Abstract
Hyperconnectivity of neuronal circuits due to increased synaptic protein synthesis is postulated to cause Autism Spectrum Disorders (ASD). The mammalian target of rapamycin (mTOR) is strongly implicated in ASD via upstream signaling. However, downstream regulatory mechanisms are ill-defined. We show that knockout (KO) of the eukaryotic translation Initiation Factor 4E-Binding Protein 2 (4E-BP2), an eIF4E-repressor downstream of mTOR, or eIF4E overexpression lead to increased translation of neuroligins, which are post-synaptic proteins that are causally linked to ASD. 4E-BP2-KO mice exhibit an increased ratio of excitatory to inhibitory synaptic inputs and autistic-like behaviors: social interaction deficits, altered communication and repetitive/stereotyped behaviors. Pharmacological inhibition of eIF4E activity or normalization of neuroligin 1, but not neuroligin 2 protein amounts, restore the normal excitation/inhibition ratio and rectify the social behavior deficits. Thus, translational control by eIF4E regulates the synthesis of neuroligins, maintaining the excitation to inhibition balance, and its dysregulation engenders ASD-like phenotypes.
Introduction
ASD encompass unique behaviors within three domains: social interactions, communication, and repetitive or restricted interests and behaviors1. Dysregulation of the molecular machinery controlling synaptic mRNA translation has been postulated to cause ASD, mainly through increased synthesis of synaptic proteins and consequently augmented connectivity2.
A key factor required for translation initiation of the vast majority of mRNAs is the eIF4F complex that includes the cap-binding protein eIF4E, the RNA helicase eIF4A and the modular scaffolding protein eIF4G that bridges the mRNA to the ribosome3. 4E-BPs repress translation initiation by disrupting eIF4F complex formation4. While most cellular mRNAs require basal amounts of eIF4E to be translated, eIF4E preferentially enhances translation of a selective group of mRNAs with extensive secondary structure at their 5’UTRs, or other sequence elements3,5.
mTOR exists in two complexes: mTORC1, which is sensitive to the drug rapamycin, and mTORC2, which is not3. mTORC1 phosphorylates 4E-BPs4, stimulating translation. Hypophosphorylated forms of 4E-BPs bind to eIF4E and inhibit translation initiation. There are three mammalian 4E-BP paralogs encoding 4E-BP1, 2 and 3. 4E-BP2 is the major form in the mammalian brain and plays an important role in long-lasting synaptic plasticity, learning, and memory6.
Upstream of mTOR, germline mutations in the phosphatase and tensin homolog on chromosome ten (PTEN) are present in about 1–5% of patients with autism7. Pten-KO mice exhibit cognitive impairment and deficits in social interactions8, which are rescued by rapamycin9. Mutations in the two Tuberous Sclerosis Complex (TSC1/2) genes cause autism in a subset of patients10. Tsc2+/− mice display synaptic plasticity and memory deficits11,12, which are rescued by rapamycin, and enhanced ultrasonic vocalizations (USVs) in mouse pups13. Deletion of Tsc1 in cerebellar Purkinje cells in mice leads to autism-like phenotypes, rescued by rapamycin14. Recently, a de novo eIF4GI mutation was reported as a component of a highly interconnected network of proteins linked with sporadic autism15. Moreover, the mTORC1/eIF4E pathway is hyperactivated in Fragile-X Syndrome (FXS) patients, diagnosed with autism16. The hypothesis that eIF4E dysregulation is causally linked to ASD is consistent with recent findings that CYFIP1 [Cytoplasmic Fragile-X mental retardation protein (FMRP) Interacting Protein-1], an eIF4E binding protein17, is associated with ASD18.
Aberrant information processing due to increased ratio of synaptic excitation to inhibition (E/I) has been proposed to cause ASD19,20. Children with ASD exhibit elevations in resting-state neuronal activity, supporting an E/I imbalance as a neurobiological feature of ASD21. Modulations of the E/I balance using optogenetics directly influence autism-like behaviors in adult mice22. Several ASD mouse models display E/I imbalances due to altered glutamatergic excitation or to altered GABA (γ-aminobutyric acid)-ergic inhibition, leading to increased23,24 or decreased25–28 E/I ratio. The balance of excitatory and inhibitory synapses is largely controlled by the expression of adhesion molecules [neuroligins (NLGNs)-neurexins] and scaffolding proteins [PSD95 (post-synaptic density protein 95), gephyrin] in neurons29. Overexpression, knockout or knock-in studies of ASD-related NLGNs mutations report ASD-like phenotypes with alterations in the E/I balance (summarized in Supplementary Table 1).
A causal link between dysregulated eIF4E-dependent translation and the development of ASD is lacking. Here we show that deletion of mouse 4E-BP2 leads to autistic-like behaviors, including social interaction deficits, altered communication and repetitive/stereotyped behaviors. We find that translation of neuroligins (Nlgn) mRNAs is enhanced in 4E-BP2-KO and eIF4E-overexpressing mice. We report that unitary excitatory and inhibitory synaptic activity is increased in 4E-BP2-KOanimals, with excitation being predominantly augmented. Moreover, pharmacological normalization of eIF4F activity corrects the E/I imbalance and rescues the social behavior deficits of 4E-BP2-KO mice. Knockdown of Nlgn1, but not Nlgn2 in 4E-BP2-KO mice reverses the defects in social interactions and rectifies the E/I imbalance. Thus, enhanced eIF4E-dependent translation of mRNAs encoding synaptic adhesion proteins, such as NLGNs, leads to an aberrant increase in excitatory synaptic function, and results in ASD-like phenotypes.
Results
Autistic-like phenotypes in 4E-BP2-KO mice
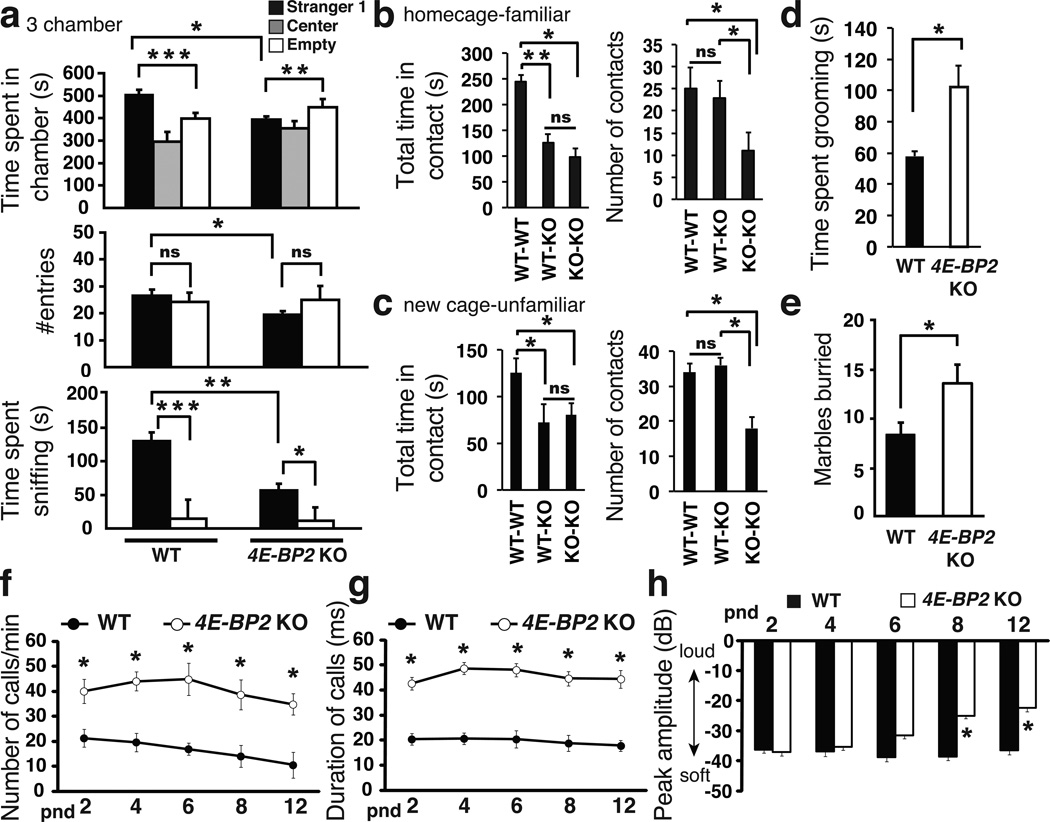
Since 4E-BP2 deletion causes increased eIF4F complex formation and activity, we postulated that it could engender ASD-related behaviors. Social interaction deficits are a salient autistic behavioral feature in humans1. To test our hypothesis, we used a three-chamber social arena32 to assess the preference of a test mouse for a social (Stranger1) over a non-social (empty wire-cage) stimulus or for social novelty (Stranger2). 4E-BP2-KO mice displayed impaired social approach behavior as depicted by the significantly reduced time spent in the Stranger1 compartment, and the decreased interaction time with Stranger1, as compared to WT littermates (Fig. 1a & Supplementary Fig. 2). Given the lack of preference for the social stimulus in the first portion of the three-chamber test, preference for social novelty was also reduced in 4E-BP2-KO mice (Supplementary Fig. 3). Because the reduced number of entries of the 4E-BP2-KO mice into the Stranger1 chamber relative to WT littermates (Fig. 1a) might underlie an anxietylike phenotype, we assessed anxiety levels. No differences between WT and 4E-BP2-KO mice were found using the Elevated Plus Maze (EPM) test following the three-chamber social approach test (Supplementary Fig. 4d). Moreover, WT and 4E-BP2-KO mice were indistinguishable in their initial exploration of the three-chamber arena (Supplementary Fig. 4a).
