Abstract
Activation induced CD154 expression on CD4 T cells is prolonged in systemic lupus erythematosus but the mechanism(s) for its dysregulation are unknown. The studies reported herein demonstrate that IL-15 is capable of prolonging CD154 expression on PHA activated CD4 T cells. Since IL-15 signals through STAT5, predicted STAT5 binding sites in the human CD154 transcriptional promoter were identified, and STAT5 binding to the proximal CD154 promoter in vitro and in vivo following primary CD4 T cell activation was demonstrated. Moreover, overexpression of wild-type(WT) STAT5 in primary human CD4 T cells augmented CD154 transcription, whereas overexpression of a dominant negative (DN) STAT5 protein inhibited CD154 transcription. Mutation of the most proximal STAT5 binding site in the CD154 promoter resulted in diminished DNA binding and reduced CD154 transcriptional activity. Interestingly, STAT5-specific siRNA inhibited CD154 surface expression at 48 but not 24 hours after T cell activation. Thus, these findings provide some of the first evidence to support a possible mechanistic link to explain how the overexpression of IL-15 observed in lupus patients may be involved in the prolonged expression of CD154 that has also been observed on lupus CD4 T cells.
Keywords: human, T cells, CD154, IL-15, STAT5, lupus
Introduction
CD154 (CD40-ligand) is a 39-kDa protein transiently expressed on the surface of activated CD4 T cells.1 Normally, CD154 expression is strictly controlled, predominantly at the level of transcription.1 However, abnormal expression of CD154 has been linked to the pathology of several autoimmune diseases, particularly systemic lupus erythematosus (SLE).2 In prior studies, we have demonstrated that lupus patients have elevated and prolonged expression of CD154 on CD4 T cells.2 In a study by Yi et al., Cyclosporine A (CsA) was found to reduce CD154 expression by inhibiting the NFAT-calcineurin pathway.3 However, in the same study, CsA was observed to lose potency after approximately 18 hours following T cell activation, suggesting that other transcription factors also contribute to prolonged expression.3 In a different study authored by Baranda et al., lupus patients were observed to have elevated serum IL-15 levels.4 From prior studies by other groups, there is published data demonstrating that IL-15 binding to its receptor on T cells triggers Jak 1 and Jak 3 to phosphorylate tyrosine residues on STAT5.5,6 Upon activation, STAT5 translocates to the nucleus and binds the γ-interferon-activated sequence (GAS), defined by the TTCN3GAA consensus sequence.7 This event ultimately leads to increased transcription of target genes. These known pathways and the independently observed associations of the aberrant expression of CD154 on T cells in lupus patients and elevated serum levels of IL-15 in lupus patients in independent studies prompted us to further explore whether CD154 expression may be regulated directly by STAT5.
Our prior work with CD154 expression and interest in understanding potential mechanisms to help identify and characterize dysregulated signaling pathways in lupus prompted us to search for potential binding sites for transcription factors which may bind to the CD154 transcriptional promoter and influence its expression. Our sequence analysis of the CD154 promoter resulted in the identification of three potential binding sites for STAT5 within and just upstream of the CD154 promoter (Figure 1). This suggests STAT5 may play a role in regulating CD154 transcription. While many cytokines are capable of signaling via STAT5, IL-2, in particular, has been shown to be decreased in lupus blood.8 By contrast, another related cytokine that signals via STAT5, IL-15, has been shown to be aberrantly overexpressed in lupus4,9–11 and may therefore contribute to CD154 hyperexpression via STAT5 signaling, especially at later time points post-T cell activation.12 To further characterize one of the potential mechanisms by which IL-15 may influence CD154 expression through STAT5, we performed the studies described within this manuscript using CD4 T cells obtained from healthy individuals. Herein, we provide clear evidence of how STAT5 plays a direct role in CD154 expression in primary CD4 T cells.
Figure 1. Consensus STAT5 binding sites in the proximal human CD154 transcriptional promoter.
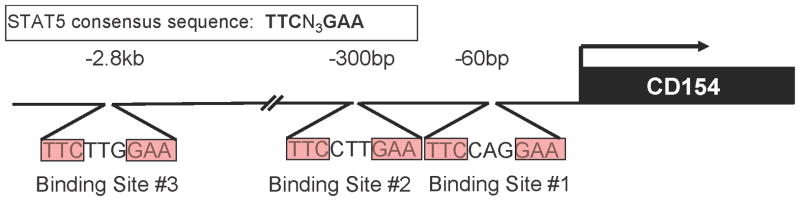
Three potential binding sites within 3kb upstream of the CD154 TSS (arrow) were identified by sequence analysis via the Transfac database and labeled as binding sites (BS)#1–3.
Results
Stimulation by IL-15 and PHA leads to persistent upregulation of CD154
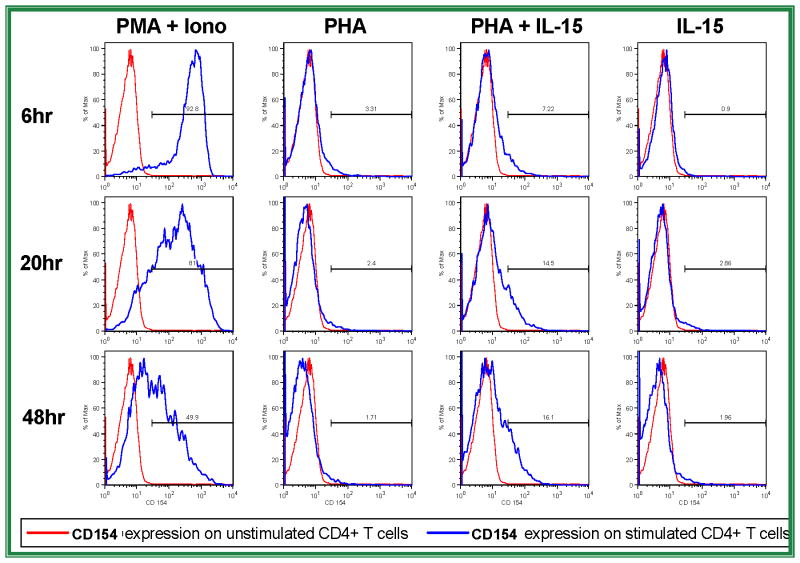
In exploring a role for IL-15 and STAT5 in CD154 regulation, the first question addressed was whether IL-15 induces or augments the expression of CD154 in primary CD4 T cells. Human peripheral blood CD4 T cells isolated by negative selection were stimulated ex vivo with PHA, IL-15, or a combination of both for 6, 20, or 48 hours. Flow cytometry data indicating CD154 surface expression levels are shown (Figure 2). Low dose PHA or IL-15 alone did not increase CD154 expression significantly, but stimulation of the primary CD4 T cells with both PHA and IL-15 led to notable CD154 expression. This CD154 expression continued to increase throughout the later time points following activation. These data support the hypothesis that IL-15 significantly augments CD154 transcription in stimulated CD4 T cells, particularly beyond the first 24 hours of stimulation. Further experiments were designed to characterize whether this effect is mediated at least in part through STAT5, a known mediator of IL-15 signaling. Subsequent experiments were performed to identify binding sites of STAT5 in the human CD154 promoter.
Figure 2. IL-15 augments late CD154 expression on suboptimally stimulated CD4 T cells.
