Abstract
Identifying drug-drug interactions is an important and challenging problem in computational biology and healthcare research. There are accurate, structured but limited domain knowledge and noisy, unstructured but abundant textual information available for building predictive models. The difficulty lies in mining the true patterns embedded in text data and developing efficient and effective ways to combine heterogenous types of information. We demonstrate a novel approach of leveraging augmented text-mining features to build a logistic regression model with improved prediction performance (in terms of discrimination and calibration). Our model based on synthesized features significantly outperforms the model trained with only structured features (AUC: 96% vs. 91%, Sensitivity: 90% vs. 82% and Specificity: 88% vs. 81%). Along with the quantitative results, we also show learned “latent topics”, an intermediary result of our text mining module, and discuss their implications.
Introduction
Drug-drug interactions (DDIs) can lead to serious adverse events, and are a major cause of morbidity and mortality. Predicting or detecting DDIs is therefore of concern to the pharmaceutical industry, drug regulatory agencies, healthcare professionals and patients [1]. Unfortunately, predicting DDIs is a nontrivial task, and is becoming increasingly challenging as more new drugs are being developed [2].
DDIs arise when the pharmacokinetics or pharmacodynamic properties of one drug are altered by other drugs. A number of DDI-prediction works focus on careful evaluation of molecular targets and metabolizing enzymes (e.g., P450 enzymes) [3] [4][5]. However, such methods rely on expensive experiments and are limited by their relatively small scale as they usually focus on a few drug pairs or a limited number of metabolizing enzymes a time. Some other works perform domain-specific studies on DDIs (e.g., for anesthesia [6], for inhibition of intestinal CYP3A4 or P-glycoprotein [7], and for calcium channel blockers [8]). Unfortunately it is difficult to generalize the knowledge to predict potential DDIs in a different context. Recently, a number of works have leveraged data mining and statistical methods to detect DDIs. These methods mine and analyze post-market data, such as spontaneous reports, insurance claim databases or electronic medical records [9][10][11]. As the time for sufficient post-market evidence to accumulate can be years, these methods do not provide timely predictions at the early stages of drug discovery and development.
There is a need for inexpensive, general and scalable DDI prediction methods that are not dependent on post-market data in order to provide early stage predictions. In this paper, we leverage large scale text mining and statistical inference techniques to achieve the above goals. We demonstrate that text mining techniques can augment existing domain knowledge (in structured format) by retrieving useful information from unstructured text data and synthesizing weak signals to provide strong evidence. The aim of this study was to develop a novel data-driven text mining technique, which is robust to noise, for predicting DDIs based on publicly available information.
Related Work
Various systems for identifying DDIs have been developed. Norén et al. implemented and evaluated a shrinkage observed-to-expected ratio for exploratory analysis of suspected drug-drug interaction in individual case safety reports, based on a comparison with an additive risk model [11]. Hu et al. presented a systematic overview of the available drug interaction information using a network approach [12]. Tari et al. integrated text mining and automated reasoning to derive DDIs [13]. Takarabe et al. investigated the relations between the drug groups and drug interaction mechanisms or symptoms using a drug interaction network based on the Anatomical Therapeutic Chemical (ATC) classification [14]. Tatonetti et al. used the FDA's Adverse Event Reporting System to build profiles and looked for pairs of drugs that match these single-drug profiles in order to predict potential interactions [15].
As opposed to approaches that only use structured data, our method is largely based on text mining. A similar work to ours is the recent paper [16] by Percha et al. to discover and explain drug-drug interactions via text mining the Medline database, a respected source of citations of peer-reviewed biomedical literature. This approach focused on the drug-gene relations and cross-linked them to obtain drug-drug interactions. We hypothesized that accurate identification of DDIs involves more factors and developed a novel method that goes beyond a single type of entity relation to explore a rich set of entity relations.
Main Proposal
We model the DDI prediction problem as a binary classification task, where a query drug pair is classified as “1” if the two drugs interact with each other, and “0” otherwise. We start by collecting DDI ground truth for model training and evaluation. In this work, we collected the ground truth data from DrugBank. Next, we collect features of each individual drug. Such features include both accurate but limited domain knowledge from structured data, and noisy but abundant information based on text mining results. Then we express a drug pair by merging the features of the two drugs into one feature vector, and train a binary classifier accordingly. At query time, given one drug pair query, the trained model predicts “1” or “0” based on the probability that the two drugs will interact.