Figure 1. Social interaction deficits, repetitive behavior and elevated USVs in4E-BP2-KO mice.
(a) Three-chamber social interaction test, showing time spent in each chamber, number of entries and time spent sniffing the wire-cage; n=12 for each group; ***p<0.001, **p<0.01, *p<0.03, two-way ANOVA with Bonferroni’s post-hoc.
(b) Homecage and (c) reciprocal social interactions tests showing total time in contact and number of contacts; n=18 per group **p<0.02, *p<0.03; one way ANOVA with Bonferroni’s post-hoc.
(d) Self-grooming test showing total time spent grooming.
(e) Marble-burying test showing marbles buried; n=12 per group *p<0.03; Student's t-test.
(f–h) Isolation induced USVs at various post-natal days (pnd) (f) number of calls/min, (g) duration of calls and (h) amplitude of USVs in pups; n=12 per group *p<0.05; two-way ANOVA, with Bonferroni’s post-hoc. Error-bars mean ±SEM.
To further characterize the social behavior of the 4E-BP2-KO mice, we performed a home cage and reciprocal social interaction test in a separate cage (Fig. 1b,c). Pairs of WT-WT, KO-WT or KO-KO mice were either recorded in their home cage, or introduced to a familiar open-field environment. The KO-WT, KO-KO pairs interacted for a shorter period of time, as compared to the WT-WT pair (Fig. 1b, c), without any difference in the total number of contacts between the WT-WT or WT-KO pairs of mice. KO-KO pairs initiated significantly fewer contacts (Fig. 1b,c). This discrepancy can be attributed to the larger number of contacts initiated by the WT mice in the WT-KO pairs (Supplementary Fig. 2h). Anxiety is not a confounding factor for these behaviors, as evidenced by the similar anxiety levels in WT and 4E-BP2-KO mice in the EPM test (Supplementary Fig. 4b, c). To assess stereotyped/repetitive behaviors, we recorded self-grooming and used a marble burying test17. 4E-BP2-KO mice displayed longer bouts and total duration of self-grooming (Fig. 1d & Supplementary Figure 2i) and buried significantly more marbles than WT littermates (Fig. 1e).
Isolation-induced USVs emitted from mouse pups separated from their mother are a quantifiable behavior considered relevant to ASD30. Tsc2+/− pups display enhanced USVs (increased number of calls, duration, peak amplitude and peak frequency)13,14. We recorded USVs in 4E-BP2-KO and WT littermate pups, following separation from their dams (Fig. 1f–h). 4E-BP2-KO pups emitted a higher number of calls (Fig. 1f), with a longer duration (Fig. 1g) at all post-natal days relative to WT littermates. Furthermore, the peak amplitude of the USVs was higher on postnatal days 8 and 12 (Fig. 1h). Taken together, our data demonstrate that genetic deletion of 4E-BP2, mimicking eIF4E upregulation, engenders ASD-like behaviors.
Increased eIF4E-dependent translation of neuroligins
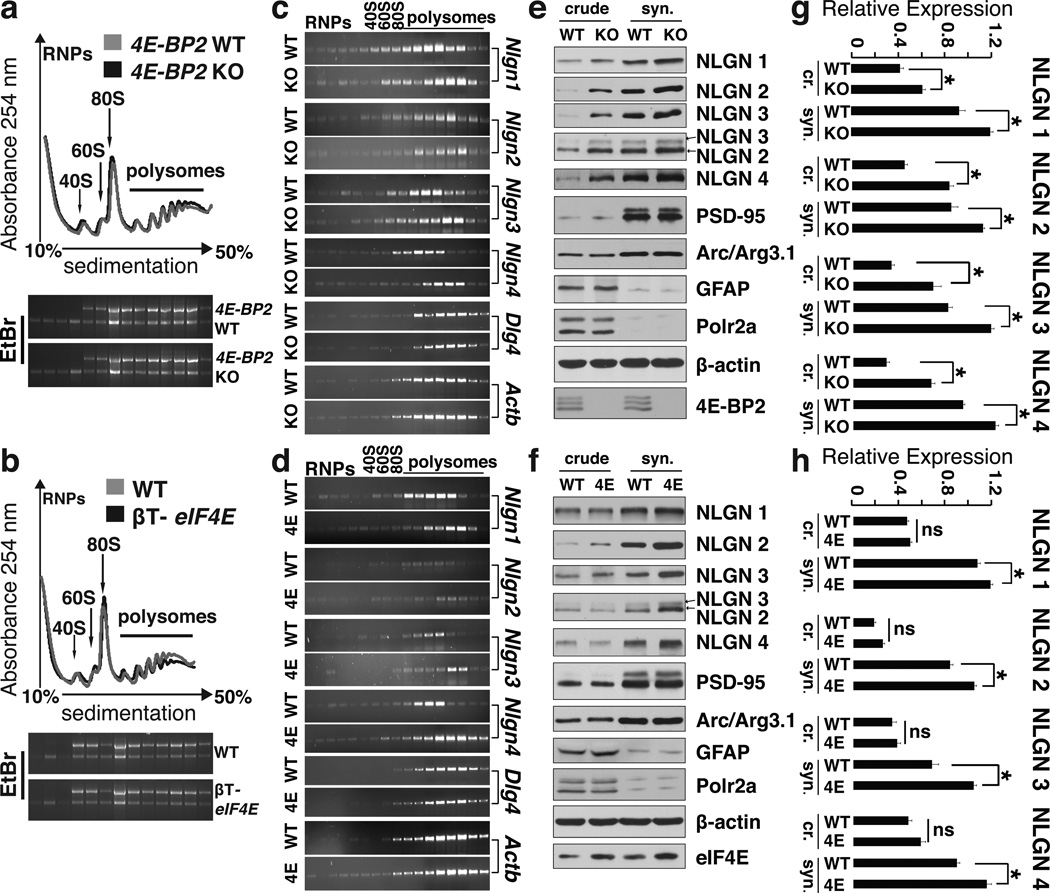
We hypothesized that the ASD-like phenotypes of the 4E-BP2-KO mice are a result of altered translation of a subset of mRNAs, whose translation initiation is stimulated by eIF4E activity. To test this hypothesis, we performed polysome profiling, which measures translation initiation rates, of hippocampal lysates from 4E-BP2-KO and eIF4E-overexpressing (βT-eIF4E)31 mice. The polysome profile was not significantly altered in 4E-BP2-KO mice as compared to WT littermates (Fig. 2a & Supplementary Fig. 5d). This is in agreement with the lack of changes in [35S] Met/Cys incorporation (Supplementary Fig. 5a, b) and previous reports32. We then examined the polysome distribution of 24 mRNAs encoding for proteins known to be associated with ASD (Supplementary Table 2)33,34. These include among others, mRNAs encoding the family of Nlgns and neurexins, shank (SH3 and multiple ankyrin repeat domains protein) 2–3 and MeCP2 (methyl-CpG-binding protein). Strikingly, out of the 24 mRNAs examined, only Nlgn1–4 mRNAs were shifted towards heavier polysome fractions in the 4E-BP2-KO animals, relative to WT littermates (Fig. 2c), indicating increased translation initiation. Neither the levels of the Nlgn1–4 mRNAs, nor those of any other mRNA were different between 4E-BP2-KO and WT mice (Supplementary Fig. 6c), thus excluding transcriptional effects. As expected, protein amounts of all four NLGNs were increased in crude and synaptosomal extracts from 4E-BP2-KO hippocampi (Fig. 2e, g). Elevated translation, but not transcription, of Nlgn mRNAs was also observed in polysome profiles of βT-eIF4E mice (Fig. 2d and Supplementary Fig. 6c) with no differences in translation or transcription of other mRNAs (Supplementary Figs. 6c, 7c). Accordingly, NLGNs protein amounts were increased in the synaptosomal fractions from βT-eIF4E mice relative to WT (Fig. 2f, h). The increased translation of NLGNs, observed only in synaptosomal fractions of βT-eIF4E mice can be explained by the mild overexpression (only 29%) of eIF4E (Fig. 2f). In contrast, no changes were observed in the translation of mRNAs encoding neurexins (Supplementary Fig. 6) or PSD95 (Dlg4 gene; Fig. 2c–f) in 4E-BP2-KO or βT-eIF4E versus WT mice. Translation of mRNAs encoding for gephyrin (Gphn), shank2, shank3 and SAPAP3 (Dlgap3) proteins, which orchestrate the formation of the post-synaptic scaffolding29 was not changed in 4E-BP2-KO or in the βT-eIF4E mice (Supplementary Fig. 7). In conclusion, relief of translational suppression by loss of 4E-BP2 or by overexpression of eIF4E selectively enhances the synthesis of neuroligins, causing an imbalance between these adhesion proteins relative to scaffolding proteins.
Figure 2. Enhanced eIF4E-dependent translation of neuroligins.
(a) Polysome profiles from hippocampal lysates of WT and 4E-BP2-KO mice and (b) WT and βT-eIF4E mice. Positions of 40S, 60S and 80S ribosome peaks and polysomes are indicated.
