Abstract
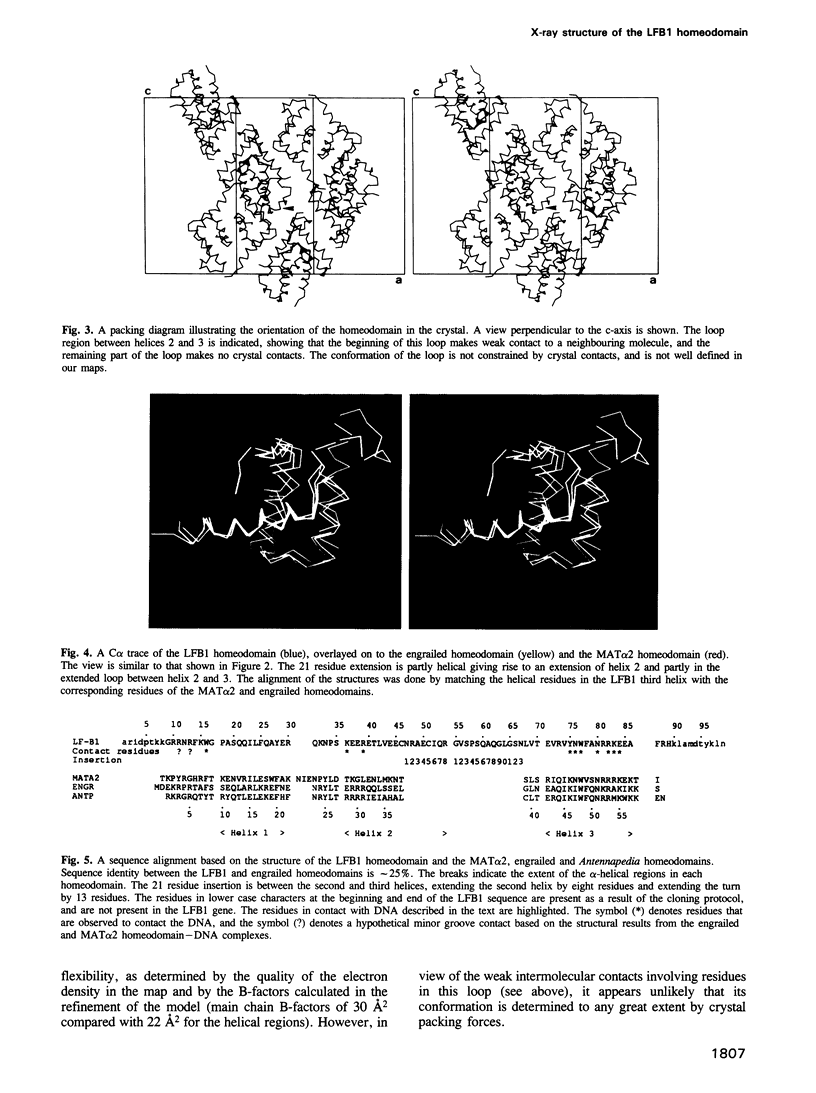
The transcription factor LFB1/HNF1 from rat liver nuclei is a 628 amino acid protein that functions as a dimer binding to the inverted palindrome GTTAATN-ATTAAC consensus site. We have crystallized a 99 residue protein containing the homeodomain portion of LFB1, and solved its structure using X-ray diffraction data to 2.8 A resolution. The topology and orientation of the helices is essentially the same as that found in the engrailed, MAT alpha 2 and Antennapedia homeodomains, even though the LFB1 homeodomain contains 21 more residues. The 21 residue insertion is found in an extension of helix 2 and consequent lengthening of the connecting loop between helix 2 and helix 3. Comparison with the engrailed homeodomain-DNA complex indicates that the mode of interaction with DNA is similar in both proteins, with a number of conserved contacts in the major groove. The extra 21 residues of the LFB1 homeodomain are not involved in DNA binding. Binding of the LFB1 dimer to a B-DNA palindromic consensus sequence requires either a conformational change of the DNA (presumably bending), or a rearrangement of the subunits relative to the DNA.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Brünger A. T., Kuriyan J., Karplus M. Crystallographic R factor refinement by molecular dynamics. Science. 1987 Jan 23;235(4787):458–460. doi: 10.1126/science.235.4787.458. [DOI] [PubMed] [Google Scholar]
- Chouard T., Blumenfeld M., Bach I., Vandekerckhove J., Cereghini S., Yaniv M. A distal dimerization domain is essential for DNA-binding by the atypical HNF1 homeodomain. Nucleic Acids Res. 1990 Oct 11;18(19):5853–5863. doi: 10.1093/nar/18.19.5853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finney M. The homeodomain of the transcription factor LF-B1 has a 21 amino acid loop between helix 2 and helix 3. Cell. 1990 Jan 12;60(1):5–6. doi: 10.1016/0092-8674(90)90708-m. [DOI] [PubMed] [Google Scholar]
- Frain M., Swart G., Monaci P., Nicosia A., Stämpfli S., Frank R., Cortese R. The liver-specific transcription factor LF-B1 contains a highly diverged homeobox DNA binding domain. Cell. 1989 Oct 6;59(1):145–157. doi: 10.1016/0092-8674(89)90877-5. [DOI] [PubMed] [Google Scholar]
- Jones T. A., Zou J. Y., Cowan S. W., Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr A. 1991 Mar 1;47(Pt 2):110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- Kissinger C. R., Liu B. S., Martin-Blanco E., Kornberg T. B., Pabo C. O. Crystal structure of an engrailed homeodomain-DNA complex at 2.8 A resolution: a framework for understanding homeodomain-DNA interactions. Cell. 1990 Nov 2;63(3):579–590. doi: 10.1016/0092-8674(90)90453-l. [DOI] [PubMed] [Google Scholar]
- Laughon A. DNA binding specificity of homeodomains. Biochemistry. 1991 Dec 3;30(48):11357–11367. doi: 10.1021/bi00112a001. [DOI] [PubMed] [Google Scholar]
