Abstract
Background
The ATP-binding cassette transporter gene ABCB1 and the glutathione S-transferase gene GSTP1 code for a multidrug resistance (MDR) protein and for a detoxifying phase II metabolic enzyme, respectively, with substrate specificities that include chemotherapy drugs commonly used to treat lung cancer.
Methods
We genotyped 11 ABCB1 and 8 GSTP1 single nucleotide polymorphisms (SNPs) in 698 white lung cancer patients (all current or former cigarette smokers) and used log-rank test statistics and proportional hazards regression to evaluate associations between SNP genotype and survival.
Results
Using data from all 698 cases, one SNP in ABCB1 (rs2235013) was statistically significantly associated with overall survival (p=0.038, log-rank test). Chemotherapy and stage jointly (p=0.025) significantly modified the association between rs2235013 and survival, with statistically significant (p=0.013, log-rank test) association observed in the subgroup of stage III-IV lung cancer patients who received chemotherapy as part of their first course of treatment (N=160; 93.1% non-small cell). Patients who inherited the minor T allele at ABCB1 rs2235013 experienced better overall and recurrence-free survival [hazard ratio, per minor T allele, (95% confidence interval): 0.66 (0.49-0.90) and 0.55 (0.31-0.95), respectively; adjusted for year of diagnosis, sex, age at diagnosis, cigarette pack years, and stage]. In addition, in the advanced-stage chemotherapy-treated subgroup, four ABCB1 SNPs (rs6949448, rs2235046, rs1128503, and rs10276036) in mutual high linkage disequilibrium with rs2235013 and an independent ABCB1 SNP (rs1045642) showed statistically significant association (p<0.05) with survival.
Conclusions
Inherited variation in ABCB1 may affect survival specifically in advanced-stage lung cancer patients who receive chemotherapy.
Introduction
The Surveillance, Epidemiology, and End Results (SEER) Program gathers population-based cancer occurrence data for the U.S. population. Among persons diagnosed between 2004 and 2009 with lung cancer as their first lifetime cancer diagnosis, SEER placed 87% in a non-small cell histologic category and 87% of these non-small cell cases in an American Joint Committee on Cancer (AJCC) 6th edition stage IIIA, IIIB, or IV group [1]. One year after diagnosis, only 31% of these persons with advanced non-small lung cancer were still alive [1]. Modern chemotherapy regimens that contain cisplatin were known, two decades ago, to somewhat improve the survival of patients with advanced non-small cell lung cancer [2]. However, treatment-related toxicities, combined with the weak survival benefits typically observed, continue to motivate efforts to identify lung cancer patients who experience particularly favorable responses to multi-agent chemotherapy. One research approach attempts to associate inherited factors, including inherited factors governing drug metabolism, with treatment response.
To evaluate inherited variation in DNA repair and cell cycle control pathway genes in relation to lung cancer risk, we designed and completed a large case-control study of white current and former cigarette smokers with and without incident lung cancer [3]. Anticipating subsequent investigation in our lung cancer case series of factors related to treatment outcome, we included ATP-binding cassette, sub-family B, member 1 gene (ABCB1) and glutathione S-transferase gene pi 1 (GSTP1) single nucleotide polymorphisms (SNPs) on a custom-designed 384-SNP microarray used to genotype all cases and controls. ABCB1 and GSTP1 code for a multidrug resistance (MDR) protein and detoxifying phase II metabolic enzyme, respectively, both agents engaged in the metabolism of chemotherapy drugs commonly used in lung cancer [4,5]. Genotyping all patients permitted study of the specificity of association between inherited variants and clinical outcome. We expected outcome associations with inherited variation in ABCB1 or GSTP1 to appear more prominently among patients exposed to chemotherapy. After screening all cases for modification by chemotherapy of the association between inherited variation and outcome, we restricted analysis to late-stage chemotherapy cases and evaluated lung cancer survival outcomes in relation to inherited variation in ABCB1 and GSTP1, genes that participate in the metabolism of two commonly used lung cancer chemotherapeutics, taxanes and platinum, respectively.
Methods
Study Population
Patients were enrolled between 1990 and 2008 within 1 year of a thoracic surgery procedure for lung cancer diagnosis, staging, or treatment at a University of Pittsburgh Medical Center (UPMC) hospital. Additional eligibility criteria for the current study included: (1) white race (as documented in the UPMC Network Cancer Registry), (2) current or ex-cigarette smoker, (3) >10 pack-year cumulative cigarette dose exposure (pack years), (4) 45-85 years old at time of lung cancer diagnosis, (5) pathologically verified lung cancer diagnosis (excluding carcinoid), and (6) DNA sample available for genotyping. In total, 782 patients fulfilled these criteria. All patients provided written informed consent and were enrolled under University of Pittsburgh Institutional Review Board-approved protocols.