(c) RT-PCR on RNA extracted from polysome fractions from WT and 4E-BP2-KO and
(d) WT and βT-eIF4E (4E) hippocampi. Representative gel images are shown; n=4 per group.
(e) Representative immunoblots of crude (cr.) and synaptosomal (syn.) fractions from hippocampal lysates of WT or 4E-BP2-KO and (f) WT or βT-eIF4E (4E) mice, probed with antibodies against the indicated proteins.
(g–h) Quantification of immunoblots from (e) and (f) for neuroligins; n=4 per group *p<0.05; Student's t-test. Error-bars mean ±SEM.
Increased synaptic activity in 4E-BP2-KO mice
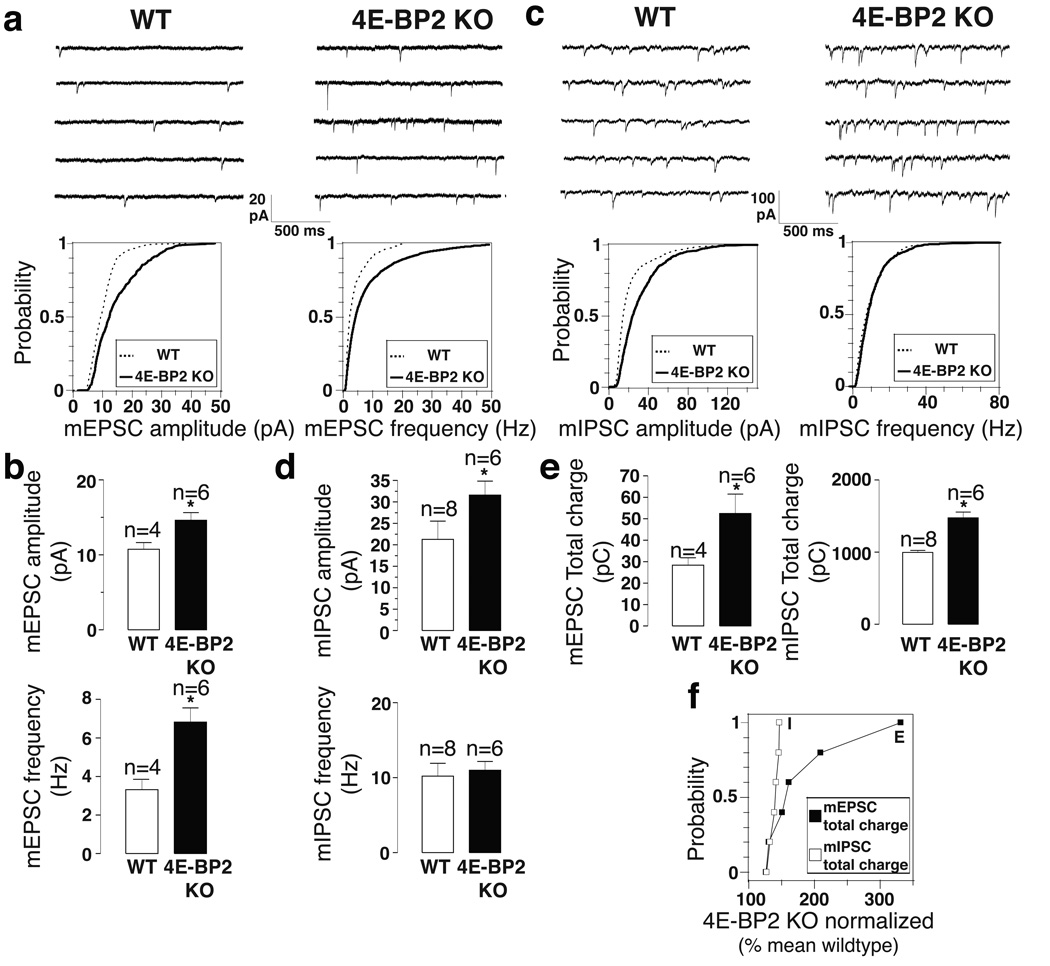
An altered E/I ratio was reported in several ASD mouse models23–28. Therefore, we examined the effects of 4E-BP2-KO on unitary excitatory and inhibitory synaptic transmission in acute hippocampal slices. AMPA (α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid) receptor-mediated miniature excitatory postsynaptic currents (mEPSCs) were increased both in amplitude and frequency in pyramidal neurons of 4E-BP2-KO relative to WT mice (Fig. 3a, b), indicating that 4E-BP2 de-repression augments transmission at excitatory synapses in acute hippocampal slices. We next examined whether transmission was similarly altered at inhibitory synapses. GABAA receptor-mediated miniature inhibitory postsynaptic currents (mIPSCs) were significantly greater in amplitude, but not in frequency (unlike mEPSCs) in pyramidal cells of 4E-BP2-KO relative to WT slices (Fig. 3 c, d). To study the differential effect on excitatory versus inhibitory transmission, we measured the total synaptic charge transfer for mEPSCs and mIPSCs over a 10 min period – a parameter reflecting both amplitude and frequency of miniature synaptic events. The total charge transfer was significantly increased for both mEPSCs and mIPSCs in 4E-BP2-KO pyramidal cells (Fig. 3e). However, the normalized increase of total charge transfer in 4E-BP2-KO relative to WT was significantly larger for mEPSCs than for mIPSCs (Fig. 3f). We also recorded minimal stimulation-evoked EPSCs and IPSCs from CA1 pyramidal neurons in WT and 4E-BP2-KO slices and observed an increase in both EPSC and IPSC amplitude, but no change in paired pulse ratio (Supplementary Fig. 8). Moreover, we observed a greater increase in Vesicular Glutamate Transporter (VGLUT) than Vesicular GABA Transporter (VGAT) puncta and protein amounts in the CA1 dendritic layer of 4E-BP2-KO hippocampal slices, relative to WT littermates (Supplementary Fig. 9a–g). Finally, spine density is increased in CA1 pyramidal cell dendrites in 4E-BP2-KO mice (Supplementary Fig. 9h, i). Thus, 4E-BP2 de-repression results in altered E/I balance.
Figure 3. Excitation is increased to a larger extent than inhibition in 4E-BP2-KO mice.
(a, c) Representative traces (top) and cumulative distribution plots (bottom) of mEPSCs (a) and mIPSCs (c) from CA1 pyramidal in acute slices from WT and 4E-BP2−/− mice, illustrating changes in amplitude and frequency.
(b, d) Bar graphs showing mEPSC (b) and (d) IPSC amplitude and frequency (n=number of cells).
(e) Bar graphs showing the synaptic total charge transfer in 4E-BP2-KO relative to WT slices. For a-e, *p<0.05; one-way ANOVA with Bonferroni’s post-hoc.
(f) Relative changes in mEPSC and mIPSC total charge transfer, normalized to the mean WT value, for each neuron from 4E-BP2-KO slices; Kolmogorov-Smirnov test, p=0.005, KS statistic=0.916. Error-bars mean ±SEM.
Pharmacological rescue of ASD-like phenotypes
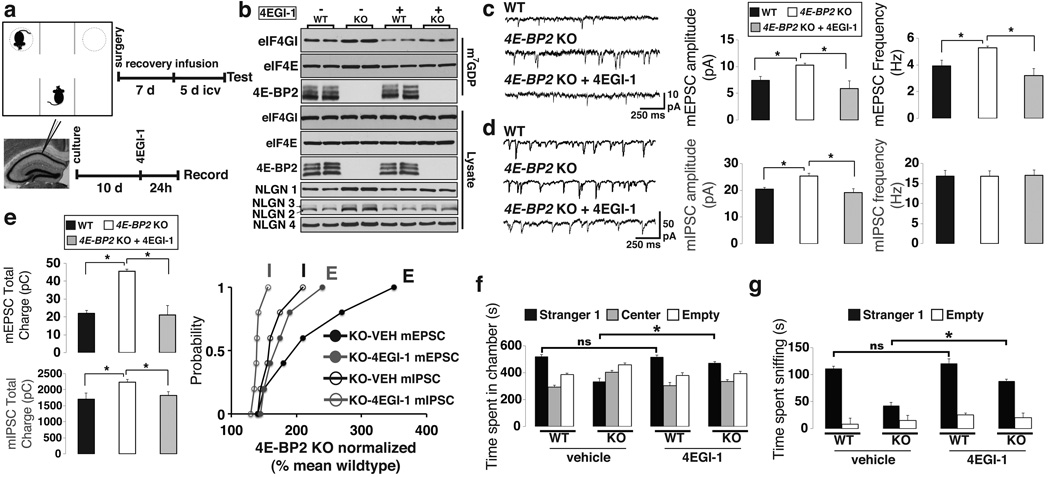
To bolster the conclusion that translation initiation through eIF4E is critical in altering synaptic function causing ASD, we used the selective inhibitor 4EGI-1, which prevents eIF4E binding to eIF4G, to test whether it would rescue autistic-like behaviors in 4E-BP2-KO mice. First, we performed cap-column pull-down assays with hippocampal lysates to assess the status of the eIF4F complex. As expected, increased amounts of eIF4G and eIF4E were bound to the m7GDP-cap analogue matrix in 4E-BP2-KO mice, as compared to WT mice (Fig. 4b). Infusion of 4EGI-1 reduced the amounts of cap-bound eIF4G and eIF4E to WT levels (Fig. 4b). Significantly, NLGNs protein levels were also reduced to WT levels following 4EGI-1 treatment (Fig. 4b).