- Nicosia A., Monaci P., Tomei L., De Francesco R., Nuzzo M., Stunnenberg H., Cortese R. A myosin-like dimerization helix and an extra-large homeodomain are essential elements of the tripartite DNA binding structure of LFB1. Cell. 1990 Jun 29;61(7):1225–1236. doi: 10.1016/0092-8674(90)90687-a. [DOI] [PubMed] [Google Scholar]
- Otting G., Qian Y. Q., Billeter M., Müller M., Affolter M., Gehring W. J., Wüthrich K. Protein--DNA contacts in the structure of a homeodomain--DNA complex determined by nuclear magnetic resonance spectroscopy in solution. EMBO J. 1990 Oct;9(10):3085–3092. doi: 10.1002/j.1460-2075.1990.tb07505.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastore A., De Francesco R., Barbato G., Castiglione Morelli M. A., Motta A., Cortese R. 1H resonance assignment and secondary structure determination of the dimerization domain of transcription factor LFB1. Biochemistry. 1991 Jan 8;30(1):148–153. doi: 10.1021/bi00215a022. [DOI] [PubMed] [Google Scholar]
- Pastore A., De Francesco R., Castiglione Morelli M. A., Nalis D., Cortese R. The dimerization domain of LFB1/HNF1 related transcription factors: a hidden four helix bundle? Protein Eng. 1992 Dec;5(8):749–757. doi: 10.1093/protein/5.8.749. [DOI] [PubMed] [Google Scholar]
- Qian Y. Q., Billeter M., Otting G., Müller M., Gehring W. J., Wüthrich K. The structure of the Antennapedia homeodomain determined by NMR spectroscopy in solution: comparison with prokaryotic repressors. Cell. 1989 Nov 3;59(3):573–580. doi: 10.1016/0092-8674(89)90040-8. [DOI] [PubMed] [Google Scholar]
- Riddihough G. Developmental biology. Homing in on the homeobox. Nature. 1992 Jun 25;357(6380):643–644. doi: 10.1038/357643a0. [DOI] [PubMed] [Google Scholar]
- Rosenfeld M. G. POU-domain transcription factors: pou-er-ful developmental regulators. Genes Dev. 1991 Jun;5(6):897–907. doi: 10.1101/gad.5.6.897. [DOI] [PubMed] [Google Scholar]
- Tomei L., Cortese R., De Francesco R. A POU-A related region dictates DNA binding specificity of LFB1/HNF1 by orienting the two XL-homeodomains in the dimer. EMBO J. 1992 Nov;11(11):4119–4129. doi: 10.1002/j.1460-2075.1992.tb05505.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verrijzer C. P., Alkema M. J., van Weperen W. W., Van Leeuwen H. C., Strating M. J., van der Vliet P. C. The DNA binding specificity of the bipartite POU domain and its subdomains. EMBO J. 1992 Dec;11(13):4993–5003. doi: 10.1002/j.1460-2075.1992.tb05606.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verrijzer C. P., van Oosterhout J. A., van Weperen W. W., van der Vliet P. C. POU proteins bend DNA via the POU-specific domain. EMBO J. 1991 Oct;10(10):3007–3014. doi: 10.1002/j.1460-2075.1991.tb07851.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolberger C., Vershon A. K., Liu B., Johnson A. D., Pabo C. O. Crystal structure of a MAT alpha 2 homeodomain-operator complex suggests a general model for homeodomain-DNA interactions. Cell. 1991 Nov 1;67(3):517–528. doi: 10.1016/0092-8674(91)90526-5. [DOI] [PubMed] [Google Scholar]