A. Data Preparation
We downloaded DrugBank data to collect our DDI ground truth for the purpose of training and evaluation. DrugBank contains 6,710 drugs, which make up the collection of drugs under consideration in this work. We randomly sample 5,000 drug pairs from a list of known interacting drug pairs to be used as positive training examples, and additional 5,000 drugs pairs that do not interact according to DrugBank as negative training examples. When evaluating the prediction accuracy of the model, we randomly sample positive and negative drug pairs from DrugBank in a similar fashion (5,000 of each), while ensuring that none of the evaluation drug pairs were used for training.
We collected Medline abstracts over a 5-year period (published from 2006 to 2010) as the corpus for mining drug-related information. This corpus contains more than 3.6 million abstracts, with over 0.7 million per year.
B. Model a Drug with Structured Information
Existing work has shown that drug targets [17] and the molecular structure similarity analysis [18][19] of drugs provide useful information in DDI prediction. We therefore choose drug targets and molecular structures as two types of structured domain knowledge explored in this work. Concretely, using a bag-of-words model (widely used in Information Retrieval) to incorporate the target information, we represent each drug as a binary-valued vector, for which every element is a unique target. The value 1 means that the target is associated with a given drug, and 0 means no association. The drug-target association information is downloaded from DrugBank. The total number of unique targets is 3,573. If the target information of a drug is not available, the corresponding drug-target vector is all 0s.
Similarly, to incorporate the molecular structure information, we use the bag-of-words model to represent each drug as a binary-valued vector, for which every element is a unique substructure1. The drug substructure information is downloaded from DrugBank. The total number of unique substructures is 309. If the substructure information of a drug is not available in DrugBank, then the corresponding drug-substructure vector is all 0s.
C. Drug-related Textual Information
To augment the aforementioned structured information, we resort to textual data to retrieve additional drug features. We extract three types of textual information from each Medline abstract. The first type of information is genes (in the form of gene names or gene symbols). We build a gene annotator based on the conditional random field (CRF) technique [20] to extract all the appearances of gene names/symbols from text. After information extraction, a normalization step follows to assign all the name/symbol variations of one gene to a unique gene ID2. We then record the number of occurrences of each unique gene in every abstract.
The second type of information is disease names. We use a dictionary lookup-based method to extract disease names. To compose a disease dictionary, we merge the PharmGKB [21] disease dictionary with the OBO disease ontology [22] that ends up with 10,397 disease names. We record the number of times that a disease is mentioned in each abstract. The synonyms are captured in the normalization step, where all the synonyms of one disease are assigned to a unique disease ID.
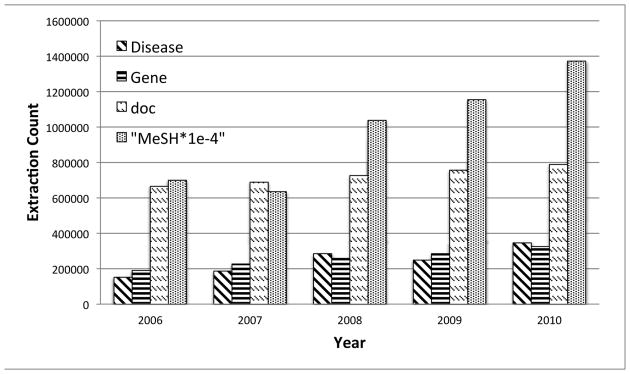
The third type of information is MeSH concepts. MeSH concepts are provided by PubMed3 along with the Medline abstracts. Such concepts are manually curated by domain experts to support various types of search over the PubMed database. We use concepts from the four MeSH subtrees (A, B, C, and D) which are most related to drug interactions. Figure 1 shows the increases in size of the Medline corpus over the five-year period and the sizes of the three types of extracted information for each year.
Fig. 1. Size of different types of extracted information (The size for MeSH is downscaled by 1e-4 to fit in the graph).
D. Model a Drug with Textual Information
We address the problem of identifying drug-entity association based on the semantics of drug-entity relations and propose a Drug-Entity-Topic (DET) model, which is an extension of the Latent Dirichlet Allocation (LDA) [23] topic model that has shown great success in the text mining domain. Concretely, DET is a generative statistical graphical model that captures the relation between drugs and other entities by explicitly modeling the latent semantics of entities.