Primary human CD4 T cells were rested (red lines) or stimulated (blue lines) with phorbol ester and ionomycin (far left column), low dose PHA alone (left middle column), PHA plus IL-15 (right middle column), or IL-15 alone (far right column) for 6 (top row), 20 (middle row), or 48 (bottom row) hours prior to analysis of cell surface CD154 expression by flow cytometry. Results are representative of 4 similar experiments.
STAT5 binds two sites in the human CD154 promoter in vitro
A search of the Transfac® database13 immediately upstream of the human CD154 transcriptional start site (TSS) yielded three potential STAT5 binding sites (BS) using the consensus binding sequence, TTCN3GAA: BS#1 (−57bp to −65bp); BS#2 (298bp to − 306bp); and BS#3 (−2807bp to −2815bp) (Figure 1). To experimentally verify whether the STAT5 transcription factor binds to these sites in vitro, nuclear extracts were generated from primary human CD4 T cells stimulated with PHA and IL-2, a cytokine known to induce STAT5 expression. These extracts were probed with radiolabeled oligonucleotides containing the putative STAT5 binding site sequences (BS#1–3) from the human CD154 promoter. Autoradiographs showing the results of electrophoretic mobility shift assays (EMSA) are shown (Figure 3, left and right panels). No detectable shifted band was seen with the BS#2 probe (data not shown). However, the radiolabeled putative STAT5 BS#1 or BS#3 probes mixed with nuclear extracts formed visible gel-retarded complexes (Figure 3, Lane 4, left panel - BS#1, right panel - BS#3) that migrated an identical distance as the one formed by STAT5 binding to the prolactin (PRL) positive control probe (Figure 3, both panels, lane 1).14,15 Addition of excess unlabeled self or PRL oligonucleotides (Figure 3, left panel, lane 5 and data not shown) or BS#3 probe (Figure 3, right panel, lanes 5 and 6) prior to adding labeled BS#1 or BS#3 probes resulted in the disappearance of this band, providing evidence of sequence-specific competition. The addition of antibody specific for the N-terminal domain of STAT5 causes the complex to supershift (lanes 3 and 7, both panels), indicating that the binding protein to PRL, BS#1, and BS#3 is STAT5. To further demonstrate specificity of STAT5 binding to the proximal human CD154 promoter, the BS#1 sequence was altered by 3 base pairs resulting in a BS#1-mut oligonucleotide probe that eliminated its ability to bind to the STAT5 protein (Figure 3, Lanes 8–11). Consistent with this observation, an excess of unlabeled BS#1-mutant probe (Figure 3, left panel, Lane 6), only minimally competed with binding of the labeled, non-mutated BS#1 probe (Figure 3, left panel, Lane 4). Thus, STAT5 binds the proximal human CD154 promoter BS#1 sequence as well as the upstream BS#3 sequence in activated primary human CD4 T cells.
Figure 3. STAT5 binds the proximal human CD154 transcriptional promoter in vitro.
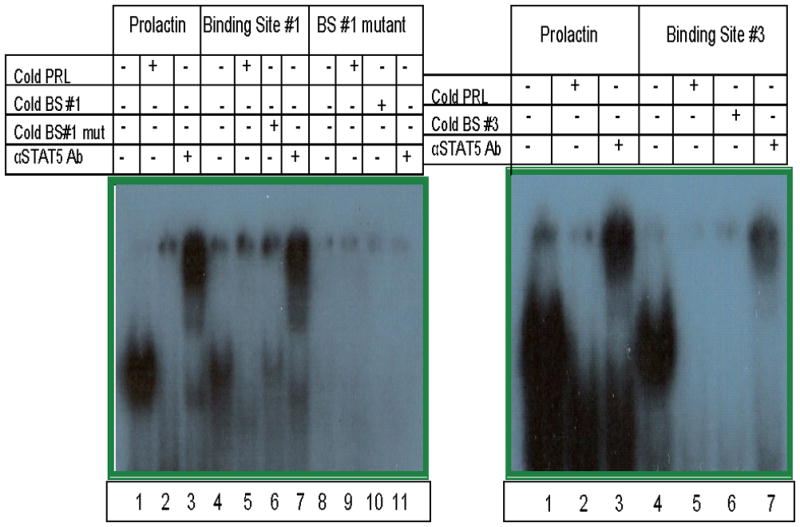
Nuclear extracts from polyclonally activated primary human CD4 T cells were incubated with radiolabeled probes corresponding to a consensus STAT5 binding site from the prolactin promoter (PRL) or predicted STAT5 binding sites #1 and #3 from the CD154 promoter, as well as a mutant BS#1 sequence predicted to disrupt STAT5 binding. Some extracts were pre-incubated with excess cold probes or STAT5 antibody prior to incubation with the specific radiolabeled probes as indicated by the plus signs. STAT5 binding was analyzed by electrophoretic mobility shift assays. Results are representative of 5 experiments.
STAT5 engagement of the proximal human CD154 promoter in vivo is important for optimal CD154 transcription
Chromatin immunoprecipitation (ChIP) was used to isolate DNA that was specifically bound to the STAT5 transcription factor in vivo following activation of primary CD4 T cells. Peripheral blood human CD4 T cells were stimulated with either PMA and ionomycin (P+I, positive control), stimulated with PHA and IL-15, or left unstimulated (negative control) before being subjected to ChIP. Real-time PCR was performed on anti-STAT5 antibody immunoprecipitated DNA fragments in order to demonstrate and quantify the degree of STAT5 binding to the CD154 proximal promoter in vivo. The bar graph (Figure 4) reveals minimal if any specific STAT5 binding in the absence of stimulation (light grey bar) compared to the isotype control antibody (black bar). However, PHA plus IL-15 stimulation (dark grey bar) resulted in a notable increase in the percentage of amplified DNA compared to total input DNA demonstrating STAT5 binding to the proximal CD154 promoter in vivo. This was more dramatic when the cells were polyclonally stimulated with phorbol ester and calcium ionophore (white bar) such that the mean STAT5 fold-binding (relative to isotype control antibody) to the proximal human CD154 promoter was 16.7-fold (16.7 ± 6.9 SEM; n=3) in vivo after 24 hr incubation of P+I stimulated primary human T cells. Thus, STAT5 binds the proximal human CD154 promoter in vivo following activation of primary CD4 T cells.
Figure 4. STAT5 binds the proximal human CD154 transcriptional promoter in vivo.