Figure 4. Rescue of excitatory/inhibitory synaptic activity imbalance and social deficits in 4E-BP2-KO by inhibiting the eIF4E-eIF4G interaction.
(a) Schematic of 4EGI-1 treatment in the three-chamber test (Top) and slice-culture recordings (Bottom).
(b) Immunoblot analysis of cap column pull-down in 4E-BP2-KO and WT mice treated with 4EGI-1 or vehicle. Total and m7GDP bound extracts were probed for eIF4GI, eIF4E and 4E-BP2; n=4 - quantification in Supplementary Fig. 16a
(c, d) Effects of 4EGI-1 on mEPSCs (c) or mIPSCs (d) amplitude and frequency in 4E-BP2-KO slices.
(e) Effects of 4EGI-1 on total charge transfer of mEPSCs (Top) and mIPSCs (Middle) in 4E-BP2-KO slices. Bottom: Relative changes in mEPSC and mIPSC total charge transfer, normalized to the mean WT value, for 4E-BP2-KO neurons. Kolmogorov-Smirnov test. For c-e, n=6 per group; *p< 0.05; one-way ANOVA with Bonferroni’s post-hoc.
(f, g) Rescue effects of 4EGI-1 infusion in the three-chamber test showing (f) time spent in chambers and (g) time spent sniffing the wire cage in 4E-BP2-KO and WT mice.
For f, g, n=12 per group; *p<0.03; two-way ANOVA, with a Bonferroni’s post-hoc. Error-bars mean ±SEM.
4EGI-1 blocks long-term memory consolidation, but not reconsolidation in mice35. In 4E-BP2-KO mice a single tetanic train elicits late-phase long-term potentiation (L-LTP)6, indicating a lower threshold for L-LTP induction compared to WT mice. Bath application of 4EGI-1 in acute hippocampal slices inhibited the facilitation of L-LTP induction in 4E-BP2-KO mice (Supplementary Fig. 10c). Moreover, application of 4EGI-1 (Fig. 4a) to hippocampal slice cultures from 4E-BP2-KO mice reversed the increase in mEPSC amplitude and frequency (Fig. 4c), and in mIPSCs amplitude (Fig. 4d), decreased the total mEPSCs and mIPSCs charge transfer (Fig. 4e), and restored the E/I balance. Next, we examined if 4EGI-1 could rescue the social behavior deficits in 4E-BP2-KO mice. We used intra-cerebroventricular (icv) cannulation to daily infuse 4EGI-1 for five days (Fig. 4a). Mice were then subjected to the three-chamber social interaction test. While a sub-threshold concentration of 4EGI-1 had no effect on WT mice (Fig. 4f and Supplementary Fig. 10b), it reversed social interaction deficits of 4E-BP2-KO mice, inasmuch as 4EGI-1-treated 4E-BP2-KO mice spent significantly more time in the Stranger1 chamber (Fig. 4f), while the number of entries for the 4E-BP2-KO mice infused with 4EGI-1 was restored to WT levels (Supplementary Fig. 12a). Also, the interaction time with Stranger1 was increased compared to vehicle-treated KO mice (Fig. 4g). These results establish causality between inactivation of the eIF4F complex and rescue of the social interaction impairments in the 4E-BP2-KO mice.
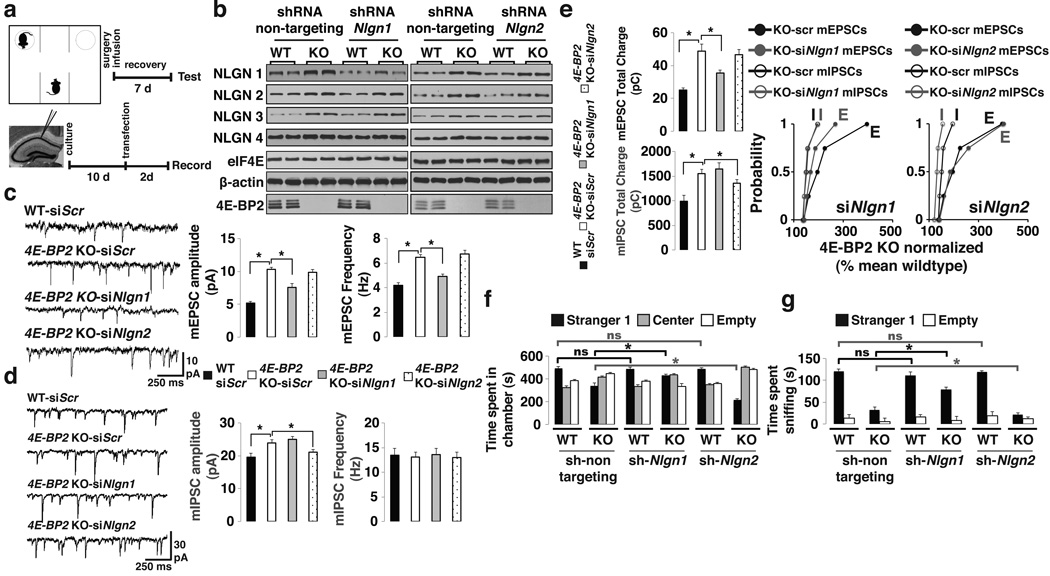
Nlgn 1 knockdown rescues ASD-like phenotypes in 4E-BP2-KO mice
NLGN1, which is present exclusively at excitatory synapses, promotes excitatory synaptic transmission36,37. In 4E-BP2-KO mice Nlgn1 mRNA translation is enhanced concomitant with increased E/I ratio (Figs. 2, 3). We therefore predicted that reduction of NLGN 1 levels in 4E-BP2-KO mice would restore the E/I balance and reverse the autism-related deficits. To investigate this, we used siRNAs against Nlgn1 in hippocampal pyramidal cells in slice cultures (Fig. 5a). Knockdown of Nlgn1 rescued the amplitude and frequency of mEPSCs in 4E-BP2-KO pyramidal cells (Fig. 5c), without affecting mIPSCs (Fig. 5d). As expected, mEPSC total charge transfer in pyramidal cells of 4E-BP2-KO mice transfected with siNlgn1 was restored to WT levels (Fig. 5e), while mIPSC total charge transfer was unaffected (Fig. 5e). Next, we knocked-down Nlgn2, a regulator of inhibitory synapses. In contrast to NLGN1, knockdown of NLGN2 rescued the increase in mIPSC amplitude in 4E-BP2-KO pyramidal cells (Fig. 5d), without affecting the amplitude or frequency of mEPSCs (Fig. 5c). The increase in total charge transfer of mIPSCs was also attenuated in siNlgn2 transfected 4E-BP2-KO pyramidal cells, while mEPSC total charge transfer remained unaltered (Fig. 5e).
Figure 5. Knockdown of neuroligin 1 rescues the excitatory/inhibitory synaptic activity imbalance and social deficits in 4E-BP2-KO mice.
(a) Schematic of the three-chamber test in mice icv-injected with lentiviruses expressing shRNAs (Top) and recordings from siRNA-transfected slice-cultures (Bottom).
(b) Western blot analysis of hippocampal lysates from WT or 4E-BP2-KO mice injected with lentiviruses expressing shRNAs against a non-targeting sequence or Nlgn1-2. Extracts were probed for NLGN1-4, β -actin, eIF4E and 4E-BP2; n=4-quantification in Supplementary Fig. 16b,c.
(c,d) Effects of Nlgn1 or Nlgn2 knockdown on mEPSCs (c) or mIPSCs (d) amplitude and frequency in transfected pyramidal cells.
(e) mEPSCs (Top) or mIPSCs (Middle) total charge transfer in Nlgn1-2 knockdown cells. Bottom: Relative changes in mEPSC and mIPSC total charge transfer, normalized to the mean WT value, for each neuron from 4E-BP2-KO slices; Kolmogorov-Smirnov test. For c-e, n=5 per group; *p<0.05; one-way ANOVA with Bonferroni’s post-hoc.
(f,g) Knockdown of Nlgn1-2 in the three-chamber test showing (f) time spent in chambers and (g) time spent sniffing the wire-cage.
For f–g, n=12 per group; *p<0.02; two-way ANOVA, with a Bonferroni’s post-hoc. Error-bars mean ±SEM.
We next subjected WT or 4E-BP2-KO mice to the three-chamber social interaction test, after icv-injection of shRNAs (Fig. 5a and Supplementary Fig. 11). Notably, NLGN1 or NLGN2 protein levels decreased to WT levels in 4E-BP2-KO mice injected with Nlgn1 or Nlgn2 shRNA, respectively, while protein amounts of the other NLGNs were not changed (Fig. 5b). We selected a concentration of lentiviral shRNA (Supplemental Fig. 11) that had no effect in WT animals (Fig. 5f, g). Importantly, this sub-threshold concentration of Nlgn1 shRNA partially rescued the social interaction impairment in 4E-BP2-KO mice as follows: 4E-BP2-KO mice injected with Nlgn1 shRNA spent significantly more time in the Stranger1 chamber (Fig. 5f), entered the chamber more times (Supplementary Fig. 12b), and interacted more with Stranger1 (Fig. 5g) relative to control KO mice. In contrast, knockdown of Nlgn2 exacerbated the social deficits in 4E-BP2-KO mice, as evidenced by the reduced time spent in the Stranger1 chamber (Fig. 5f), and the decreased amount of time spent interacting with Stranger1 (Fig. 5g) as compared to control KO mice. We recapitulated these findings with additional shRNAs against Nlgn1 or Nlgn2 (Supplementary Fig. 13). In summary, NLGN1 knockdown reverses changes at excitatory synapses and partially rescues the social interaction deficits in 4E-BP2-KO mice, while NLGN2 knockdown weakens inhibitory synapses and exacerbates the phenotype, thus establishing a strong link between eIF4E-dependent translational control of NLGNs, E/I balance and the development of ASD-like phenotypes.