Let us assume disease is our focus entity for easy presentation. The same technique is applicable to genes and MeSH concepts. For each Medline document dori, we build an artificial document, d = {ad, ed}|entity (e.g., entity = disease) that contains two sections: a subject section ad and a content section ed. A “word” is the basic component of the document. A word in section ad is a drug name that is extracted from the original Medline document dori. The word repeats n times if the drug is mentioned n times in dori. A word in section ed is a disease name that is extracted from dori. Similarly, the word repeats the same number of times as the number of times that the disease is found in dori. Conceptually, we consider the artificial document d to be talking about drugs, but the content of d is entirely conveyed by diseases.
In a DET model, given a document d, the collection of drugs in ad determines the document content that is observable as a list of diseases. We assume the existence of K disease topics that summarize the latent semantics of diseases, and use variable z to represent the topics. Each drug is associated with a statistical distribution over the topics. We treat latent topics as drug features, so that our goal is then to learn the probability distribution of the drugs given the latent topics p(drug|z). Note that by Bayes's rule:
| (1) |
The right-hand side can be directly estimated in the DET model. Interested readers are referred to [24] Appendix B for technical details.
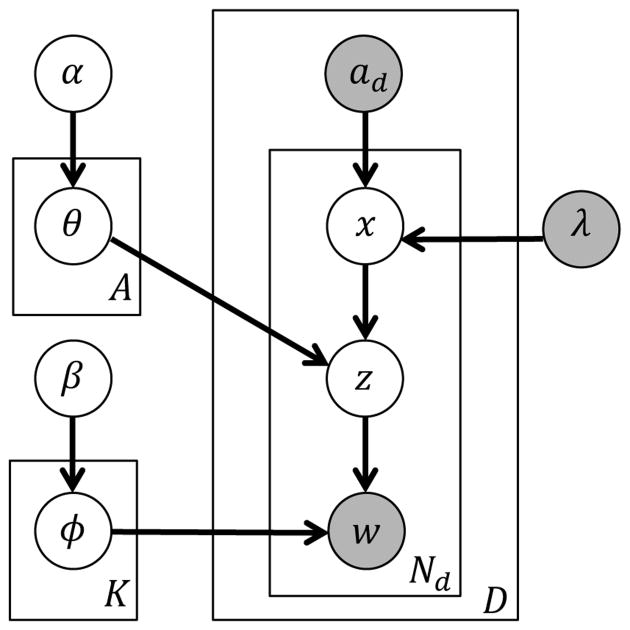
This generative process of the DET model can be represented as the hierarchical Bayesian model shown in Figure 2 using plate notations. Interested readers are referred to [23] for details of plate notation of topic models. In the model, latent variables are light-colored while the observed ones are shadowed. D is the number of documents of a corpus. w denotes a specific disease word observed in document d and Nd is the number of disease words in d. K and A represent the number of topics and drugs. ad is the observed set of drugs for d. In the DET model, each drug x is a probabilistic multinomial distribution over topics parameterized by θ. The observable parameter λ controls the sampling of drugs. Given a drug, each topic z is a multinomial distribution over disease words parameterized by ϕ. The prior distributions of topics and disease words follow Dirichlets parameterized respectively by α and β. To “generate” a document d, a drug x is sampled from ad, a topic z is then chosen based on the drug-topic distribution θ, and finally a “disease word” w is chosen based on the topic-word distribution ϕ corresponding to the chosen topic.
Fig. 2. Drug-Entity-Topic model (DET).
In order to learn the probability p(drug|z), we need to estimate model parameter θ. To accomplish this task we use Gibbs sampling, which is a an algorithm to approximate the joint distribution of multiple variables by drawing a sequence of samples.
E. Model Drug-Drug Relation
Given a pair of drugs q and r, we measure five types of relations. The structure relation is expressed as R(q, r)struct = {qstruct, rstruct}, where the structure vectors of the two drugs are concatenated with a fixed order. Similarly, the target relation is expressed as R(q, r)target = {qtarget, rtarget}. Drug-drug relations based on topic features can be handled in the same way. For example, by concatenating the disease-topic (dz) vectors of two drugs, we get the disease-topic relation expressed as R(q, r)dz = {p(q|z1) … p(q|zK), p(r|z1) … p(r|zK)}|disease. Drug-drug relations based on gene topics (gz) and MeSH concepts topics (mz) are constructed in the same way. The five types of relations are treated as five types of candidate features to model and predict DDIs. One can use all the features or a subset of the features in the predictive model as desired. Now the question is how to fit R(q, t)x x ∈ (struct, target, dz, gz, mz) features into a predictive model. We adopt the Binary Logic Regression model for this task. Let be an n-dimensional vector that represents the relation between drugs q and r. Logistic regression is able to learn a mapping of the form f: x → y, where y is a binary prediction. For example, in our problem, y = 1 means drugs q and r have interactions and y = 0 means no interaction between the two drugs is predicted.
