resolution. Two octamer duplexes of the non-cognate DNA, stacked end-to-end, are bound to the dimeric enzyme in B-DNA-like conformations. The protein--DNA interactions of this complex are prototypic for non-specific DNA binding. In contrast, only one cognate decamer duplex is bound and deviates considerably from canonical B-form DNA. Most notably, a kink of approximately 50 degrees is observed at the central TA step with a concomitant compression of the major groove. Base-specific hydrogen bonds between the enzyme and the recognition base pairs occur exclusively in the major groove. These interactions appear highly co-operative as they are all made through one short surface loop comprising residues 182-186. Numerous contacts with the sugar phosphate backbone extending beyond the recognition sequence are observed in both types of complex. However, the total surface area buried on complex formation is > 1800 A2 larger in the case of cognate DNA binding. Two acidic side chains, Asp74 and Asp90, are close to the reactive phosphodiester group in the cognate complex and most probably provide oxygen ligands for binding the essential cofactor Mg2+. An important role is also indicated for Lys92, which together with the two acidic functions appears to be conserved in the otherwise unrelated structure of EcoRI endonuclease. The structural results give new insight into the physical basis of the remarkable sequence specificity of this enzyme.
Full text
PDF








Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Beese L. S., Steitz T. A. Structural basis for the 3'-5' exonuclease activity of Escherichia coli DNA polymerase I: a two metal ion mechanism. EMBO J. 1991 Jan;10(1):25–33. doi: 10.1002/j.1460-2075.1991.tb07917.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett S. P., Halford S. E. Recognition of DNA by type II restriction enzymes. Curr Top Cell Regul. 1989;30:57–104. doi: 10.1016/b978-0-12-152830-0.50005-0. [DOI] [PubMed] [Google Scholar]
- Bernstein F. C., Koetzle T. F., Williams G. J., Meyer E. F., Jr, Brice M. D., Rodgers J. R., Kennard O., Shimanouchi T., Tasumi M. The Protein Data Bank: a computer-based archival file for macromolecular structures. J Mol Biol. 1977 May 25;112(3):535–542. doi: 10.1016/s0022-2836(77)80200-3. [DOI] [PubMed] [Google Scholar]
- Bougueleret L., Schwarzstein M., Tsugita A., Zabeau M. Characterization of the genes coding for the Eco RV restriction and modification system of Escherichia coli. Nucleic Acids Res. 1984 Apr 25;12(8):3659–3676. doi: 10.1093/nar/12.8.3659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brünger A. T., Krukowski A., Erickson J. W. Slow-cooling protocols for crystallographic refinement by simulated annealing. Acta Crystallogr A. 1990 Jul 1;46(Pt 7):585–593. doi: 10.1107/s0108767390002355. [DOI] [PubMed] [Google Scholar]
- Connolly B. A., Eckstein F., Pingoud A. The stereochemical course of the restriction endonuclease EcoRI-catalyzed reaction. J Biol Chem. 1984 Sep 10;259(17):10760–10763. [PubMed] [Google Scholar]
- D'Arcy A., Brown R. S., Zabeau M., van Resandt R. W., Winkler F. K. Purification and crystallization of the EcoRV restriction endonuclease. J Biol Chem. 1985 Feb 25;260(4):1987–1990. [PubMed] [Google Scholar]
- Delcourt S. G., Blake R. D. Stacking energies in DNA. J Biol Chem. 1991 Aug 15;266(23):15160–15169. [PubMed] [Google Scholar]
- Fliess A., Wolfes H., Seela F., Pingoud A. Analysis of the recognition mechanism involved in the EcoRV catalyzed cleavage of DNA using modified oligodeoxynucleotides. Nucleic Acids Res. 1988 Dec 23;16(24):11781–11793. doi: 10.1093/nar/16.24.11781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freemont P. S., Lane A. N., Sanderson M. R. Structural aspects of protein-DNA recognition. Biochem J. 1991 Aug 15;278(Pt 1):1–23. doi: 10.1042/bj2780001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glusker J. P. Structural aspects of metal liganding to functional groups in proteins. Adv Protein Chem. 1991;42:1–76. doi: 10.1016/s0065-3233(08)60534-3. [DOI] [PubMed] [Google Scholar]
