Abstract
Recurrent chromosomal deletions in cancer are typically thought to harbor tumor suppressors. In a recent publication in Nature, Northcott and colleagues identify a novel region of structural variation in medulloblastoma that leads to oncogenic activation of GFI1B and GFI1 by repositioning these genes next to super-enhancers.
Genomic instability is one of the enabling hallmarksof cancer and can lead to extensive chromosomal abnormalities(Hanahan and Weinberg, 2011). Sites of recurrent genomic aberrations have long been thought to harbor genes important for tumor development; indeed, oncogenes such as MYC and ERBB2 are found in amplifications, the BCR-ABLfusion gene results from a chromosomal translocation, and tumor suppressors such as RB1, PTEN, and TP53 are frequently lost in deleted regions.
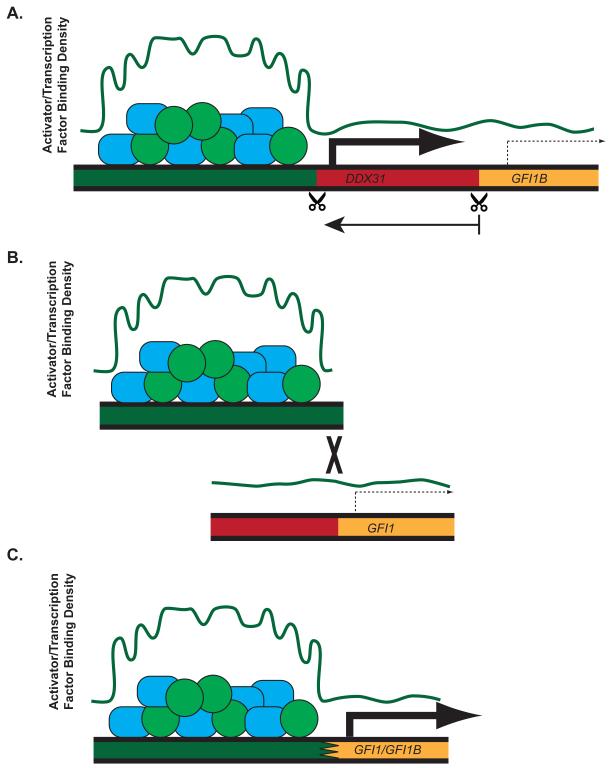
Medulloblastoma is the most common malignant pediatric brain tumor, with large-scale genomic and transcriptomic analyses identifying four distinct molecular subgroups (Taylor et al., 2012). Northcott, Lee, Zichner,and coworkers(Northcott et al., 2014)recently analyzed whole-genome sequencing of primary group 3 and 4 medulloblastomasamples for somatic structural variants (SVs). Rather than limiting their search torecurrent amplifications or deletions, they analyzed all chromosomal breakpoints and identifyied a novel region of interest spanning over 400kb on chromosome 9q34.13.A single gene at this locus, growth factor independence 1B (GFI1B), was found to be overexpressed concomitant with proximal SV. Despite the variety of observed SVs ranging from deletions (Figure 1A), tandem duplication, and/or inversions, these SVs resulted in repositioning GFI1B next to super-enhancers (Figure 1).Additionally, after observing mutually exclusive expression patterns within group 3 tumors between GFI1B and its paralog, growth factor independence 1 (GFI1), the authorsidentified that GFI1 was also subject to SV, similarly varied between interchromosomal translocations and tandem duplications (Figure 1B). These also led tosimilar displacement of GFI1to regions adjacent to other enhancers or super-enhancers (Figure 1C).
Figure 1. GFI1 and GFI1B hijack enhancers via structural variation.
In medulloblastoma, GFI1B (as shown in A) and GFI1 (as shown in B) normally reside in heterochromatic regions with little to no expression. Structural variation including deletions (A) or interchromosomal translocations (B) can lead to juxtaposition of these genes next to enhancers or super-enhancers (as shown in C) which drive oncogenic expression due to their increased concentrations of bound activators and transcription factors (plotted in green and represented by circles and rectangles).
Enhancers are short stretches of genomic DNA that serve to bind activators and function in cis to drive transcription of nearby genes(Ong and Corces, 2011). Recently, super-enhancers have been identified as exceptionally large euchromatic regions that serve as a concentrated site of activatorand transcription factor binding and stimulate higher transcriptional activity than typical enhancers. While enhancers are highly prevalent throughout the genome, super-enhancers are scattered sparsely at a few hundred sites, residing at key cell-identity genes where they define cell types by driving specific expression patterns(Hnisz et al., 2013; Whyte et al., 2013). Super-enhancers are also thought to be dynamic,forming at essential oncogenes during tumorigenesis where they remain exquisitely sensitive to bromodomain inhibition(Lovén et al., 2013).