Experiments
F. Experiment Setup, Parameter Setting and Evaluation Metrics
With data prepared as described in Section-A, for each year, we conduct 5 independent runs of the experiments and report the average of the 5 runs as the result. Experiment results over the five-year period are pretty consistent. We therefore report results of years 2006 and 2010 only (the boundary years) to be concise.
For the DET model, we set the number of topics for each entity type as K = 50. Following the convention for topic models, we set the hyper-parameters as α = 50/K, β = 0.01. Typically, the value of hyper-parameters does not influence model performance much.
We adopt sensitivity, specificity, accuracy, ROC curves and Area Under the Curve (AUC) to evaluate the performance of our proposed method. Sensitivity evaluates a method's ability to identify positives, which in our case are pairs of drugs that interact with each other. Specificity on the other hand measures a method's ability to identify negatives, pairs of drugs that have no interaction. Accuracy measures the percentage of correct predictions combining both the positives and negatives. All three metrics take values in [0, 1]. The ROC curve and AUC are widely used measurements for evaluating predictive models. For a better performing model the ROC curve will be closer to the upper-left corner in the ROC plot, and the AUC value will be closer to 1.
G. Examples of Topic and Drug Distributions
Tables I and II illustrate 6 topic examples that are learned by DET from the 2010 Medline corpus. Each topic is illustrated with: (a) the top 10 (most likely) entities conditioned on the topic, and (b) the top 10 drugs conditioned on the topic. Every result is reported with the probability value determined by DET. The three disease topics are about kidney disease, diabetes and depression. DET does a good job of identifying the latent semantics in entities. For example, topic 17 reveals a strong relation between vitamin D and kidney disease, which is correct. The topic also indicates there is a relation between kidney disease and hyperparathyroidism. Checking domain knowledge shows that (secondary) hyperparathyroidism is a consequence of having end-stage renal disease, which is kidney related. For each topic, the top 10 most likely drugs are also reasonably identified. For example, cinacalcet is used to treat secondary hyperparathyroidism, saxagliptin treats high blood sugar value in patients with type-2 diabetes, and desvenlafaxine is used for major depressive disorder. These validation demonstrates DET's power in identifying the latent semantics in large corpora. The three MeSH concept topics are about ecosystems, tomography and infection. Note that, as we mentioned before, text mining results are typically noisy. For example, we see disease “blind” ranked No. 4 in the topic about depression. However, as we will show later, our prediction solution is robust and can still benefit from such topic features.
Table I. Examples of disease topics (3 out of 50) and drugs learned from the 2010 Medline corpus.
| Topic 17 | Topic 30 | Topic 32 | |||
|---|---|---|---|---|---|
|
| |||||
| DISEASE | PROB. | DISEASE | PROB. | DISEASE | PROB. |
| kidney disease | 0.0906 | diabetes | 0.3285 | depression | 0.2310 |
| disease | 0.0643 | diabetes mellitus | 0.0661 | relapse | 0.0447 |
| vitamin d deficiency | 0.0411 | hyperglycemia | 0.0605 | major depressive disorder | 0.0385 |
| death | 0.0282 | type 2 diabetes mellitus | 0.0404 | blind | 0.0362 |
| stone | 0.0264 | hypoglycemia | 0.0380 | major depression | 0.0333 |
| end-stage renal disease | 0.0240 | diabetic nephropathy | 0.0230 | mental disorders | 0.0271 |
| renal failure | 0.0220 | proteinuria | 0.0210 | stress | 0.0264 |
| secondary hyperparathyroidism | 0.0220 | hypokalemia | 0.0173 | suicide | 0.0256 |
| hypocalcemia | 0.0219 | hyperglycaemia | 0.0160 | drug abuse | 0.0234 |
| hyperparathyroidism | 0.0211 | type 1 diabetes mellitus | 0.0149 | anxiety disorders | 0.0229 |
|
| |||||
| DRUG | PROB. | DRUG | PROB. | DRUG | PROB. |
|
| |||||
| Cinacalcet | 0.0626 | Streptozocin | 0.0730 | Fluoxetine | 0.0618 |
| Preotact | 0.0548 | Glucagon recombinant | 0.0458 | Escitalopram | 0.0589 |
| Sevelamer | 0.0473 | Exenatide | 0.0434 | Paroxetine | 0.0562 |
| Calcium carbonate | 0.0409 | Glyburide | 0.0419 | Citalopram | 0.0550 |
| Calcitriol | 0.0387 | Insulin Glargine | 0.0404 | Agomelatine | 0.0519 |
| Tamsulosin | 0.0327 | Insulin Aspart | 0.0394 | Sertraline | 0.0509 |
| Paricalcitol | 0.0270 | Saxagliptin | 0.0326 | Cocaine | 0.0502 |
| Cholecalciferol | 0.0236 | Sitagliptin | 0.0325 | Desvenlafaxine | 0.0492 |
| Ergocalciferol | 0.0221 | Glimepiride | 0.0300 | Nicotine | 0.0461 |
| Iohexol | 0.0195 | Acarbose | 0.0297 | Venlafaxine | 0.0434 |
Table II. Examples of MeSH topics (3 out of 50) and drugs learned from the 2010 Medline corpus.