- Grasby J. A., Connolly B. A. Stereochemical outcome of the hydrolysis reaction catalyzed by the EcoRV restriction endonuclease. Biochemistry. 1992 Sep 1;31(34):7855–7861. doi: 10.1021/bi00149a016. [DOI] [PubMed] [Google Scholar]
- Harrison S. C. A structural taxonomy of DNA-binding domains. Nature. 1991 Oct 24;353(6346):715–719. doi: 10.1038/353715a0. [DOI] [PubMed] [Google Scholar]
- Heitman J. How the EcoRI endonuclease recognizes and cleaves DNA. Bioessays. 1992 Jul;14(7):445–454. doi: 10.1002/bies.950140704. [DOI] [PubMed] [Google Scholar]
- Jeltsch A., Alves J., Maass G., Pingoud A. On the catalytic mechanism of EcoRI and EcoRV. A detailed proposal based on biochemical results, structural data and molecular modelling. FEBS Lett. 1992 Jun 8;304(1):4–8. doi: 10.1016/0014-5793(92)80576-3. [DOI] [PubMed] [Google Scholar]
- Jenkins J., Janin J., Rey F., Chiadmi M., van Tilbeurgh H., Lasters I., De Maeyer M., Van Belle D., Wodak S. J., Lauwereys M. Protein engineering of xylose (glucose) isomerase from Actinoplanes missouriensis. 1. Crystallography and site-directed mutagenesis of metal binding sites. Biochemistry. 1992 Jun 23;31(24):5449–5458. doi: 10.1021/bi00139a005. [DOI] [PubMed] [Google Scholar]
- Jones T. A. Diffraction methods for biological macromolecules. Interactive computer graphics: FRODO. Methods Enzymol. 1985;115:157–171. doi: 10.1016/0076-6879(85)15014-7. [DOI] [PubMed] [Google Scholar]
- Kabsch W., Sander C. Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers. 1983 Dec;22(12):2577–2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
- Kennard O., Hunter W. N. Oligonucleotide structure: a decade of results from single crystal X-ray diffraction studies. Q Rev Biophys. 1989 Aug;22(3):327–379. doi: 10.1017/s0033583500002997. [DOI] [PubMed] [Google Scholar]
- Kim Y. C., Grable J. C., Love R., Greene P. J., Rosenberg J. M. Refinement of Eco RI endonuclease crystal structure: a revised protein chain tracing. Science. 1990 Sep 14;249(4974):1307–1309. doi: 10.1126/science.2399465. [DOI] [PubMed] [Google Scholar]
- Lavery R., Sklenar H. Defining the structure of irregular nucleic acids: conventions and principles. J Biomol Struct Dyn. 1989 Feb;6(4):655–667. doi: 10.1080/07391102.1989.10507728. [DOI] [PubMed] [Google Scholar]
- Mazzarelli J., Scholtissek S., McLaughlin L. W. Effects of functional group changes in the EcoRV recognition site on the cleavage reaction catalyzed by the endonuclease. Biochemistry. 1989 May 30;28(11):4616–4622. doi: 10.1021/bi00437a016. [DOI] [PubMed] [Google Scholar]
- Newman P. C., Williams D. M., Cosstick R., Seela F., Connolly B. A. Interaction of the EcoRV restriction endonuclease with the deoxyadenosine and thymidine bases in its recognition hexamer d(GATATC). Biochemistry. 1990 Oct 23;29(42):9902–9910. doi: 10.1021/bi00494a021. [DOI] [PubMed] [Google Scholar]
- Record M. T., Jr, deHaseth P. L., Lohman T. M. Interpretation of monovalent and divalent cation effects on the lac repressor-operator interaction. Biochemistry. 1977 Nov 1;16(22):4791–4796. doi: 10.1021/bi00641a005. [DOI] [PubMed] [Google Scholar]
- Roberts R. J., Macelis D. Restriction enzymes and their isoschizomers. Nucleic Acids Res. 1992 May 11;20 (Suppl):2167–2180. doi: 10.1093/nar/20.suppl.2167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz S. C., Shields G. C., Steitz T. A. Crystal structure of a CAP-DNA complex: the DNA is bent by 90 degrees. Science. 1991 Aug 30;253(5023):1001–1007. doi: 10.1126/science.1653449. [DOI] [PubMed] [Google Scholar]