Discussion
We show that 4E-BP2 deficiency in mice causes autistic-like phenotypes that strikingly resemble behavioral hallmarks of ASD. 4E-BP2-KO mice display a previously undocumented deficit in social interactions and exhibit repetitive/stereotyped behaviors. Communication between 4E-BP2-KO pups and their dams is altered, as evidenced by enhanced ultrasonic vocalizations. This phenotype is similar to that reported for TSC2+/− 13 or conditional TSC2-KO14 mice, which could conceivably be explained by increased eIF4E activity. These newly discovered autistic-like phenotypes of 4E-BP2-KO mice are neither confounded by anxiety, nor by the previously reported long-term memory deficits6,38, as short-term memory circuits are intact. Thus, 4E-BP2-KO mice constitute a novel model for mTORC1/eIF4E-dependent autism-like phenotypes due to dysregulated translational control. In accordance with this model, a point mutation in the promoter region of the EIF4E gene is associated with increased activity of eIF4E in autistic siblings from two unrelated families39.
In two models of enhanced eIF4E activity (4E-BP2-KO and eIF4E-overexpression mice), we show that Nlgn mRNAs are translated more efficiently. These results highlight the important role of 4E-BP2 and eIF4E in post-synaptic translational control of neuronal mRNAs. Increased mTOR signaling to its downstream effectors promotes the preferential translation of a subset of mRNAs referred to as “eIF4E sensitive”, harboring extensive secondary structure elements in their 5’ UTRs5,40. Reporter mRNAs containing the full-length 5’UTRs of Nlgn1 and Nlgn2, but not Dlg4 or Actb, fused to luciferase, are better translated in cells with elevated mTORC1 signaling (Pten+/−, Tsc2 KO) and/or enhanced cap-dependent translation (4E-BP2-KO, eIF4E overexpression) as compared to parental cells (Supplementary Fig. 14). Interestingly, the 5'UTRs of Nlgn mRNAs possess a repeated structural element that is absent from the other mRNA 5’UTRs (Supplementary Fig. 15). These secondary structure repeats might account for the preferential increase in translation of neuroligins in response to increased eIF4F activity.
Mutations and copy number variation in NLGNs have been associated with ASD41. In 4E-BP2-KO mice, increased amounts of NLGN1–4 lead to a disproportionate ratio of NLGNs to PSD95 (in excitatory synapses) or to gephyrin (in inhibitory synapses) because the general composition of the post-synaptic scaffold (other than NLGNs) is not altered by 4E-BP2 deletion or similarly overexpression of eIF4E. An altered E/I synaptic ratio is a prominent feature in ASD and in mouse models of autism19,21. The relative impact of 4E-BP2-KO on excitation is greater than on inhibition, thus engendering a shift in the E/I balance. In 4E-BP2-KO mice pharmacological inhibition of the eIF4E-eIF4G interaction using the selective inhibitor 4EGI-1 reduces eIF4F activity to wild-type, restores the L-LTP threshold and rescues the E/I imbalance phenotype. In 4E-BP2-KO mice, knockdown of Nlgn1 restores the E/I balance and improves social approach behavior to WT levels. In contrast, Nlgn2 knockdown increases the E/I imbalance and exacerbates the social behavior deficits. These results establish a link between eIF4E translational control of Nlgns, E/I balance and ASD-like phenotypes (Supplementary Fig. 1).
This novel regulatory mechanism should provide an impetus to further investigate the role of eIF4E-mediated translation in ASD and to explore therapeutic avenues for the pharmacological modulation of eIF4F activity (e.g. with 4E-GI-1 and its derivatives) or the function of neuroligins.
Methods Summary
All procedures are in accordance with the collaborating institutions Animal Care Committee guidelines. Experimenters were blinded to the genotype during testing and scoring. All data are presented as mean ±SEM (error bars). Statistical results, along with tests used are summarized in Supplementary Table 4 Statistical significance was set a priori at 0.05.
Full Methods are available in the online version of the paper at www.nature.com/nature
Methods Section
Knockout and Knock-in Mice
4E-BP2-KO and βT-eIF4E mice were previously described10,34. Where mentioned, WT mice are littermates of the corresponding genotype KO or knock-in mice. For all experiments littermates from heterozygote crossings were used. Mice were backcrossed for more than 10 generations to C57Bl/6J mice. Food and water were provided ad libitum, and mice were kept on a 12 h light/dark cycle. Pups were kept with their dams until weaning at postnatal day 21. After weaning, mice were group housed (maximum of 4 per cage) by sex, or housed in pairs (for homecage social interaction). Cages were maintained in ventilated racks in temperature (20–21°C) and humidity (~55%) controlled rooms, on a 12-hour circadian cycle (7am-7pm light period). Standard bedding was used for housing with the addition of Enviro-Dri (Shepherd Specialty Paper) and a small cardboard house. All procedures were in compliance with the Canadian Council on Animal Care guidelines and were approved by McGill University, UCSF and Université de Montréal Animal Care Committees. For all behavioral assays, mice were handled for 3 consecutive days prior to experimental testing.
Three-Chamber Social Approach and Preference for Social Novelty Tests
A three-chamber arena was used to assess social approach and preference for social novelty32. Test mice (2 month old males) were placed in the middle chamber and allowed to explore all the empty chambers of the apparatus freely for 10 min. Immediately after habituation, an unfamiliar mouse (Stranger1, male C57BL/6 age matched) was introduced into one of the two side chambers, enclosed in a wire cage allowing only for the test mouse to initiate any social interaction. An identical empty wire cage was placed in the other side chamber. Following placement, the side doors were opened simultaneously and the test mouse was allowed to explore the whole three-chamber arena for 20 min. At the end of the 20 min sociability test, a new unfamiliar mouse (Stranger2, male C57BL/6 age matched) was placed in the previously unoccupied wire cage, and test mice were examined for an additional 20 min to assess preference for social novelty. The location of the empty wire cage was alternated between side chambers for different subjects. The time spent in each compartment, the number of transitions between chambers and the time spent sniffing the Stranger1, Stranger2 or empty wire cages, were manually scored.
Homecage Social Interaction Test
WT or KO 4E-BP2 male littermate mice were weaned and housed in pairs of WT-WT, KO-KO or WT-KO. At the age of 2 months, mice were recorded for 20 min to assess homecage social interactions. The cage lid, grid and cardboard nest were removed for the recording session and time in close contact [huddling, sniffing (nose-to-nose, anogenital sniffing) and allogrooming] was scored for each mouse. A small mark with an odorless, permanent marker and an ear punch (both marked the day before the test), were used to identify each genotype. Tagging was alternated randomly between WT and KO mice in the various sessions.
Reciprocal Social Interaction Test
Subjects were unfamiliar 4E-BP2-KO or WT mice, originating from separate litters. The social interaction in pairs (WT-WT, WT-KO or KO-KO) test was performed in a neutral cage (gray Plexiglas box, 30×30×30 cm). Mice were first habituated for 2 days in 10 min sessions and on day 3 pairs of unfamiliar mice with the same genotype were placed into the neutral cage for 10 min. The time spent in close contact (indicative of social interaction) was registered. Close contact is defined as close huddling, sniffing (nose-to-nose, anogenital sniffing) or allogrooming by the target and stimulus mouse. A small mark with an odorless, permanent marker and an ear punch (both marked the day before the test), were used to identify each genotype. Tagging was alternated randomly between WT and KO mice in the various sessions.
Self-Grooming Test
WT or 4E-BP2-KO mice were placed in a new Plexiglas cage with fresh bedding and no nest or cardboard material. Self-grooming behavior was recorded for 10 min following an initial 10 min habituation phase.
Marble Burying Task
Mice were individually placed in Plexiglas cages containing 5 cm deep of fresh bedding, with 20 black marbles pre-arranged in 5×4 evenly spaced rows. Testing was conducted for 20-min. After the test period, unburied marbles were counted. Marbles were considered buried if they were at least one-half covered with bedding.
Isolation Induced USVs in Mouse Pups
A microphone (Knowles Acoustics FG-3329C) was fixed inside the top of an anechoic glass chamber wrapped with a heated mat maintaining a constant temperature of 21°C. The microphone was connected to a bat detector (Ultrasonic Detector D 230; Pettersson Elktronic AB; Uppsala, Sweden), which was set for frequency division mode so that the entire frequency range (10–120 kHz) was divided by 10 and thus made audible to the human ear. The frequency division system of the D 230 retains the original input amplitude. The signal from the bat detector was amplified through a MIDI interface (US-122; Tascam TEAC Professional Division) and fed to a standard PC for recording using a digital recording system (MITSYN, WLH, Version 2005.7.0.0, Belmont, Mass 02478). Newborn pups (postnatal days 2, 4, 6, 8 and 12) were carefully separated from their dams and placed individually in the recording chamber. USVs were recorded for 5 min. Recordings were analyzed blind to genotype with MITSYN (audio-editing) software.