| Topic 5 | Topic 7 | Topic 44 | |||
|---|---|---|---|---|---|
|
| |||||
| MeSH | PROB. | MeSH | PROB. | MeSH | PROB. |
| Animals | 0.0871 | Male | 0.0465 | Microbial Sensitivity Tests | 0.0531 |
| Ecosystem | 0.0196 | Humans | 0.0391 | Humans | 0.0419 |
| Seasons | 0.0167 | Tomography, X-Ray Computed | 0.0381 | Anti-Bacterial Agents | 0.0347 |
| Time Factors | 0.0114 | Animals | 0.0310 | Drug Resistance, Bacterial | 0.0218 |
| Environmental Monitoring | 0.0106 | Magnetic Resonance Imaging | 0.0249 | Staphylococcal Infections | 0.0141 |
| Species Specificity | 0.0093 | Female | 0.0248 | Male | 0.0138 |
| Insecticides | 0.0076 | Sensitivity and Specificity | 0.0192 | Antifungal Agents | 0.0123 |
| Plasmodium falciparum | 0.0075 | Diagnosis, Differential | 0.0159 | Methicillin-Resistant Staphylococcus aureus | 0.0103 |
| Photosynthesis | 0.0074 | Reproducibility of Results | 0.0139 | Cross Infection | 0.0088 |
| Biodiversity | 0.0071 | Contrast Media | 0.0125 | Drug Resistance, Multiple, Bacterial | 0.0087 |
|
| |||||
| DRUG | PROB. | DRUG | PROB. | DRUG | PROB. |
|
| |||||
| Sulfadoxine | 0.0470 | Gadobenate Dimeglumine | 0.0544 | Tigecycline | 0.0606 |
| Permethrin | 0.0422 | Gadofosveset trisodium | 0.0497 | Fluconazole | 0.0559 |
| Chlorophyll A | 0.0412 | Gadopentetate dimeglumine | 0.0457 | Linezolid | 0.0500 |
| Artemether | 0.0391 | Gadodiamide | 0.0398 | Daptomycin | 0.0499 |
| Nifedipine | 0.0359 | Gadobutrol | 0.0378 | Piperacillin | 0.0489 |
| Amodiaquine | 0.0319 | Diatrizoate | 0.0283 | Amikacin | 0.0478 |
| Astemizole | 0.0319 | Bretylium | 0.0268 | Ceftazidime | 0.0470 |
| Mefloquine | 0.0305 | Hydroxyurea | 0.0259 | Oxacillin | 0.0469 |
| Chloroquine | 0.0265 | Iohexol | 0.0226 | Ciprofloxacin | 0.0463 |
| Primaquine | 0.0256 | Ethiodized oil | 0.0216 | Levofloxacin | 0.0458 |
H. Performance Evaluation
We train eight DDI prediction models. The first (Target) and second (Structure) models use chemical structure information and known drug target information collected from DrugBank, respectively, as features. The third model (Structure+Target) uses both types of features in prediction. These models are based on structured domain knowledge only and do not use any text mining results. The fourth (Gene), fifth(Disease) and sixth(MeSH) models use gene topics, disease topics and MeSH concepts topics, respectively, as features. These three models use only a single type of text mining data as features without the guidance of any domain knowledge. The seventh (Combined text features) model uses all three types of textual features in prediction. In the last model (Combined all data), we combine domain knowledge and multiple types of textual features. The performance of each model is reported in Table III. Note that we used 0.5 as the cutoff for all eight models to calculate evaluation metrics. As the results illustrate, models using individual features generate less balanced prediction results. For example, the model based on drug target domain knowledge achieves high specificity (≈ 91%) but low sensitivity (≈ 47%). The model based on combined text mining data performs comparably to (a little better than) the model based on combined structured data. This shows that text mining is able to automatically extract useful information embedded in large corpora without relying on any domain knowledge or costly manual examination. As one might expect, the best DDI prediction is achieved when combining both domain knowledge in the form of structured data and text mining data. The best model achieves high prediction accuracy (8% more accurate than without using textual data) and produces highly balanced results (sensitivity=90%, specificity=88%).