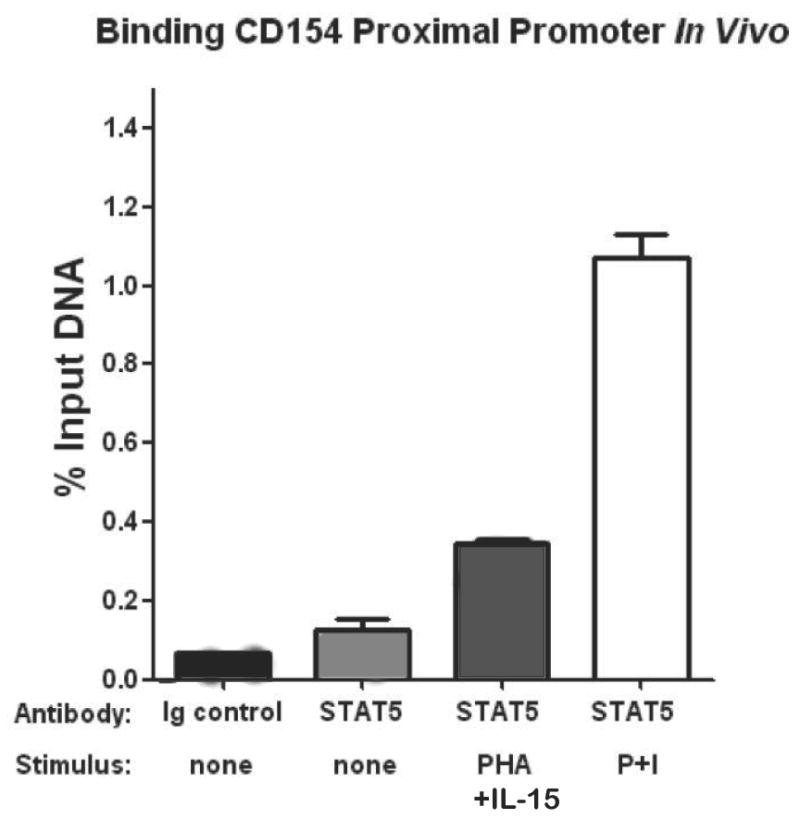
Primary human CD4 T cells were rested or stimulated with PHA and IL-15 or with P+I for 2 hours as per the Materials and Methods. ChIP was carried out using anti-STAT5 or IgG isotype control antibodies as described in the Materials and Methods. Immunoprecipitated DNA was amplified by real-time PCR using oligonucleotide primers designed to amplify the proximal human CD154 promoter containing BS#1. Results are shown as a percentage of the input DNA. Data are means ±SEM of duplicate samples and are representative of 5 individual experiments.
The presence of STAT5 influences transcriptional activity of the CD154 promoter
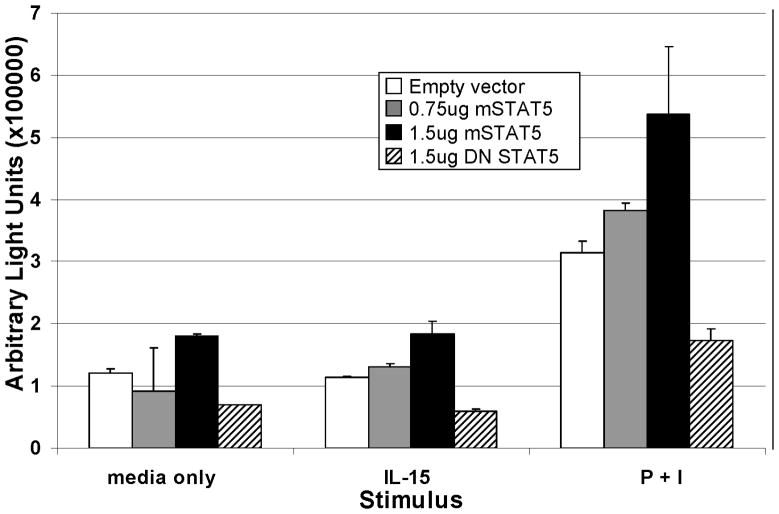
To determine the functional relevance of STAT5 binding to the proximal CD154 promoter, an ex vivo dual-luciferase reporter assay was employed. Negatively selected primary human CD4 T cells were cotransfected with a human CD154 promoter-driven luciferase reporter vector that contains the BS#1 STAT5 binding site, and either a WT STAT5 expression vector, or a dominant negative STAT5 (DN STAT5)16,17 expression vector. After culturing for 6 hours with P+I, or IL-15, cells were assayed for luciferase expression. IL-15 alone did not appear to augment the transcriptional expression of CD154, as the arbitrary light units (ALU) values of the IL-15-stimulated samples were similar to cultures with media alone (Figure 5). However, luciferase expression in samples stimulated with P+I increased CD154 transcription with increasing doses of WT STAT5 plasmid in a dose-dependent fashion. Moreover, transfection with the DN STAT5 plasmid dramatically diminished ALU levels compared to controls. This finding suggests that overexpression of WT STAT5 augments CD154 promoter activity via direct binding of STAT5 to the CD154 promoter, although IL-15 stimulation alone is not sufficient to induce CD154 transcription. Interestingly, endogenous STAT5 appears important for CD154 transcriptional activity as supported by the observation that transfection of the DN STAT5 construct inhibited CD154 promoter reporter gene activity.
Figure 5. STAT5 binding to the proximal CD154 promoter influences CD154 transcription.
Primary human CD4 T cells were transiently transfected with a CD154 promoter driven luciferase reporter plasmid and co-transfected with either an empty vector (control), increasing amounts of a STAT5 expression plasmid, or a dominant negative STAT5 expression plasmid. CD4 T cells were rested and stimulated with nothing (left bars), IL-15 (middle bars), or P+I (right bars) for 6 hours prior to luciferase analysis. Results are representative of 4 similar experiments and SEM bars are depicted.
Mutation of binding site #1 decreases transcriptional activity of the CD154 promoter reporter gene
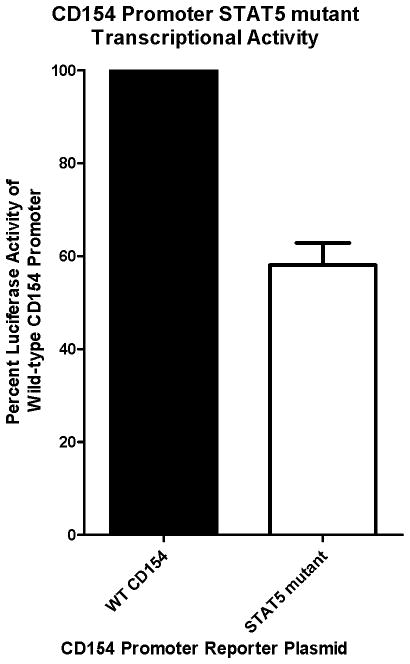
To provide further evidence that STAT5 binding to BS#1 is important for CD154 transcriptional promoter activity, we constructed a CD154 luciferase expression plasmid containing a mutation within the BS#1 site in the proximal promoter. The original pCD154-Luc plasmid18 was modified by site-directed mutagenesis, introducing the same three base pair mutation as was used in the gel shift experiments with the BS#1-mut oligonucleotide that disrupted STAT5 binding to the human CD154 proximal promoter (Figure 3). Primary human CD4 T cells were transiently cotransfected with a human CD154 transcriptional reporter gene, either WT control or BS#1 mutant, along with a control Renilla luciferase expression vector to control for transfection efficiency. As expected, the BS#1 mutant CD154 luciferase expression plasmid demonstrated a decrease of expression by more than 40% (Figure 6), providing additional functional evidence of the importance and specificity of endogenous STAT5 binding to BS#1 of the CD154 promoter to promote CD154 transcription.
Figure 6. Mutation of the most proximal STAT5 binding site in the CD154 promoter diminishes CD154 transcription.