Elevated Plus Maze (EPM) Test
For the elevated plus maze apparatus we constructed a cruciform platform consisting of: two open arms (30 cm × 5 cm) assembled opposite each other and two enclosed arms (30 cm × 5 cm × 15 cm) also facing each other. A center platform (5 cm × 5 cm) was used as an anchor point for all arms and the entire apparatus was elevated 40 cm off the floor. Low light and white noise conditions were used in the test room. Each mouse was placed in the center of the maze facing an open arm. During the testing period of 5 min, the total time spent in each arm was recorded.
Contextual Fear Conditioning Test
WT or 4E-BP2-KO mice were cannulated, as described in the stereotaxic surgery section. During acquisition, two foot shocks of 0.5 mA for 2 s separated by 60 s were administered after the 2-min period of context exploration. Immediately after acquisition, mice were infused with different concentrations of 4EGI-1. Twenty four h after training, mice were tested for contextual fear memory, as assessed by % freezing in the conditioning context for a 5-min period, in 5-s intervals, either “freezing” or “not freezing”. Freezing (%) indicates the number of intervals where freezing was observed divided by total number of 5-s intervals.
Polysome Profile Analysis
Polysome profile analysis was carried out as previously described40. Intact hippocampi were washed with ice-cold PBS containing 100 µg/ml cycloheximide and subsequently lysed in a hypotonic lysis buffer (5 mM Tris-HCl (pH 7.5), 2.5 mM MgCl2, 1.5 mM KCl, 100 µg/ml cycloheximide, 2 mM DTT, 0.5% Triton X-100, and 0.5% sodium deoxycholate). Lysate concentration was balanced using a Bradford-assay (BIORAD) and by measuring total RNA concentration using a NANODROP2000 spectrophotometer (Thermo Scientific). Lysates were loaded onto 10–50% sucrose density gradients (20 mM HEPES-KOH (pH 7.6), 100 mM KCl, 5 mM MgCl2) and centrifuged at 35,000 rpm for 2 hours at 4°C. Gradients were fractionated and the optical density (OD) at 254 nm was continuously recorded using an ISCO fractionator (Teledyne ISCO; Lincoln, NE). Total RNA from each fraction was isolated using Trizol (Invitrogen) and reverse transcribed using the Superscript III kit (Invitrogen). Polysome to monosome ratio was calculated as the area under the A254 absorbance curve, using the function describing the absorbance values, processed with the definite integral command in MATLAB.
qRT-PCR and RT-PCR
Total or polysome reverse transcribed RNA was analyzed using a Biorad iQ SYBR Green Supermix kit as previously described in49. For all experiments n=4 and the primers used are summarized in Supplementary Table 3 For RT-PCR results depicted on agarose gels, products were amplified for 20–25 cycles, to remain within the linear range of the SYBR Green reaction. Results are presented in arbitrary units as relative amounts using serial dilutions of cortical or hippocampal RNA as qRT-PCR concentration standards.
Western Blotting and Antibodies
All tissues and cells were dissociated in RIPA buffer (unless otherwise specified). Western Blotting was previously described49. Antibodies against indicated proteins were: Neuroligin 1 and Neuroligin 2/3, Neuroligin 2, Neuroligin 3, VGAT, VGLUT (Synaptic Systems); Neuroligin 4 (Abcam); PSD95, GFAP, 4E-BP2, β-actin, eIF4E and eIF4GI (Cell Signaling); Neurexin1 (α, β), Neurexin2 (α, β), Neurexin3 (α, β), SAPAP3, Shank2, Shank3, Gephyrin, Arc/Arg3.1 and Polr2a (Santa-Cruz) and β -tubulin (SIGMA); secondary anti-mouse and anti-rabbit (GE Healthcare) and anti-goat (Santa-Cruz). Quantification of immunoblots was performed using ImageJ (NIH). Values were normalized to β-actin or another control where specified.
Synaptosome Preparation
Intact hippocampi from WT, 4E-BP2 or βT-eIF4E mice were dissected in ice cold PBS and homogenized in 320 mM sucrose, 1 mM EDTA, 5 mM Tris-HCl, pH 7.4 and 25 µM DTT. Synaptosomes were isolated on a discontinuous Percoll (GE Healthcare) gradient50. The fraction between 15% and 23% Percoll was isolated and resuspended in 2x SDS-PAGE sample buffer. Polr2a (RNA polymerase II large subunit) is used as a nuclear marker, GFAP (Glial Fibrillary Acidic Protein) as a glial marker and Arc/Arg3.1 (activity-regulated cytoskeleton-associated protein) as a marker for synaptosomes.
m7GDP Pull-down
Dorsal hippocampi were dissected in cold PBS, and homogenized in lysis buffer containing 40mM HEPES-KOH (pH 7.5), 120mM NaCl, 1mM EDTA, 0.1 mM GDP, 10mM pyrophosphate, 10mM β -glycerophosphate, 50mM NaF, and 0.3% CHAPS. Extracts were mixed with 30 µl of m7GDP-agarose (GE Healthcare) for 1.5 hr at 4°C. The resins were washed four times and proteins were eluted with 2x SDS-PAGE sample buffer.
Golgi Staining and Immunostaining
Hippocampal slices (50 µm) were prepared from whole brains of 2 month-old mice after fixation (4% PFA). Golgi staining of neuronal somata and processes was carried out using the FD Rapid Golgi Stain Kit (FD NeuroTechnologies) following the manufacturer’s protocol. Images of dendritic spines from hippocampal CA1 neurons were acquired using a 63x oil lens on a Zeiss Axioscope. For immunostaining, 2 month-old mice were perfused with 4% PFA, intact brains were removed and hippocampal slices were prepared. Slices were briefly permeabilized with 50 mM NH4Cl and 0.2% (w/v) saponin (SIGMA) in Phosphate Buffered Saline (PBS), blocked in 5% goat serum, incubated with primary antibodies (1:200) for 3 hours, washed with PBS, incubated with Alexa Fluor 488 or 594 (Molecular Probes), washed with PBS and mounted on microscope coverslips with Vectashield mounting medium (Vector Labs). Images were acquired with a Zeiss confocal microscope using a 63x oil-immersion lens.
Field EPSP Recordings
Transverse hippocampal slices (400 µm) were prepared from 6 week-old WT or 4E-BP2-KO littermates as described earlier49. Briefly, slices were submerged and superfused with an oxygenated artificial cerebrospinal fluid (ACSF) containing: 124 mM NaCl, 2.5 mM KCl, 1.25 mM NaH2PO4, 1.3 mM MgSO4, 2.5 mM CaCl2, 26 mM NaHCO3 and 10 mM glucose at 28 °C. Slices were allowed to recover at least 2 h before recording was initiated. A concentric bipolar tungsten stimulating electrode placed in the middle of CA1 stratum radiatum was used to stimulate Schaffer collateral afferents and elicit field extracellular field potentials (fEPSPs). fEPSPs were recorded with ASCF filled glass electrode (2–3 MΩ) placed in CA1 stratum radiatum ~200 mm from the stimulating electrode. Baseline stimulation was applied at 0.033 Hz by delivering 0.1 ms pulses that evoked 35% of maximal fEPSPs. LTP was induced with a single train at 100Hz for 1 s. 4EGI-1 (50 µM, Calbiochem) was applied 30 min before the onset of tetanic stimulation and for 1 h.
Miniature EPSC and IPSC Recordings
For whole-cell recordings acute slices were prepared as described above but from younger mice (19 to 22 day-old). Organotypic hippocampal slice cultures were prepared and biolistically transfected as previously described42. siRNAs against Nlgn1, 2 or scrambled (mix of Nlgn1:UCUGGGAAUAUCUGGUCCAUGCUACUU, Nlgn2: GUCCUCACUCUCUCCAGAGUCCUCCAC or scrambled siRNA (NC1 negative control) were designed using siRNA-design Software (IDT). An Enhanced Yellow Fluorescent Protein (EYFP)-expressing plasmid was co-transfected with all siRNAs (pcDNA3.1-EYFP). Whole-cell recordings were obtained from CA1 pyramidal neurons using borosilicate pipettes (3–6 MΩ). The intracellular solution for recording EPSCs contained: 130 mM CsMeSO3, 5 mM CsCl, 2 mM MgCl2, 5 mM diNa-phosphocreatine, 10 mM HEPES, 2 mM ATP-Tris, 0.4 mM GTP-Tris, and 0.1% biocytin (pH 7.2 to 7.3; 275 – 285 mOsmol). Recordings were made in the voltage-clamp mode using a Multiclamp 700A amplifier (Molecular Devices, Foster City, CA, USA). Holding potential was maintained at −60 mV and series resistance was routinely monitored. Recorded signals were low-pass filtered at 2 kHz, digitized at 20 kHz and stored on a PC. Data acquisition and off-line analyses were performed using 1322A Digidata acquisition board, and pClamp 10 software (Molecular Devices, Foster City, CA, USA). Data were only included if the holding current was stable or if series resistance varied < 25% of initial value. Miniature EPSCs (mEPSCs) or IPSCs (mIPSCs) were recorded in the presence of TTX (0.5 µM; Alomone Labs). AMPA receptor-mediated mEPSCs were isolated by co-application of DL-2-amino-5-phosphonovaleric acid (AP5; 50 µM; Tocris Biosciences) and gabazine (5 µM; Tocris Biosciences), to block NMDA and GABAA receptors respectively. GABAA receptor-mediated mIPSCs were recorded using a CsCl-based intracellular solution which contained: 135 mM CsCl, 10 mM HEPES, 2 mM MgCl2, and 4 mM MgATP (pH 7.25). In these experiments, 6,7-dinitroquinoxaline-2,3-dione (DNQX; 5 mM) and AP5 (50 mM) were present to block excitatory transmission via AMPA and NMDA receptors. For analysis, mEPSCs or mIPSCs were detected on a running template (mean of ~ 20 events) with a well-defined baseline, using pClamp10 software. Detection threshold was set at 3 pA and 150–200 events were sampled per neuron. Total charge was calculated by summing the charge transfer of all individual events (mEPSCs or mIPSCs) detected over a 10 min acquisition period for each neuron.