Table III. Performance evaluation.
| 2006 | sensitivity | specificity | accuracy |
|---|---|---|---|
| Structure | 83.11% | 78.84% | 80.98% |
| Target | 46.29% | 91.10% | 68.69% |
| Structure+Target | 82.37% | 81.70% | 82.03% |
| Gene | 77.37% | 75.63% | 76.50% |
| Disease | 84.13% | 74.47% | 79.30% |
| MeSH | 74.47% | 87.85% | 81.16% |
| Combined text feature | 79.25% | 86.85% | 83.05% |
| Combined all data | 90.29% | 88.61% | 89.45% |
|
| |||
| 2010 | sensitivity | specificity | accuracy |
|
| |||
| Structure | 82.67% | 80.32% | 81.50% |
| Target | 47.47% | 91.38% | 69.43% |
| Structure+Target | 83.58% | 81.83% | 82.71% |
| Gene | 84.71% | 72.10% | 78.40% |
| Disease | 90.45% | 71.45% | 80.95% |
| MeSH | 73.31% | 92.54% | 82.93% |
| Combined text feature | 80.16% | 88.24% | 84.20% |
| Combined all data | 91.19% | 87.85% | 89.52% |
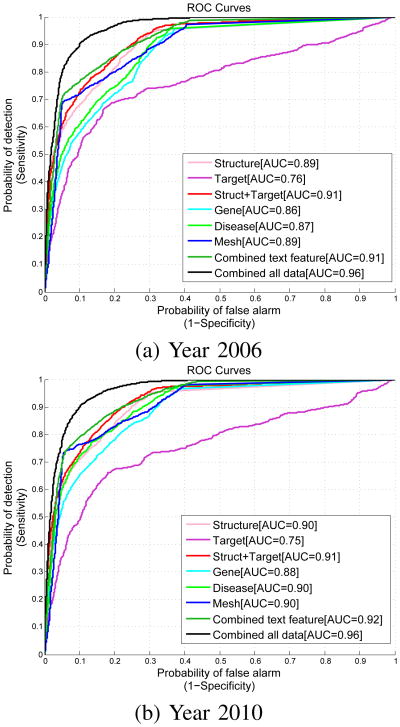
Because accuracy, sensitivity and specificity are calculated based on a single cutoff value, they might not truly reflect the performance of a predictive model in a comprehensive manner. We therefore evaluate the predictive models' discrimination and calibration performance by plotting ROC curves and calculate AUC to evaluate discrimination. Figure 3 shows the plots with AUC values reported. For all the evaluations, the model augmented with textual information (Combined all data) significantly outperforms all the other models. It outperforms the model using structured data only (Struct+Target) with a large margin near the upper-left corner. This observation validates our motivation for this work that text mining results, although noisy, can benefit DDI prediction significantly, as long as they are handled properly.
Fig. 3. Performance comparision between models trained on features retrieved from various sources. The model trained using combined data demonstrates significant advantages in both cases.
Discussion
Structured domain knowledge about drugs is limited in comparison to the abundant information available in text. In this paper, we introduce a novel solution to exploit a large amount of unstructured text data, identify intrinsic patterns, and use text mining to augment limited domain knowledge in predicting drug-drug interactions. Although noise is inevitable in automatically generated text mining data, our method is robust and capable of leveraging textual information to significantly improve DDI prediction performance. Our DET model is also able to extract the latent semantics (statistically) to model the relations between drugs and entities like diseases, genes, and MeSH concepts, providing a new way to explore large corpora in the biomedical domain.