Primary human CD4 T cells were transfected with a WT, or BS#1 STAT5-mutant version, CD154 promoter-driven firefly luciferase reporter plasmid. After a brief rest period, the CD4 T cells were activated with P+I overnight, and cell lysates were analyzed for firefly luciferase activity controlled for transfusion efficiency with a co-transfected control Renilla luciferase reporter plasmid. Results are representative of 4 similar experiments and SEM bars are depicted.
siSTAT5 inhibits CD154 expression
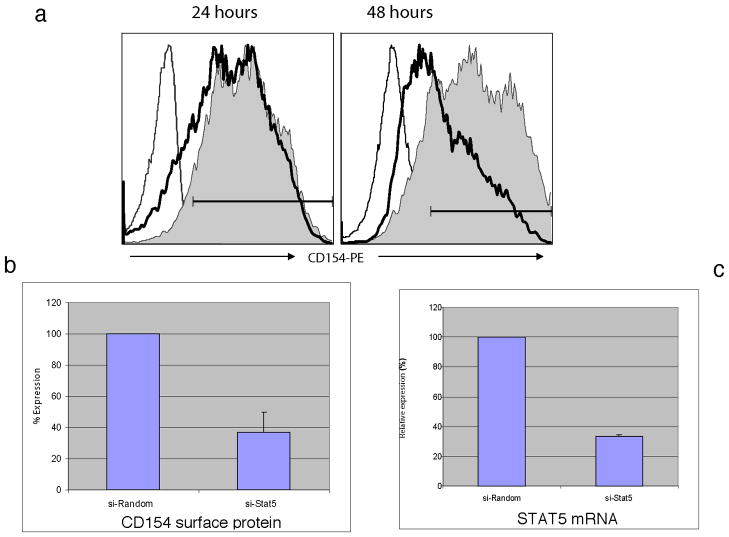
The essential role of STAT5 augmenting CD154 promoter transcription was further demonstrated by utilizing small interfering RNA (siRNA) technology. Primary human CD4 T cells isolated from peripheral blood were electroporated with expression constructs containing either a negative control random sequence (siRandom) or STAT5 siRNA sequences (see Methods). Electroporated cells were rested overnight and then stimulated with P+I before analyzing cells for CD154 expression by flow cytometry at 24 and 48 hours. No significant changes in the level of CD154 surface expression were observed at 6 hours (data not shown) or 24 hours (Figure 7a, left) post-stimulation as measured by percent CD154 positive cells or CD154 mean fluorescent intensity (13% decreased percent positive; 12% decreased MFI). However, in samples from nine of twelve donors, CD4 T cells transfected with the siSTAT5 vector showed significant inhibition (>50%) of CD154 surface expression at 48 hours (Figure 7a, right) following stimulation compared to the control using the means of data from multiple experiments (Figure 7b, 57% decreased MFI). By 72 hours after stimulation, CD154 expression became too low to reliably determine any differences between cells transfected with siSTAT5 vector and the control vector (data not shown).
Figure 7. Endogenous STAT5 in CD4 T cells is critical to optimal CD154 expression at 48 hours post-activation.
(a) Primary human CD4 T cells were transfected with random sequence (control) or STAT5-specific si-RNA. After 24 (left) and 48 (right) hours of stimulation with P+I, cells were evaluated for CD154 expression by flow cytometry (far left line – Ig isotype control; solid-si-Stat5; shadowed area - non-coding sequence control). Results are representative of 7 similar experiments.
(b) Average (n=12) inhibition of surface CD154 protein by transfected si-STAT5 following 48 hours of CD4 T cell stimulation.
(c) Effect of si-RNA was confirmed by evaluation of STAT5 mRNA expression by Real Time RT-PCR. Expression of STAT5 mRNA was normalized to 18S RNA. Data are presented as relative STAT5 mRNA expression in the presence of STAT5 siRNA compared to random non-coding sequence (n=6).
To verify that the diminished CD154 surface expression could be attributed to STAT5 siRNA interference with STAT5 mRNA levels, we performed real-time RT-PCR experiments with similarly prepared donor CD4 T cells at 48 hrs following stimulation with either P+I, or PHA plus IL-15, demonstrating a greater than 60% decrease in STAT5 mRNA levels in the presence of STAT5 siRNA (Figure 7c). Interestingly, the siRNA only inhibited CD154 expression at the later time point suggesting STAT5 engagement and upregulation of CD154 occurs later after T cell activation and may reflect the role of IL-15 in vivo, particularly in regards to CD154 overexpression at later time points as is seen in ex vivo experiments with T cells from SLE patients19,20 (also known to over-express IL-15 in the blood4,10). Thus, STAT5 engagement of the human CD154 promoter and upregulation of CD154 expression occurs later (~24–48 hours) after the initial T cell stimulation.
IL-15 is elevated in SLE
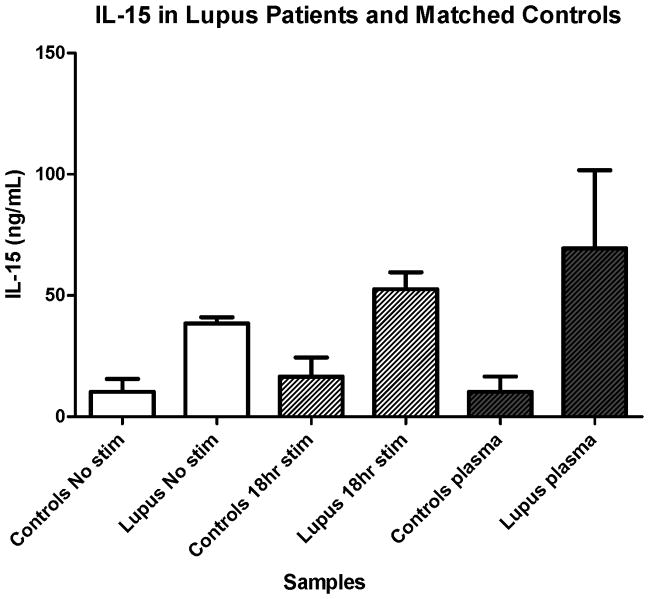
We confirmed increased IL-15 expression in SLE ex vivo and post activation in vitro similar to our other assays. IL-15 was elevated in lupus patients (n=2) compared to age-, gender-, and race-matched controls (n=2) as measured in plasma levels (Figure 8, black bars). Patients also had notably elevated levels of IL-15 in CD4 T cell culture supernatent after 18 hours of incubation, either with stimulation with anti-CD3Ab/anti-CD28Ab-coated beads (Figure 8, grey bars), or resting without any stimulation (Figure 8, white bars). Although we do not have sufficient numbers of patient and matched controls at this time to show statistical significance, this observation is consistent with our hypothesis that IL-15 is dysregulated in patients with lupus. Of note, both lupus patients had highly active disease when blood was obtained at the time of initial diagnosis. Interestingly, even though one of the two lupus patients had received high dose steroids (2 days of oral prednisone and 1 dose of 1 gm of intravenous methylprednisolone), IL-15 plasma levels still correlated with severity of disease at time of testing in this small sample of patients (data not shown).
Figure 8. Plasma IL-15 and CD4 cell culture supernatants either with stimulation or without stimulation are elevated in SLE compared to matched controls.
Primary human CD4+ T cells from SLE patients (n=2) or matched healthy controls (n=2) were cultured either with anti-CD3Ab/anti-CD28Ab-coated beads (grey hatched bars) or without any stimulation (white bars); supernatants were collected after 18 hrs in culture and assayed by ELISA for IL-15 levels. Plasma samples (black bars) from both SLE patients and matched controls were also assayed for IL-15 levels by ELISA. SEM bars are depicted.