Evoked EPSCs and IPSCs
Putative single-fiber EPSCs or IPSCs were evoked using minimal stimulation. Stimulus strength was adjusted to a value that generated ~ 50% successes (range 40- 60%). Amplitudes of mean EPSC or IPSC were extracted from 5 min bins over a 10–20 min period.
Metabolic labeling
Acute hippocampal slices were prepared from WT or 4E-BP2-KO or βT-eIF4E mice as described above and 30 mCi/ml [35S]Met/Cys was added for 3 h. Slices were homogenized in 25mM Tris-HCl pH 7.4, 150mM NaCl, 1mM EDTA, 1% NP-40 and 5% glycerol and lysates were centrifuged at 16,000g. Total extracts were resolved with SDS-PAGE electrophoresis and transferred onto nitrocellulose membranes, which were then exposed onto autoradiography film (KODAK) (1 week) and developed. For total protein calculations, lysates were analyzed with SDS-PAGE electrophoresis and gels stained with GelCode Blue Stain Reagent (Pierce) according to the manufacturer’s protocol. Signals were quantified using ImageJ.
Cloning of 5’UTR luciferase constructs
Full length 5’ UTRs of mouse Nlgn1 (NCBI reference NM_001163387.1), Nlgn2 (NM_198862.2), Actb (β-actin) (NM_007393.3) and Dlg4 (PSD95) (NM_007864.3), were amplified from mouse genomic DNA, using specific primers and cloned into pGL4.13 firefly luciferase vector (Promega) upstream of the FF luciferase gene.
MEF and transient transfection
4E-BP2-KO, eIF4E overexpressing, Tsc2 KO and Pten+/− Mouse Embryonic Fibroblasts were prepared as previously described43. Cells were maintained in DMEM medium (Wisent) supplemented with 10% FBS (Wisent) and 5% penicillin/streptomycin (Wisent) in a humidified, 37°C, 5% CO2 incubator. Transient transfections were carried out for 3 hours with lipofectamine 2000 (Invitrogen) as per the manufacturer’s recommendations (using a 3:1 ratio of Firefly-expressing (pGL4.13/Renilla expressing vectors). Renilla and Firefly luciferase signals were detected 24 hours post-transfection with a STOPnGlo kit (Promega), using a Fluostar OPTIMA luminometer (BMG Labtech).
Stereotaxic Surgery, Cannulation and Infusion
Two month-old mice were deeply anaesthetized with 1ml/kg of body weight of a mixture of Ketamine (55 mg per kg), Xylazine (3 mg per kg) and Medetomidine hydrochloride (0.3 mg per kg) injected intraperitoneally. Mice were placed in a Kopf stereotactic frame. A small hole was made 1 mm rostral of the bregma and 2 mm right of midline and stainless steel 22-gauge cannulas were implanted at these coordinates. Obturators inserted in the guides prevented blocking. Dental cement was applied to stabilize the implants. After surgery, an intramuscular injection of analgesic (carprofen, 5 mg per kg) was given. An intraperitoneal injection of Antisedan (0.3 mg per kg) suspended anesthesia. Mice were then allowed to recover from surgery for 7 days and were handled daily during the recovery period. Infusions were carried out daily for 5 days at a depth of 2 mm. 4EGI-1 (Calbiochem) was prepared in 50% DMSO in 100 mM Tris-Cl (pH 7.2) and infused daily at a rate of 0.1 µl/min at a final concentration of 20 µg/µl; this sub-threshold concentration had no effect in WT mice. At the end of the experiment cannula placement and injection site accuracy were checked by infusion of 1 µl of Eosin Y (Sigma). For lentivirus injections, the same coordinates were used for a single injection without cannulas. A total of 3 µl of lentiviral particles at 1.4×10E7 TU/ml were injected. Animals were returned to their homecage for 7 days (and handled during the last 3) before behavioral testing.
Lentiviral transduction and titration
Lentiviruses were produced by co-transfection of HEK293T cells with transfer vector (pLKO.1), packaging plasmid psPAX2 and envelope plasmid pMD2.G as described in www.lentiweb.com. For mouse Nlgn1 knockdown, shRNA#1 was TRCN0000032022, shRNA#2 was TRCN0000032020, mouse Nlgn2 knockdown shRNA#1 was TRCN0000180497 and shRNA#2 was TRCN0000184441, the non-targeting shRNA was SHC002 (all constructs from Mission SIGMA). N2A cells used for validation of shRNAs and siRNAs or MEFs used for titration were maintained in DMEM (Gibco) supplemented with 10% FBS and 1% penicillin/streptomycin (Gibco) at 37°C, 5%CO2. Viral titer (TU/ml) was calculated using puromycin selection of MEFs infected with different viral particle dilutions and stained with 0.2% crystal violet and 20% methanol solution. Colonies were measured using the CellCount plugin of ImageJ (NIH). Viral titers were adjusted to 1.4×10E7 TU/ml for in vivo injections.
Statistical Analysis and Behavioral Scoring
Experimenters were blinded to the genotype during testing and scoring. All data are presented as mean ±SEM (error bars). Statistical results, along with tests used are summarized in Supplementary Table 4 Statistical significance was set a priori at 0.05 (ns: non-significant). For most behavioral assays we used a two-way ANOVA with a Bonferroni Multiple Comparison post-hoc test unless otherwise specified in the figure legend or Supplementary Table 4 For quantification of Western Blots for 4EGI-1 or knockdown experiments we used a two-way ANOVA with Bonferroni’s post hoc. To compare changes in group means in amplitude or frequency of mEPSCs or mIPSCs, a Student's t-test for independent measures was used. A Kolmogorov-Smirnov test was used to compare cumulative distributions of mEPSC and mIPSC parameters between groups (150 events per neuron) and the relative change in cumulative sum of mEPSC or mIPSC total charge transfer of 4E-BP2-KO (expressed as % of mean WT). For fear conditioning statistical analyses were done by Student’s t tests and two-way ANOVA followed by between-group comparisons using Tukey’s posthoc test. For field potential recordings, we used a two-way ANOVA test.
Supplementary Material
Acknowledgements
This work was supported by the Canadian Institutes of Health Research (N.S.; MOP-114994, J-C.L.; MOP-10848, P.D. and F.M.; MOP-93679, and P.L. and N.S; MOP-44050), Autism Speaks (Grant #7109 to N.S.), the Fonds de la Recherche en Santé du Québec (J-C.L. FRSQ; Groupe de Recherche sur le Système Nerveux Central). J-C.L. is the recipient of the Canada Research Chair in Cellular and Molecular Neurophysiology. I.R. was supported by a Fellowship of the Savoy Foundation. We thank Y. Svitkin, A. Parsyan, E. Petroulakis, R. Karni and V. Polunovski for advice; K. Gamache, A. Sylvestre, S. Perreault, C. Lister and I. Harvey for technical assistance, T. Alain for assistance with lentiviral titration, W. Sossin and P. Skehel for critical reading of the manuscript.
Footnotes
Supplementary information is linked to the online version of the paper at www.nature.com/nature
Author contributions
C.G.G. and N.S. conceived and designed this study, wrote the manuscript, supervised and coordinated the project. C.G.G: behavioral, biochemical and imaging experiments, data and statistical analysis. C.G.G., A.K., I.R., D.B.W. and C.V: electrophysiology experiments and data analysis; T.N., S.Y: biochemical experiments and data analysis; E.R: statistical analysis; P.D.&F.M: bioinformatics analysis; M.T.&D.R. provided the eIF4E transgenic mice; P.L. supervised the project and edited the manuscript and; K.N. contributed to the design of behavioral experiments, edited the manuscript and supervised the project; J-C.L. supervised, conceived and designed the electrophysiological experiments, edited the manuscript and supervised the project.
The authors wish to declare that they have no competing financial interests.
Contributor Information
Christos G. Gkogkas, Email: christos.gkogkas@mcgill.ca.
Jean-Claude Lacaille, Email: jean-claude.lacaille@umontreal.ca.