Tumor Histology, Cancer Stage, Treatment, and Outcome
To determine primary tumor histology, cancer stage, first course of treatment, and survival outcomes, we linked research patient data to the Cancer Registry. Analyses classed primary tumor ICD-O-3 histologic type codes [6] into four broad groupings representing adenocarcinoma and bronchioloalveolar carcinoma, squamous cell carcinoma, small cell carcinoma, and other non-small cell types, including non-small cell carcinoma (not otherwise specified), large cell carcinoma, and adenosquamous carcinoma. Analyses defined the anatomic extent of disease at diagnosis according to AJCC pathologic TNM stage grouping, if defined, or clinical TNM stage grouping, otherwise, in effect at the time of diagnosis (3rd, 4th, 5th, and 6th editions for cases diagnosed in 1990-1991, 1992-1997, 1998-2002, and 2003-2008 [7], respectively). Cancer Registry results coded according to North American Association of Central Cancer Registries (NAACCR) standards [8,9] identified patients administered pre- or post-adjuvant chemotherapy as part of the first course of treatment, before disease progression or recurrence. To identify affirmatively the subset of patients who received platinum (cisplatin or carboplatin) or taxane (paclitaxel or docetaxel) chemotherapy, we decoded the free-text field used by cancer registrars to document chemotherapy. Overall survival and recurrence-free survival were calculated as time from initial diagnosis to death and time from initial diagnosis to first recurrence, respectively, with follow-up censored on date of last contact. Analyses of recurrence-free survival excluded patients who never became disease-free after diagnosis.
Patients without a matching lung tumor record in the Cancer Registry (N=10, 1.3% of 782) and patients with a matching lung tumor record that lacked values for lung tumor histology, stage, and/or treatment (N=29, 3.7% of 782) were excluded. Histologies other than adenocarcinoma or squamous cell carcinoma were more frequent among excluded than included patients. Excluded and included patients were statistically similar (p>0.15) with respect to sex, age at diagnosis, year of diagnosis, pack years, and stage.
SNP Selection
Priority was given to SNPs with the following attributes: (1) functional significance as determined by amino acid substitution; (2) potential for regulatory change; (3) predicted alteration in protein stability; (4) evolutionary conservation across species; (5) disease association reported in the literature, and (6) genotyping feasible with Illumina® GoldenGate technology. In total, we selected 12 SNPs in ABCB1 and 9 in GSTP1.
DNA Isolation and Genotyping
Genomic DNA was isolated using Gentra Systems, Inc. (Minneapolis, MN) isolation kits and genotyped using an Illumina® GoldenGate custom-designed 384-SNP microarray. Analyses serially excluded: (1) assigned genotypes by Illumina® GenomeStudio 2010.1 using a GenTrain score <0.45 or a ClusterSep score <0.25, (2) individuals with genotype call rates <95% (calculated using all SNPs covered by the 384-SNP microarray, including SNPs in genes other than ABCB1 and GSTP1), (3) SNPs that failed in more than 2% of the samples, and 4) SNPs with minor allele frequency (MAF) <5%. After excluding 45 patients (6.1% of 743 patients with complete Cancer Registry data) for call rates <95%, 698 patients remained for analysis. Women were excluded more often than men (7.9 vs. 4.5%, p=0.0548) because of low call rates. Excluded and included patients were statistically similar (p>0.45) with respect to age at diagnosis, year of diagnosis, pack years, stage, and exposure to any chemotherapy as part of first planned course of treatment. After excluding one ABCB1 SNP (rs1015415) that failed in >2% of samples and one GSTP1 SNP (rs8191438) with MAF <5%, 11 ABCB1 and 8 GSTP1 SNPs remained. We assessed these 11 ABCB1 and 8 GSTP1 SNPs for association with survival even though twoGSTP1 SNPs departed (p<0.01) from Hardy-Weinberg Equilibrium (HWE) when evaluated separately in our 698 study patients and in 929 white control subjects from our earlier study (Supplemental Table 1) [3]. The 11 SNPs in ABCB1 and 8 SNPs in GSTP1 captured (at r2 > 0.80) 58% (52 of 89 alleles) and 72% (18 of 25 alleles), respectively, of the common-variant CEU SNPs in these genes (MAF > 5% in HapMap Genome Browser release #28 (phases 1, 2, and 3), ABCB1 – chr7:86,950,884..87,190,500, GSTP1 – chr11:67,087,862..67,120,699; http://hapmap.ncbi.nlm.nih.gov/). All SNP alleles in this manuscript are reported in forward orientation with respect to the genome.