- Selent U., Rüter T., Köhler E., Liedtke M., Thielking V., Alves J., Oelgeschläger T., Wolfes H., Peters F., Pingoud A. A site-directed mutagenesis study to identify amino acid residues involved in the catalytic function of the restriction endonuclease EcoRV. Biochemistry. 1992 May 26;31(20):4808–4815. doi: 10.1021/bi00135a010. [DOI] [PubMed] [Google Scholar]
- Steitz T. A. Structural studies of protein-nucleic acid interaction: the sources of sequence-specific binding. Q Rev Biophys. 1990 Aug;23(3):205–280. doi: 10.1017/s0033583500005552. [DOI] [PubMed] [Google Scholar]
- Taylor J. D., Badcoe I. G., Clarke A. R., Halford S. E. EcoRV restriction endonuclease binds all DNA sequences with equal affinity. Biochemistry. 1991 Sep 10;30(36):8743–8753. doi: 10.1021/bi00100a005. [DOI] [PubMed] [Google Scholar]
- Taylor J. D., Halford S. E. Discrimination between DNA sequences by the EcoRV restriction endonuclease. Biochemistry. 1989 Jul 25;28(15):6198–6207. doi: 10.1021/bi00441a011. [DOI] [PubMed] [Google Scholar]
- Thielking V., Alves J., Fliess A., Maass G., Pingoud A. Accuracy of the EcoRI restriction endonuclease: binding and cleavage studies with oligodeoxynucleotide substrates containing degenerate recognition sequences. Biochemistry. 1990 May 15;29(19):4682–4691. doi: 10.1021/bi00471a024. [DOI] [PubMed] [Google Scholar]
- Thielking V., Selent U., Köhler E., Landgraf A., Wolfes H., Alves J., Pingoud A. Mg2+ confers DNA binding specificity to the EcoRV restriction endonuclease. Biochemistry. 1992 Apr 21;31(15):3727–3732. doi: 10.1021/bi00130a001. [DOI] [PubMed] [Google Scholar]
- Thielking V., Selent U., Köhler E., Wolfes H., Pieper U., Geiger R., Urbanke C., Winkler F. K., Pingoud A. Site-directed mutagenesis studies with EcoRV restriction endonuclease to identify regions involved in recognition and catalysis. Biochemistry. 1991 Jul 2;30(26):6416–6422. doi: 10.1021/bi00240a011. [DOI] [PubMed] [Google Scholar]
- Vermote C. L., Halford S. E. EcoRV restriction endonuclease: communication between catalytic metal ions and DNA recognition. Biochemistry. 1992 Jul 7;31(26):6082–6089. doi: 10.1021/bi00141a018. [DOI] [PubMed] [Google Scholar]
- Vermote C. L., Vipond I. B., Halford S. E. EcoRV restriction endonuclease: communication between DNA recognition and catalysis. Biochemistry. 1992 Jul 7;31(26):6089–6097. doi: 10.1021/bi00141a019. [DOI] [PubMed] [Google Scholar]
- Wang B. C. Resolution of phase ambiguity in macromolecular crystallography. Methods Enzymol. 1985;115:90–112. doi: 10.1016/0076-6879(85)15009-3. [DOI] [PubMed] [Google Scholar]
- Wilmot C. M., Thornton J. M. Analysis and prediction of the different types of beta-turn in proteins. J Mol Biol. 1988 Sep 5;203(1):221–232. doi: 10.1016/0022-2836(88)90103-9. [DOI] [PubMed] [Google Scholar]
- Winkler F. K., D'Arcy A., Blöcker H., Frank R., van Boom J. H. Crystallization of complexes of EcoRV endonuclease with cognate and non-cognate DNA fragments. J Mol Biol. 1991 Jan 20;217(2):235–238. doi: 10.1016/0022-2836(91)90536-f. [DOI] [PubMed] [Google Scholar]
- Zebala J. A., Choi J., Barany F. Characterization of steady state, single-turnover, and binding kinetics of the TaqI restriction endonuclease. J Biol Chem. 1992 Apr 25;267(12):8097–8105. [PubMed] [Google Scholar]
- van Tilbeurgh H., Jenkins J., Chiadmi M., Janin J., Wodak S. J., Mrabet N. T., Lambeir A. M. Protein engineering of xylose (glucose) isomerase from Actinoplanes missouriensis. 3. Changing metal specificity and the pH profile by site-directed mutagenesis. Biochemistry. 1992 Jun 23;31(24):5467–5471. doi: 10.1021/bi00139a007. [DOI] [PubMed] [Google Scholar]