Our study has limitations. First, the SVD-based feature reduction technique makes it difficult to interpret the results (i.e., the significance of the attributes) because reduced-dimension features do not preserve physical meanings. Second, we only focus on scientific literature in this study. For some commercial drugs that were not revealed to the academic community, only limited information can be mined. Third, we evaluated our results with a limited database containing only a fraction of confirmed drug-drug interactions. Despite these limitations, this pilot work still shows good promise to better detect DDIs with text mining techniques.
Conclusion and Future work
We demonstrated a novel approach of leveraging text-mining augmented features to build a logistic regression model with improved prediction performance (in terms of discrimination and calibration). There are several paths we would like to explore in our future work. We would like to investigate other drug-related textual data sources, such as patents. Due to the specialization in drug development research, some of the new drugs and new compounds are reported in patents only. We expect a performance improvement when patent information is considered. Second, some of the drug domain knowledge is semi-structured, such as a paragraph that describes the pharmacodynamics or mechanism of action, protein binding, or experimental properties of a drug in the DrugBank database. It is currently not clear how to best make use of such information automatically in building predictive models, and we plan to extend our method to incorporate such semi-structured data.
Acknowledgments
Xiaoqian Jiang is partially supported by 4R00LM011392 and U54HL108460.
Footnotes
The substructures of drug “Abacavir” include “Hydroxy Compounds”, “Alkanes and Alkenes”, “Aliphatic and Aryl Amines” etc.
E.g.: “RHO”, “rhodopsin”, “RP4”, “retinitis pigmentosa 4, autosomal dominant”, “opsin 2, rod pigment”, and “OPN2” are variants of one gene and are assigned to a single ID.
PubMed is a free database that provides access to the Medline database and other services. http://www.ncbi.nlm.nih.gov/pubmed
Contributor Information
Su Yan, Email: syan@us.ibm.com, IBM Almaden Research Lab, San Jose, CA, USA.
Xiaoqian Jiang, Email: xiaoqian.jiang@gmail.com, Division of Biomedical Informatics, University of California at San Diego, USA.
Ying Chen, Email: yingchen@us.ibm.com, IBM Almaden Research Lab, San Jose, CA, USA.
References
- 1.Obach RS. Drug-drug interactions: an important negative attribute in drugs. Drugs of Today. 2003;39(5):301–338. doi: 10.1358/dot.2003.39.5.799456. [DOI] [PubMed] [Google Scholar]
- 2.Pham PA. Drug-drug interaction programs in clinical practice. Clinical Pharmacology & Therapeutics. 2008;83(3):396–398. doi: 10.1038/sj.clpt.6100504. [DOI] [PubMed] [Google Scholar]
- 3.Zhang L, Zhang YD, Zhao P, Huang SM. Predicting drug-drug interactions: an FDA perspective. The AAPS journal. 2009;11(2):300–306. doi: 10.1208/s12248-009-9106-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Almond LM, Yang J, Jamei M, Tucker GT, Rostami-Hodjegan A. Towards a quantitative framework for the prediction of DDIs arising from cytochrome p450 induction. Current Drug Metabolism. 10(4):420–432. doi: 10.2174/138920009788498978. [DOI] [PubMed] [Google Scholar]
- 5.Lu C, Hatsis P, Berg C, Lee F, Balani S. Prediction of pharmacokinetic drug-drug interactions using human hepatocyte suspension in plasma and cytochrome p450 phenotypic data. ii. in vitro-in vivo correlation with ketoconazole. Drug Metab Dispos. 2008;36(7):1255–60. doi: 10.1124/dmd.107.018796. [DOI] [PubMed] [Google Scholar]
- 6.Dexter F. Statistical analysis of drug interactions in anesthesia. Journal of Theoretical Biology. 1995;172(4):305–314. doi: 10.1006/jtbi.1995.0027. [DOI] [PubMed] [Google Scholar]
- 7.Tachibana T, Kato M, Watanabe T, Mitsui T, Sugiyama Y. Method for predicting the risk of drug-drug interactions involving inhibition of intestinal CYP3A4 and P-glycoprotein. Xenobiotica the Fate of Foreign Compounds in Biological Systems. 2009;39(6):430–43. doi: 10.1080/00498250902846252. [DOI] [PubMed] [Google Scholar]