References
- 1.Fombonne E. Epidemiology of pervasive developmental disorders. Pediatric research. 2009;65:591–598. doi: 10.1203/PDR.0b013e31819e7203. [DOI] [PubMed] [Google Scholar]
- 2.Kelleher RJ, Bear MF., 3rd The autistic neuron: troubled translation? Cell. 2008;135:401–406. doi: 10.1016/j.cell.2008.10.017. [DOI] [PubMed] [Google Scholar]
- 3.Hay N, Sonenberg N. Upstream and downstream of mTOR. Genes Dev. 2004;18:1926–1945. doi: 10.1101/gad.1212704. [DOI] [PubMed] [Google Scholar]
- 4.Pause A, et al. Insulin-dependent stimulation of protein synthesis by phosphorylation of a regulator of 5'-cap function. Nature. 1994;371:762–767. doi: 10.1038/371762a0. [DOI] [PubMed] [Google Scholar]
- 5.Koromilas AE, Lazaris-Karatzas A, Sonenberg N. mRNAs containing extensive secondary structure in their 5' non-coding region translate efficiently in cells overexpressing initiation factor eIF-4E. EMBO J. 1992;11:4153–4158. doi: 10.1002/j.1460-2075.1992.tb05508.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Banko JL, et al. The translation repressor 4E-BP2 is critical for eIF4F complex formation, synaptic plasticity, and memory in the hippocampus. J Neurosci. 2005;25:9581–9590. doi: 10.1523/JNEUROSCI.2423-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhou J, Parada LF. PTEN signaling in autism spectrum disorders. Current opinion in neurobiology. 2012 doi: 10.1016/j.conb.2012.05.004. [DOI] [PubMed] [Google Scholar]
- 8.Kwon CH, et al. Pten regulates neuronal arborization and social interaction in mice. Neuron. 2006;50:377–388. doi: 10.1016/j.neuron.2006.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhou J, et al. Pharmacological inhibition of mTORC1 suppresses anatomical, cellular, and behavioral abnormalities in neural-specific Pten knock-out mice. J Neurosci. 2009;29:1773–1783. doi: 10.1523/JNEUROSCI.5685-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jeste SS, Sahin M, Bolton P, Ploubidis GB, Humphrey A. Characterization of autism in young children with tuberous sclerosis complex. Journal of child neurology. 2008;23:520–525. doi: 10.1177/0883073807309788. [DOI] [PubMed] [Google Scholar]
- 11.Auerbach BD, Osterweil EK, Bear MF. Mutations causing syndromic autism define an axis of synaptic pathophysiology. Nature. 2011;480:63–68. doi: 10.1038/nature10658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ehninger D, et al. Reversal of learning deficits in a Tsc2+/− mouse model of tuberous sclerosis. Nat Med. 2008;14:843–848. doi: 10.1038/nm1788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Young DM, Schenk AK, Yang SB, Jan YN, Jan LY. Altered ultrasonic vocalizations in a tuberous sclerosis mouse model of autism. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:11074–11079. doi: 10.1073/pnas.1005620107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tsai PT, et al. Autistic-like behaviour and cerebellar dysfunction in Purkinje cell Tsc1 mutant mice. Nature. 2012 doi: 10.1038/nature11310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.O'Roak BJ, et al. Sporadic autism exomes reveal a highly interconnected protein network of de novo mutations. Nature. 2012;485:246–250. doi: 10.1038/nature10989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hoeffer CA, et al. Altered mTOR signaling and enhanced CYFIP2 expression levels in subjects with fragile × syndrome. Genes, brain, and behavior. 2012;11:332–341. doi: 10.1111/j.1601-183X.2012.00768.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Napoli I, et al. The fragile × syndrome protein represses activity-dependent translation through CYFIP1, a new 4E-BP. Cell. 2008;134:1042–1054. doi: 10.1016/j.cell.2008.07.031. [DOI] [PubMed] [Google Scholar]
- 18.Nowicki ST, et al. The Prader-Willi phenotype of fragile × syndrome. J Dev Behav Pediatr. 2007;28:133–138. doi: 10.1097/01.DBP.0000267563.18952.c9. [DOI] [PubMed] [Google Scholar]
- 19.Rubenstein JL, Merzenich MM. Model of autism: increased ratio of excitation/inhibition in key neural systems. Genes, brain, and behavior. 2003;2:255–267. doi: 10.1034/j.1601-183x.2003.00037.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Uhlhaas PJ, Singer W. Neuronal Dynamics and Neuropsychiatric Disorders: Toward a Translational Paradigm for Dysfunctional Large-Scale Networks. Neuron. 2012;75:963–980. doi: 10.1016/j.neuron.2012.09.004. [DOI] [PubMed] [Google Scholar]
- 21.Cornew L, Roberts TP, Blaskey L, Edgar JC. Resting-State Oscillatory Activity in Autism Spectrum Disorders. Journal of autism and developmental disorders. 2011 doi: 10.1007/s10803-011-1431-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yizhar O, et al. Neocortical excitation/inhibition balance in information processing and social dysfunction. Nature. 2011;477:171–178. doi: 10.1038/nature10360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Luikart BW, et al. Pten knockdown in vivo increases excitatory drive onto dentate granule cells. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2011;31:4345–4354. doi: 10.1523/JNEUROSCI.0061-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bateup HS, Takasaki KT, Saulnier JL, Denefrio CL, Sabatini BL. Loss of Tsc1 in vivo impairs hippocampal mGluR-LTD and increases excitatory synaptic function. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2011;31:8862–8869. doi: 10.1523/JNEUROSCI.1617-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Peca J, et al. Shank3 mutant mice display autistic-like behaviours and striatal dysfunction. Nature. 2011;472:437–442. doi: 10.1038/nature09965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schmeisser MJ, et al. Autistic-like behaviours and hyperactivity in mice lacking ProSAP1/Shank2. Nature. 2012;486:256–260. doi: 10.1038/nature11015. [DOI] [PubMed] [Google Scholar]
- 27.Won H, et al. Autistic-like social behaviour in Shank2-mutant mice improved by restoring NMDA receptor function. Nature. 2012;486:261–265. doi: 10.1038/nature11208. [DOI] [PubMed] [Google Scholar]
- 28.Chao HT, et al. Dysfunction in GABA signalling mediates autism-like stereotypies and Rett syndrome phenotypes. Nature. 2010;468:263–269. doi: 10.1038/nature09582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Levinson JN, El-Husseini A. Building excitatory and inhibitory synapses: balancing neuroligin partnerships. Neuron. 2005;48:171–174. doi: 10.1016/j.neuron.2005.09.017. [DOI] [PubMed] [Google Scholar]
- 30.Scattoni ML, Crawley J, Ricceri L. Ultrasonic vocalizations: a tool for behavioural phenotyping of mouse models of neurodevelopmental disorders. Neurosci Biobehav Rev. 2009;33:508–515. doi: 10.1016/j.neubiorev.2008.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ruggero D, et al. The translation factor eIF-4E promotes tumor formation and cooperates with c-Myc in lymphomagenesis. Nature medicine. 2004;10:484–486. doi: 10.1038/nm1042. [DOI] [PubMed] [Google Scholar]
- 32.Graff JR, et al. Therapeutic suppression of translation initiation factor eIF4E expression reduces tumor growth without toxicity. J Clin Invest. 2007;117:2638–2648. doi: 10.1172/JCI32044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Glessner JT, et al. Autism genome-wide copy number variation reveals ubiquitin and neuronal genes. Nature. 2009;459:569–573. doi: 10.1038/nature07953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang K, et al. Common genetic variants on 5p14.1 associate with autism spectrum disorders. Nature. 2009;459:528–533. doi: 10.1038/nature07999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hoeffer CA, et al. Inhibition of the interactions between eukaryotic initiation factors 4E and 4G impairs long-term associative memory consolidation but not reconsolidation. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:3383–3388. doi: 10.1073/pnas.1013063108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chubykin AA, et al. Activity-dependent validation of excitatory versus inhibitory synapses by neuroligin-1 versus neuroligin-2. Neuron. 2007;54:919–931. doi: 10.1016/j.neuron.2007.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dahlhaus R, et al. Overexpression of the cell adhesion protein neuroligin-1 induces learning deficits and impairs synaptic plasticity by altering the ratio of excitation to inhibition in the hippocampus. Hippocampus. 2010;20:305–322. doi: 10.1002/hipo.20630. [DOI] [PubMed] [Google Scholar]
- 38.Banko JL, et al. Behavioral alterations in mice lacking the translation repressor 4E-BP2. Neurobiol Learn Mem. 2007;87:248–256. doi: 10.1016/j.nlm.2006.08.012. [DOI] [PubMed] [Google Scholar]
- 39.Neves-Pereira M, et al. Deregulation of EIF4E: a novel mechanism for autism. J Med Genet. 2009;46:759–765. doi: 10.1136/jmg.2009.066852. [DOI] [PubMed] [Google Scholar]
- 40.Larsson O, et al. Distinct perturbation of the translatome by the antidiabetic drug metformin. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:8977–8982. doi: 10.1073/pnas.1201689109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Levy D, et al. Rare de novo and transmitted copy-number variation in autistic spectrum disorders. Neuron. 2011;70:886–897. doi: 10.1016/j.neuron.2011.05.015. [DOI] [PubMed] [Google Scholar]
- 42.Bidinosti M, et al. Postnatal deamidation of 4E-BP2 in brain enhances its association with raptor and alters kinetics of excitatory synaptic transmission. Mol Cell. 2010;37:797–808. doi: 10.1016/j.molcel.2010.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Petroulakis E, et al. p53-dependent translational control of senescence and transformation via 4E-BPs. Cancer Cell. 2009;16:439–446. doi: 10.1016/j.ccr.2009.09.025. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.