Discussion
Stimulation of primary human T cells with PHA in the presence of IL-15 results in upregulation of CD154 that begins at 24 hrs and persists at 48 hrs following stimulation, implicating a role for IL-15 in CD154 expression. A novel binding site for the STAT5 transcriptional activator was located 56–65 bp upstream of the human CD154 TSS and was experimentally verified through EMSA and ChIP. Functional evidence for STAT5 binding to the CD154 promoter was demonstrated using transcriptional reporter genes and mRNA analysis. The importance of endogenous STAT5 in primary human CD4 T cells to CD154 expression was also demonstrated using siRNA technology to knockdown STAT5 mRNA.
In an effort to determine the relative significance of our findings with regard to STAT5 binding on the CD154 promoter, we conducted a search of the ENCODE dataset. Our query affirmed the novelty of our findings. There are only 2 ENCODE cell lines (GM12878--a B lymphocyte lymphoblastoid human cell line transformed by Ebstein Barr virus and K562--a myelogenous leukemia human cell line) with transcription factor binding data for STAT5. However, neither of the experiments with these cell lines yielded any data regarding STAT5 transcription factor binding to any part of the CD154 gene, including the promoter region (https://genome.ucsc.edu/cgi-bin/hgTracks?hgS_doOtherUser=submit&hgS_otherUserName=Rmjlowe&hgS_otherUserSessionName=CD40LG%2DSTAT5%2Dtfbn). Thus, STAT5 was identified as a novel CD154 transcriptional regulator that acts via engagement of the proximal human CD154 promoter.
These results serve to further extend prior published work by ourselves and others that have helped create a model for understanding the transcriptional regulation of CD154 expression in CD4 T cells. By showing how CD154 transcription is directly regulated by STAT5 and also influenced by IL-15 signaling, we have helped demonstrate important mechanistic links to well characterized intracellular signaling pathways that may be further characterized and exploited as therapeutic targets for diseases such as systemic lupus erythematosus.
One of the more interesting observations made in these studies is that CD154 expression involving stimulation with IL-15 begins around 24 hours and persists at 48 hours after T cell activation. The typical CD154 expression pattern involves a rapid rise in CD154 expression on CD4 T cells after activation with a consequent drop in expression by 48 to 72 hours. However, when primary human T cells were exposed to IL-15 at the time of activation, we did not observe the expected decrease in expression by 48 hrs after cell activation. This suggests that IL-15 is involved in prolonging CD154 expression after the initial upregulation following T cell activation. Interestingly, lupus patients have both elevated serum levels of IL-154,10 (Figure 8) and abnormally excessive and prolonged CD154 expression.19,20 Yi and colleagues demonstrated that CD154 expression was prolonged in lupus T cells which could be abrogated early (first 18 hrs) following stimulation by treating with cyclosporine A that blocks the calcineurin-dependent NFAT pathway but not at later time points.3 Studies by Skov et al. demonstrated that previously activated T cells will respond to IL-15 by upregulating CD154 expression without further stimulation.21 This data further supports the hypothesis that elevated levels of IL-15 may play an important role in maintaining a pathogenic excessive and prolonged expression of CD154 on CD4 T cells. Based on these studies and the data presented herein, we can postulate that perhaps the inhibition of excess IL-15 will disrupt prolonged CD154 expression following T cell activation in patients with lupus. Since directly blocking CD154 expression was found to be likely effective but have unacceptable toxicities in patients, a strategy for controlling lupus disease activity by indirectly suppressing CD154 expression by blocking IL-15 is a potentially attractive alternative therapeutic strategy.
In summary, in addition to providing novel, direct evidence for the role of STAT5 binding to the CD154 promoter and thereby augmenting its expression, the current study also provides some additional indirect evidence that IL-15 may signal in part via STAT5 engagement of the proximal CD154 transcriptional promoter. Even though elevated levels of IL-15 in patients with SLE has been documented in the current study and by others,10 we have yet to directly link elevated IL-15 expression levels in lupus patients to increased STAT5 activity and its consequent effects on CD154 expression on CD4 T cells. However, we are encouraged to pursue this line of research by data generated by other groups studying the pathogenic mechanisms in other autoimmune diseases, such as rheumatoid arthritis. Mottonen et al. published data showing that IL-15 up-regulates the expression of CD154 on synovial fluid T cells.22
Future research, in addition to improving our understanding of how IL-15 signaling influences CD154 gene expression in health and disease, will also expand to other potentially important mechanisms of CD154 gene regulation including epigenetic modifiers of expression.23 These include DNA methylation and its effects on CD154 transcriptional activity,24 as well as micro RNA species that alter CD154 translational efficiency.25 This will hopefully lead to a further improved understanding of how we may therapeutically manipulate CD154 expression in T cells to create novel therapies for the treatment of lupus and other autoimmune diseases.
Materials and methods
Patients and matched controls for IL-15 studies
Peripheral venous blood was drawn from two SLE patients with lupus nephritis (an African-American 13yo female and a Caucasian 13yo female) and two matched controls (an African-American 12yo female and a Caucasian 13yo female). All subjects gave informed assent, and informed consent was given by one of each child’s parents. Both SLE patients had blood samples drawn at the time of diagnosis. At this time, lab tests performed were consistent with highly active lupus: ESR > 100, proteinuria with urine protein to creatinine ratio > 2.0, ANA with a titer of 1:640 or higher, and abnormally elevated Smith and/or double-stranded DNA antibodies. One SLE patient did not receive any steroids or treatment prior to blood draw; the other SLE patient had received 2 days of oral prednisone and 1 gram of methylprednisolone on the day prior to the blood draw. Institutional review board approval was obtained prior to recruiting of volunteers for the collection of blood samples.
CD4 T cell isolation and stimulation
Primary human CD4 T cells were isolated from the peripheral blood of healthy adult donors, healthy matched pediatric donors, and patients with lupus by negative selection according to the manufacturer’sprotocol (RosetteSep, StemCell Technologies, Inc., Vancouver, British Columbia) as described.1 Flow cytometry26 confirmed 96–98% CD4 T cell purity of the resulting cell populations (data not shown). Cells were cultured for 6 hours at 37 °C in 5% CO2 in complete RPMI 1640 medium containing 10% FCS, 2 mM L-glutamine, 100 U ml−1 penicillin and 100 μg ml−1 streptomycin alone, or also in the presenceof 10 ng ml−1 Phorbol myristate acetate (PMA) and 1.5 μM Ionomycin, (Sigma-Aldrich, St.. Louis, MO, USA). In the IL-15 and CD154 expression studies, cells were stimulated with 1.5 μg ml−1 PHA (Sigma-Aldrich), or 10 U ml−1IL-2, 75 ng ml−1 IL-15 alone (R&D Systems, Minneapolis, MN), or a combination of 1.5 μg ml−1 PHA and 75 ng ml−1rIL-15. Stimulation time was 6, 24, or 48 hours. Cells from pediatric lupus patients and healthy matched controls were cultured for IL-15 production either with anti-CD3Ab/anti-CD28Ab-coated beads at a 1:1 bead to cell ratio (Dynabeads Human T-Activator CD3/CD28, Life Technologies, Inc, Carlsbad, CA) or without any stimulation for 18hrs before culture supernatants were collected and stored at −80°C until assayed for IL-15 concentration as detailed below.