- 8.Imaura M, Ohno Y, Nakajima K, Suzuki H. Clinically significant drug-drug and drug-food interactions associated with calcium channel blockers. Clinical Calcium. 2005;15(10):1709–1716. [PubMed] [Google Scholar]
- 9.Tatonetti NP, Fernald GHH, Altman RB. A novel signal detection algorithm for identifying hidden drug-drug interactions in adverse event reports. Journal of the American Medical Informatics Association: JAMIA. 2012 Jan;19(1):79–85. doi: 10.1136/amiajnl-2011-000214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.van Puijenbroek E, Egberts A, Heerdink E, Leufkens H. Detecting drug-drug interactions using a database for spontaneous adverse drug reactions: an example with diuretics and non-steroidal anti-inflammatory drugs. Eur J Clin Pharmacol. 2000;56(9-10):733–8. doi: 10.1007/s002280000215. [DOI] [PubMed] [Google Scholar]
- 11.Norén GN, Sundberg R, Bate A, Edwards IR. A statistical methodology for drug-drug interaction surveillance. Statistics in Medicine. 2008;27(16):3057–3070. doi: 10.1002/sim.3247. [DOI] [PubMed] [Google Scholar]
- 12.Hu TM, Hayton WL. Architecture of the drug-drug interaction network. Journal of Clinical Pharmacy and Therapeutics. 2011;36(2):135–143. doi: 10.1111/j.1365-2710.2009.01103.x. [DOI] [PubMed] [Google Scholar]
- 13.Tari L, Anwar S, Liang S, Cai J, Baral C. Discovering drug-drug interactions: a text-mining and reasoning approach based on properties of drug metabolism. Bioinformatics. 2010;26(18):i547–53. doi: 10.1093/bioinformatics/btq382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Takarabe M, Shigemizu D, Kotera M, Goto S, Kanehisa M. Characterization and classification of adverse drug interactions. Genome Informatics International Conference on Genome Informatics. 2010;22:167–175. no. Japic Id. [PubMed] [Google Scholar]
- 15.Tatonetti NP, Fernald GH, Altman RB. A novel signal detection algorithm for identifying hidden drug-drug interactions in adverse event reports. Journal of the American Medical Informatics Association. 2011;2:79–85. doi: 10.1136/amiajnl-2011-000214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Percha B, Garten Y, Altman RB. Discovery and explanation of drug-drug interactions via text mining. Pacific Symposium On Biocomputing. 2012:410–21. no. Ddi. [PMC free article] [PubMed] [Google Scholar]
- 17.Huang J, Niu C, Green CD, Yang L, Mei H, Han JDD. Systematic prediction of pharmacodynamic drug-drug interactions through protein-protein-interaction network. PLoS computational biology. 2013 Mar;9(3):e1002 998+. doi: 10.1371/journal.pcbi.1002998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vilar S, Harpaz R, Uriarte E, Santana L, Rabadan R, Friedman C. Drug-drug interaction through molecular structure similarity analysis. JAMIA. 2012;19(6):1066–1074. doi: 10.1136/amiajnl-2012-000935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vilar S, Uriarte E, Santana L, Tatonetti NP, Friedman C. Detection of Drug-Drug Interactions by Modeling Interaction Profile Fingerprints. PLoS ONE. 2013 Mar;8:58321. doi: 10.1371/journal.pone.0058321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lafferty JD, McCallum A, Pereira FCN. Conditional random fields: Probabilistic models for segmenting and labeling sequence data. Proceedings of the Eighteenth International Conference on Machine Learning; 2001; pp. 282–289. [Google Scholar]
- 21.Hewett M, Oliver DE, Rubin DL, Easton KL, Stuart JM, Altman RB, Klein TE. Pharmgkb: the pharmacogenetics knowledge base. Nucleic Acids Research. 2002;30:163–165. doi: 10.1093/nar/30.1.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Smith B, Ashburner M, Rosse C, Bard J, Bug W, Ceusters W, Goldberg L, Eilbeck K, Ireland A, Mungall C, Leontis N, Rocca-Serra P, Ruttenberg A, Sansone S, Scheuermann R, Shah N, Whetzel P, Lewis S. The obo foundry: coordinated evolution of ontologies to support biomedical data integration. Nat Biotechnol. 2007;25(11):1251–5. doi: 10.1038/nbt1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Blei DM, Ng AY, Jordan MI. Latent dirichlet allocation. the Journal of Machine Learning Research. 2003;3:993–1022. [Google Scholar]
- 24.Rosen-Zvi M, Chemudugunta C, Griffiths T, Smyth P, Steyvers M. Learning author-topic models from text corpora. ACM Trans Inf Syst. 2010 Jan;28(1):4:1–4:38. [Google Scholar]