This idea of dynamic super-enhancers works in concert with data showingepigenetic heterogeneity and plasticity play integral roles in the acquisition of drug resistance in cancer(Sharma et al., 2010). With a low somatic mutation rate (0.52 per Mb) and frequent alteration of chromatin modifiers across all subgroups (Jones et al., 2012), medulloblastoma may be the perfect candidate to observe this phenomenon. Northcott et al., however, describe “enhancer hijacking”asa mechanism in which genomic instability leads to the utilization of existing epigenetic structure to drive oncogene expression. Thus, as epigenetic plasticity represents a complementary approach to the acquisition of somatic mutations in the pathogenesis of cancer, this study leads to intriguing questions about the state of the epigenome in medulloblastoma.Is the utiltization of enhancer hijacking (rather than dynamic generation of a new super-enhancer) simply due to the enrichment of somatic copy number aberrations in these subgroups (Northcott et al., 2012)? Are there fundamental differences between medulloblastoma and other cancers that lead to a relatively static epigenome in medulloblastoma, or are super-enhancers not so readily plastic or dynamic?
Another question that remains to be explored is whether there is an underlying function within the GFI1 gene family that promotes enhancer hijacking. GFI1 and GFI1B are highly homologous transcriptional repressors that are expressed in the hematopoetic compartment and are known proto-oncogenes in leukemia and lymphoma. Since bothGFI1andGFI1Bare subjected to transcriptional autoregulationand are able to repress each other’s expression, does the underlying SV represent the only mechanism of escape from a complex net of feedback loops? Northcott et al. show that GFI1/GFI1B cooperate with MYC to drive medulloblastoma in an orthotopic xenograft mouse model, despite the fact that neither alonewas able to promote tumorigenesis in this model. Curiously, GFI1 activation,but not GFI1B, correlated with MYC expression in medulloblastoma, whereas both cooperated with MYC in this preclinical in-vivo model. Whether and how they cooperate to make a permissive environment for such SV remains to be determined.
Each of the four medulloblastoma subgroups differ in age distribution, gender, and outcome. Two of thesesubgroups (groups 1 and 2) are driven predominantly by a single prominent signaling pathway (WNT or SHH), while group 3 and group 4 tumors show more complex genetics and signaling. Group 3 tumors generally have the poorest prognosis, andby identifying these super-enhancer activated oncogenes, the authors identify a possible therapeutic avenue using bromodomain inhibitors. Furthermore, this study was successful in identifying oncogenes at regions containing, in part, common deletions. Despite knowing for 15 years that translocations in Burkitt’s lymphoma can lead to activation of MYC by juxtaposing it next to the IGλ enhancer, the current paradigm stillfocuses on the assumption that the type of SV defines the function of the gene of interest: gains representing oncogenic loci and losses representing tumor suppressor loci.While large efforts have been spent to identify tumor suppressors in commonly deleted regions,Northcott, Lee, Zichner, and colleagues ask us to rethink these strategies for tumors where SVs have clearly delineated and recurrent breakpoints, especially if studies to date have not yielded strong candidates.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- Hnisz D, Abraham BJ, Lee TI, Lau A, Saint-André V, Sigova AA, Hoke HA, Young RA. Super-enhancers in the control of cell identity and disease. Cell. 2013;155:934–947. doi: 10.1016/j.cell.2013.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones DT, Jäger N, Kool M, Zichner T, Hutter B, Sultan M, Cho Y-J, Pugh TJ, Hovestadt V, Stütz AM, et al. Dissecting the genomic complexity underlying medulloblastoma. Nature. 2012;488:1–6. doi: 10.1038/nature11284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovén J, Hoke HA, Lin CY, Lau A, Orlando DA, Vakoc CR, Bradner JE, Lee TI, Young RA. Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell. 2013;153:320–334. doi: 10.1016/j.cell.2013.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Northcott PA, Shih DJ, Peacock J, Garzia L, Morrissy AS, Zichner T, Stütz AM, Korshunov A, Reimand J, Schumacher SE, et al. Subgroup-specific structural variation across 1,000 medulloblastoma genomes. Nature. 2012;488:1–8. doi: 10.1038/nature11327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Northcott PA, Lee C, Zichner T, Stütz AM, Erkek S, Kawauchi D, Shih DJH, Hovestadt V, Zapatka M, Sturm D, et al. Enhancer hijacking activates GFI1 family oncogenes in medulloblastoma. Nature. 2014 doi: 10.1038/nature13379. advance on. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ong C-T, Corces VG. Enhancer function: new insights into the regulation of tissue-specific gene expression. Nat. Rev. Genet. 2011;12:283–293. doi: 10.1038/nrg2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma SV, Lee DY, Li B, Quinlan MP, Takahashi F, Maheswaran S, McDermott U, Azizian N, Zou L, Fischbach MA, et al. A Chromatin-Mediated Reversible Drug-Tolerant State in Cancer Cell Subpopulations. Cell. 2010;141:69–80. doi: 10.1016/j.cell.2010.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor MD, Northcott PA, Korshunov A, Remke M, Cho Y-J, Clifford SC, Eberhart CG, Parsons DW, Rutkowski S, Gajjar A, et al. Molecular subgroups of medulloblastoma: the current consensus. Acta Neuropathol. 2012;123:465–472. doi: 10.1007/s00401-011-0922-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whyte WA, Orlando DA, Hnisz D, Abraham BJ, Lin CY, Kagey MH, Rahl PB, Lee TI, Young RA. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell. 2013;153:307–319. doi: 10.1016/j.cell.2013.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]