EMSA
Nuclear proteins from stimulated primary human CD4 T cells were extracted as previously described.27 Synthetic oligonucleotides (IDT, Coralville, IA, USA), used as 32P-labeled probes, or as unlabeled competitors were as follows: Binding site-1 (BS#1): 5′ – CAC ATT TTC CAG GAA GTG TGG -3′ (sense) and 5′ – CCA CAC TTC CTG GAA AAT GTG -3′ (anti-sense); binding site- 2 (BS#2): 5′ – GTC ACT TTC CTT GAA ACT GGT -3′ (sense), and ACC AGT TTC AAG GAA AGT GAC -3′(anti-sense); binding site-3 (BS#3): 5′-ATA CAT TTC TTG GAA ACT AAG-3′ (sense); 5′-CTT AGT TTC CAA GAA ATG TAT-3′ (anti-sense). The following oligonucleotide was used as a probe for the mutated binding site-1 (anti-sense): 5′– CAC ATT TTC CAC CTA GTG TGG – 3′. Consensus STAT5 binding sites are in bold letters. Underlined residues have been mutated.
In brief, 2 μg of nuclear protein, 25 000 cpm of 32P-labeled probe, and EMSA binding buffer (500 mM Tris-HCL, pH 7.5; 500mM KCL; 15 mM MgCl2; 10 mg ml−1 BSA; 50% glycerol; 5 mg ml−1 single strand sperm DNA; 1 mM DTT) were mixed and incubated for 20 minutes on ice. For specificity determinations, nuclear protein was reacted with excess (200-fold) unlabeled probe, or anti-STAT5 antibody (Santa Cruz Biotechnology, Santa Cruz, CA, USA), 30 min before the addition of labeled probe. The mixtures were then electrophoresed on 6% TBE gradient pre-poured DNA retardation gels (Invitrogen, Carlsbad, CA, USA). Radioactive bands were detected by conventional autoradiography as previously described.18
Site-directed mutagenesis
The WT promoter-driven luciferase reporter plasmid, pCD154-Luc, was changed by introducing a three base pair mutation in BS#1 in order to match the mutant oligonucleotide that no longer binds STAT5 by gel shift assay (Figure 3). We used commercial reagents (QuickChange Site-Directed Mutagenesis Kit, Agilent Technologies, Santa Clara, CA, USA), and followed the manufacturer’s protocol as previously described.28 Briefly, mutant primers for STAT5 BS#1, located approximately 60 bp upstream of the CD154 TSS: 5′- GCC CAT TTT GGC TCT CAT CTT CAT TTT CCT TCA GTG GAA CTA AGG-3′ (sense), and 5-′CCT TAG TTC CAC TGA AGG AAA ATG AAG ATG AGA GCC AAA ATG GGC -3′ (antisense) primers were selected and synthesized. Twenty-five ng ml−1 of reporter plasmid was used as a template in polymerase chain reactions (PCRs) with selected mutant primers in the final concentration 1 μg ml−1, and 1 μl/reaction of PfuTurbo DNA polymerase. WT supercoiled DNA was then digested with 1 μl of Dpn I restriction enzyme, and 50 μl of XL1-Blue Supercompetent cells were transformed with 1 μl of digested DNA. Mutations in plasmid DNA purified from selected colonies were confirmed by DNA sequencing analysis.
Luciferase Reporter Gene assay and Plasmid DNA
Primary human peripheral blood CD4 T cells, 5–10 × 106 per electroporation cuvette, were transiently co-transfected by Amaxa as previously described1 with pCD154-Luc (5 μg, contains a 1.3 Kb portion of the human CD154 gene (−1227 to +67), numbers are given with respect to the TSS18), pRL-null (0.5 μg, Promega), and pcDNA control (Sigma Aldrich, St. Louis, MO) vectors, or expression plasmids pSTAT5B (5 μg)16 (a kind gift from Dr. N. Selliah, Humigen), dominant negative (DN) STAT5b17 (pXM-Stat5b, a kind gift from Dr. N. Selliah); or constitutively active STAT5b (Stat5b-CA, a kind gift from Dr. M. Farrar, University of Minnesota).29,30 After electroporation and a 2-hour rest at 37°C in a CO2 incubator, CD4 T cells were stimulated in vitro, with PMA (25 ng ml−1) and ionomycin (1.5 μM), with or without IL-2 (100 U ml−1) for 6 h at 37°C. Cell lysates were generated with Passive Lysis Buffer (Promega Corporation, Madison, WI, USA) as previously described.26 Firefly luciferase reporter activity was determined in duplicate and correlated for transfection efficiency based on Renilla luciferase activity (Dual Luciferase Assay Kit, Promega) using a Lumat LB 9507 luminometer (EE and G Berthold, Bad Wildbad, Germany) as previously detailed.31,32 pRL null, a Renilla luciferase plasmid was cotransfected for the purpose of data correction. An empty expression vector was cotransfected as a control where appropriate.
Chromatin immunoprecipitation (ChIP) assays
ChIP assay used reagents from Upstate Biotechnology, Inc. (Lake Placid, NY) with modifications to the manufacturer protocol as previously described.33–35 Nuclear proteins and chromatin were crosslinked, sonicated, and extracts from 1–5 million cells were precipitated overnight at 4°C with 5–10 μg of each test antibody: anti-STAT5b (N-20) rabbit polyclonal, anti-NFκB p50 (NLS) rabbit polyclonal, or their respective IgG isotype control antibody (Santa-Cruz Labs, Santa Cruz, CA, USA) and Protein G agarose. DNA extracted from each precipitate was probed by TaqMan® real time PCR (Applied Biosystems, Grand Island, NY) using the following oligonucleotide primers upstream of the human CD154 TSS: 5′– TTT GCT GGG AGA GAA GAC TAC GA -3′, and 5′– TGG CCA CCT TAC TCA GGA TTA GTT A -3′. Fold differences between specific antibody and isotype control immunoprecipitations were calculated using the formula: 2−ΔCt, where ΔCt = Ct sample − Ctcontrol
STAT5 Knockdown by siRNA
STAT5 knockdown on CD154 protein expression was studied on primary human CD4 T cells through the use of transient transfection of small interfering RNA35 (siRNA, Custom SMART pool, Dharmacon, ThermoScientific, Lafayette, CO, USA). The siRNA cocktail used was a mixture of four different synthetic sequences targeting STAT5 mRNA (J-005169-11, -12, -13 and -14). A non-targeting siRNA pool (D-001810-10-05, Thermo Scientific) was used as a negative control. Primary human CD4 T cells were transfected according to the manufacturer’s protocol (AMAXA, Koeln, Germany) on program U-024 with 4.2 μg per transfection of either STAT5 siRNA or an equal amount of scrambled sequence siRNA. Transfection efficiency was confirmed using the provided pmaxGFP control plasmid. After transfection, cells were incubated without stimulation overnight, and then cultured in the presence of stimulation reagents, or culture medium alone. CD154 protein surface expression was measured by flow cytometry after 24, 48 and 72 hours of stimulation with P+I (Sigma-Aldrich). STAT5 mRNA was measured at 48 hours post-stimulation. Data for this assay was only evaluated if the siRNA transfection efficiency was 70% or greater.
Flow cytometry and antibodies
The following mouse anti-human antibodies were purchased (BD Biosciences, San Jose, CA, USA): FITC- or PE-conjugated anti-CD4, (clone SK3, IgG1, κ); anti-CD25, (clone 2A3, IgG1, κ); and anti-CD69 (clone FN50, IgG1, κ). The following corresponding isotype control antibodies were also purchased (BD Biosciences Pharmingen, San Diego, CA, USA): PE-conjugated anti-CD154 (clone TRAP1, mouse IgG1, κ) and PE-conjugated anti-CD4 (clone RM4-5, IgG2a, κ). Stained cells were analyzed on a FACSCalibur (BD Biosciences) and analyzed on FlowJo software (Tree star, Inc., Ashland, OR, USA) as described previously.26
Enzyme-linked immunosorbent assay (ELISA) for IL-15
Plasma and CD4 cell culture supernatants were tested for IL-15 from SLE patients (n=2) and matched controls (n=2) using a commercially available human IL-15 ELISA Kit (Invitrogen Corp) following manufacturer instructions. All samples were tested in duplicate with the mean value used. The detection threshold for this assay was <11 pg ml−1. All values below the detection threshold given by the manufacturer were assigned a value of 1 pg ml−1. The coefficient of variation for intra-assay precision was 4–5% and the coefficient of variation for inter-assay precision was 5–9% as documented by the manufacturer. Recovery of Hu IL-15 added to serum averaged 81%. The recovery of Hu IL-15 added to tissue culture medium containing 1% fetal bovine serum averaged 107%.
Quantitative real-time RT-PCR
Total RNA was extracted from CD4 T cells by TRIzol reagent (Invitrogen Corp) according to the manufacturer’s protocol; about 2.0 μg of total RNA was reverse transcribed using iScript Synthesis Kit (Bio-Rad Laboratories, Hercules, CA, USA) per the manufacturer’s instructions. Quantitative real-time RT-PCR experiments were performed in duplicate using the IQ5 iCycler and iQ™ Sybr Green Supermix (Bio-Rad Laboratories). The same cDNA served as template for the genes of interest, and as an internal control, for 18S RNA. Total reaction volume 25 μl, in which 0.5 μl of cDNA, and 300–900 pmol of primers were used. A nontemplate control was routinely performed in duplicate for each primer pair. Amplification curves were analyzed using iQ-5 Optical System Software (Bio-Rad Laboratories). A melting curve program was run at the end of each PCR cycle to verify the amplification of single product. The relative expression levels of STAT5 were calculated as the comparative threshold cycle 2−ΔCT method, where ΔCT is the difference between the mean threshold cycle value of duplicates test samples and endogenous 18S RNA.
Acknowledgments
We would like to acknowledge Dr. Michael Farrar for the kind gift of the constitutively active STAT5b plasmid (Stat5b-CA), Dr. Selliah for the kind gift of the mSTAT5b plasmid, and Audrey Daggan for her technical contributions.
We would like to acknowledge the importance and valuable contribution of our Cell Cytometry Core at UAB in completing the experiments described in this manuscript. We would also like to acknowledge that funding for this research was provided by the Lupus Foundation (Cron), National Institutes of Health R01-AR-48527 (Cron), R21-AR-49335 (Cron), Arthritis Foundation (Lowe), and University of Alabama at Birmingham Pediatrics Child Health Research Center (Lowe).
Footnotes
Conflicts of Interest
The authors have no financial interests to disclose.
References
- 1.Cron RQ. CD154 transcriptional regulation in primary human CD4 T cells. Immunol Res. 2003;27(2–3):185–202. doi: 10.1385/IR:27:2-3:185. [DOI] [PubMed] [Google Scholar]
- 2.Cron RQ. CD154 and lupus. Pediatr Rheumatol Online J. 2003;1:172–181. [Google Scholar]
- 3.Yi Y, McNerney M, Datta SK. Regulatory defects in cbl and mitogen-activated protein kinase (Extracellular signal-related kinase) pathways cause persistent hyperexpression of CD40 ligand in human lupus T cells. J Immunol. 2000;165(11):6627–6634. doi: 10.4049/jimmunol.165.11.6627. [DOI] [PubMed] [Google Scholar]
- 4.Baranda L, de la Fuente H, Layseca-Espinosa E, Portales-Perez D, Nino-Moreno P, Valencia-Pacheco G, et al. IL-15 and IL-15R in leucocytes from patients with systemic lupus erythematosus. Rheumatology (Oxford) 2005;44(12):1507–1513. doi: 10.1093/rheumatology/kei083. [DOI] [PubMed] [Google Scholar]
- 5.Waldmann TA, Tagaya Y. The multifaceted regulation of interleukin-15 expression and the role of this cytokine in NK cell differentiation and host response to intracellular pathogens. Annu Rev Immunol. 1999;17:19–49. doi: 10.1146/annurev.immunol.17.1.19. [DOI] [PubMed] [Google Scholar]
- 6.Tagaya Y, Bamford RN, DeFilippis AP, Waldmann TA. IL-15: a pleiotropic cytokine with diverse receptor/signaling pathways whose expression is controlled at multiple levels. Immunity. 1996;4(4):329–336. doi: 10.1016/s1074-7613(00)80246-0. [DOI] [PubMed] [Google Scholar]
- 7.Schmitt-Ney M, Doppler W, Ball RK, Groner B. Beta-casein gene promoter activity is regulated by the hormone-mediated relief of transcriptional repression and a mammary-gland-specific nuclear factor. Mol Cell Biol. 1991;11(7):3745–3755. doi: 10.1128/mcb.11.7.3745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lieberman LA, Tsokos GC. The IL-2 defect in systemic lupus erythematosus disease has an expansive effect on host immunity. J Biomed Biotechnol. 2010;2010:740619. doi: 10.1155/2010/740619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Park YB, Kim DS, Lee WK, Suh CH, Lee SK. Elevated serum interleukin-15 levels in systemic lupus erythematosus. Yonsei Med J. 1999;40(4):343–348. doi: 10.3349/ymj.1999.40.4.343. [DOI] [PubMed] [Google Scholar]
- 10.Aringer M, Stummvoll GH, Steiner G, Koller M, Steiner CW, Hofler E, et al. Serum interleukin-15 is elevated in systemic lupus erythematosus. Rheumatology (Oxford) 2001;40(8):876–881. doi: 10.1093/rheumatology/40.8.876. [DOI] [PubMed] [Google Scholar]
- 11.Clark DN, Markham JL, Sloan CS, Poole BD. Cytokine inhibition as a strategy for treating systemic lupus erythematosus. Clin Immunol. 2013;148(3):335–343. doi: 10.1016/j.clim.2012.11.001. [DOI] [PubMed] [Google Scholar]
- 12.Chae DW, Nosaka Y, Strom TB, Maslinski W. Distribution of IL-15 receptor alpha-chains on human peripheral blood mononuclear cells and effect of immunosuppressive drugs on receptor expression. J Immunol. 1996;157(7):2813–2819. [PubMed] [Google Scholar]
- 13.Matys V, Kel-Margoulis OV, Fricke E, Liebich I, Land S, Barre-Dirrie A, et al. TRANSFAC and its module TRANSCompel: transcriptional gene regulation in eukaryotes. Nucleic Acids Res. 2006;34(Database issue):D108–110. doi: 10.1093/nar/gkj143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.DaSilva L, Rui H, Erwin RA, Howard OM, Kirken RA, Malabarba MG, et al. Prolactin recruits STAT1, STAT3 and STAT5 independent of conserved receptor tyrosines TYR402, TYR479, TYR515 and TYR580. Mol Cell Endocrinol. 1996;117(2):131–140. doi: 10.1016/0303-7207(95)03738-1. [DOI] [PubMed] [Google Scholar]
- 15.Mayr S, Welte T, Windegger M, Lechner J, May P, Heinrich PC, et al. Selective coupling of STAT factors to the mouse prolactin receptor. Eur J Biochem. 1998;258(2):784–793. doi: 10.1046/j.1432-1327.1998.2580784.x. [DOI] [PubMed] [Google Scholar]
- 16.Selliah N, Zhang M, DeSimone D, Kim H, Brunner M, Ittenbach RF, et al. The gammac-cytokine regulated transcription factor, STAT5, increases HIV-1 production in primary CD4 T cells. Virology. 2006;344(2):283–291. doi: 10.1016/j.virol.2005.09.063. [DOI] [PubMed] [Google Scholar]
- 17.Ahonen TJ, Xie J, LeBaron MJ, Zhu J, Nurmi M, Alanen K, et al. Inhibition of transcription factor Stat5 induces cell death of human prostate cancer cells. J Biol Chem. 2003;278(29):27287–27292. doi: 10.1074/jbc.M304307200. [DOI] [PubMed] [Google Scholar]
- 18.Schubert LA, King G, Cron RQ, Lewis DB, Aruffo A, Hollenbaugh D. The human gp39 promoter. Two distinct nuclear factors of activated T cell protein-binding elements contribute independently to transcriptional activation. J Biol Chem. 1995;270(50):29624–29627. doi: 10.1074/jbc.270.50.29624. [DOI] [PubMed] [Google Scholar]
- 19.Koshy M, Berger D, Crow MK. Increased expression of CD40 ligand on systemic lupus erythematosus lymphocytes. J Clin Invest. 1996;98(3):826–837. doi: 10.1172/JCI118855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Desai-Mehta A, Lu L, Ramsey-Goldman R, Datta SK. Hyperexpression of CD40 ligand by B and T cells in human lupus and its role in pathogenic autoantibody production. J Clin Invest. 1996;97(9):2063–2073. doi: 10.1172/JCI118643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Skov S, Bonyhadi M, Odum N, Ledbetter JA. IL-2 and IL-15 regulate CD154 expression on activated CD4 T cells. J Immunol. 2000;164(7):3500–3505. doi: 10.4049/jimmunol.164.7.3500. [DOI] [PubMed] [Google Scholar]
- 22.Mottonen M, Isomaki P, Luukkainen R, Toivanen P, Punnonen J, Lassila O. Interleukin-15 up-regulates the expression of CD154 on synovial fluid T cells. Immunology. 2000;100(2):238–244. doi: 10.1046/j.1365-2567.2000.00023.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang Y, Zhao M, Sawalha AH, Richardson B, Lu Q. Impaired DNA methylation and its mechanisms in CD4(+)T cells of systemic lupus erythematosus. J Autoimmun. 2013;41:92–99. doi: 10.1016/j.jaut.2013.01.005. [DOI] [PubMed] [Google Scholar]
- 24.Lu Q, Wu A, Tesmer L, Ray D, Yousif N, Richardson B. Demethylation of CD40LG on the inactive X in T cells from women with lupus. J Immunol. 2007;179(9):6352–6358. doi: 10.4049/jimmunol.179.9.6352. [DOI] [PubMed] [Google Scholar]
- 25.Chen T, Li Z, Jing T, Zhu W, Ge J, Zheng X, et al. MicroRNA-146a regulates the maturation process and pro-inflammatory cytokine secretion by targeting CD40L in oxLDL-stimulated dendritic cells. FEBS Lett. 2011;585(3):567–573. doi: 10.1016/j.febslet.2011.01.010. [DOI] [PubMed] [Google Scholar]
- 26.Cron RQ, Bort SJ, Wang Y, Brunvand MW, Lewis DB. T cell priming enhances IL-4 gene expression by increasing nuclear factor of activated T cells. J Immunol. 1999;162(2):860–870. [PubMed] [Google Scholar]
- 27.Schreiber E, Matthias P, Muller MM, Schaffner W. Rapid detection of octamer binding proteins with ‘mini-extracts’, prepared from a small number of cells. Nucleic Acids Res. 1989;17(15):6419. doi: 10.1093/nar/17.15.6419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Selliah N, Zhang M, White S, Zoltick P, Sawaya BE, Finkel TH, et al. FOXP3 inhibits HIV-1 infection of CD4 T-cells via inhibition of LTR transcriptional activity. Virology. 2008;381(2):161–167. doi: 10.1016/j.virol.2008.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Burchill MA, Goetz CA, Prlic M, O’Neil JJ, Harmon IR, Bensinger SJ, et al. Distinct effects of STAT5 activation on CD4+ and CD8+ T cell homeostasis: development of CD4+CD25+ regulatory T cells versus CD8+ memory T cells. J Immunol. 2003;171(11):5853–5864. doi: 10.4049/jimmunol.171.11.5853. [DOI] [PubMed] [Google Scholar]
- 30.Kirken RA, Erwin RA, Wang L, Wang Y, Rui H, Farrar WL. Functional uncoupling of the Janus kinase 3-Stat5 pathway in malignant growth of human T cell leukemia virus type 1-transformed human T cells. J Immunol. 2000;165(9):5097–5104. doi: 10.4049/jimmunol.165.9.5097. [DOI] [PubMed] [Google Scholar]
- 31.Behre G, Smith LT, Tenen DG. Use of a promoterless Renilla luciferase vector as an internal control plasmid for transient co-transfection assays of Ras-mediated transcription activation. Biotechniques. 1999;26(1):24–26. 28. doi: 10.2144/99261bm03. [DOI] [PubMed] [Google Scholar]
- 32.Cron RQ, Bartz SR, Clausell A, Bort SJ, Klebanoff SJ, Lewis DB. NFAT1 enhances HIV-1 gene expression in primary human CD4 T cells. Clin Immunol. 2000;94(3):179–191. doi: 10.1006/clim.1999.4831. [DOI] [PubMed] [Google Scholar]
- 33.Torgerson TR, Genin A, Chen C, Zhang M, Zhou B, Anover-Sombke S, et al. FOXP3 inhibits activation-induced NFAT2 expression in T cells thereby limiting effector cytokine expression. J Immunol. 2009;183(2):907–915. doi: 10.4049/jimmunol.0800216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brunner M, Zhang M, Genin A, Ho IC, Cron RQ. A T-cell-specific CD154 transcriptional enhancer located just upstream of the promoter. Genes Immun. 2008;9(7):640–649. doi: 10.1038/gene.2008.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cron RQ, Bandyopadhyay R, Genin A, Brunner M, Kersh GJ, Yin J, et al. Early growth response-1 is required for CD154 transcription. J Immunol. 2006;176(2):811–818. doi: 10.4049/jimmunol.176.2.811. [DOI] [PMC free article] [PubMed] [Google Scholar]