Statistical Analysis
We used X2tests to evaluate independence between genotype and clinical factors, such as treatment and stage, the Kaplan-Meier method to estimate survival functions, and the log-rank test with a Wilcoxon weight (the number at risk ni at failure time ti) to compare survival differences according to genotype. Using SAS 9.2 (PROC PHREG), we fit Cox proportional hazards models to control genotype-outcome associations for potential confounders and applied a log-likelihood ratio test to genotype by cofactor interaction terms to screen for effect modification. To detect departures from the proportional hazards assumption, we examined Schoenfeld residual plots and tested the statistical significance of terms representing the time-dependent interaction between genotype and the natural logarithm of survival time. We fit additive genetic models with the common allele as reference.
Haplotype-based analyses used the Expectation-Maximization (EM) algorithm implemented in SAS Genetics (PROC HAPLOTYPE) to estimate group-level haplotype frequencies and to generate subject-level haplotype probability weights.1 We used these haplotype probability weights and Cox regression to identify haplotypes independently associated with survival.
Results
Characteristic of the study population (N=698) are presented in Table 1. Our study population included N=160 stage III-IV patients who received chemotherapy, the subgroup most expected to experience different outcomes according to variation in ABCB1 or GSTP1. Though diverse with respect to sex, age, and pack years, the patients in this subgroup were characterized by diagnosis between 2000 and 2008, non-small cell histology, and initial treatment with a chemotherapeutic regimen that included both platinum and taxane (Table 1). Our study population also included N=470 stage I-II patients (N=362 not given chemotherapy and N=108 given chemotherapy) and N=68 stage III-IV patients not given chemotherapy.
Table 1. Characteristics of the study population (N=698), by stage and chemotherapy administration.
| Characteristic | Stage I-II | Stage III-IV | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
|||||||||
| No chemotherapy | Yes chemotherapy | No chemotherapy | Yes chemotherapy | Overall | ||||||
|
|
|
|
|
|
||||||
| N | % | N | % | N | % | N | % | N | % | |
| All | 362 | 100.0 | 108 | 100.0 | 68 | 100.0 | 160 | 100.0 | 698 | 100.0 |
| Sex | ||||||||||
| men | 183 | 50.6 | 67 | 62.0 | 44 | 64.7 | 88 | 55.0 | 382 | 54.7 |
| women | 179 | 49.4 | 41 | 38.0 | 24 | 35.3 | 72 | 45.0 | 316 | 45.3 |
| Age (years) at diagnosis | ||||||||||
| <55 | 21 | 5.8 | 11 | 10.2 | 2 | 2.9 | 29 | 18.1 | 63 | 9.0 |
| 55-64 | 86 | 23.8 | 44 | 40.7 | 17 | 25.0 | 44 | 27.5 | 191 | 27.4 |
| 65-69 | 91 | 25.1 | 23 | 21.3 | 14 | 20.6 | 30 | 18.8 | 158 | 22.6 |
| 70-74 | 70 | 19.3 | 21 | 19.4 | 15 | 22.1 | 28 | 17.5 | 134 | 19.2 |
| 75+ | 94 | 26.0 | 9 | 8.3 | 20 | 29.4 | 29 | 18.1 | 152 | 21.8 |
| Year of diagnosis | ||||||||||
| 1990-1999 | 147 | 40.6 | 14 | 13.0 | 40 | 58.8 | 27 | 16.9 | 228 | 32.7 |
| 2000-2008 | 215 | 59.4 | 94 | 87.0 | 28 | 41.2 | 133 | 83.1 | 470 | 67.3 |
| Pack years | ||||||||||
| <35 | 99 | 27.3 | 30 | 27.8 | 15 | 22.1 | 36 | 22.5 | 180 | 25.8 |
| 35-49 | 57 | 15.7 | 18 | 16.7 | 14 | 20.6 | 40 | 25.0 | 129 | 18.5 |
| 50-74 | 108 | 29.8 | 33 | 30.6 | 24 | 35.3 | 47 | 29.4 | 212 | 30.4 |
| 75+ | 98 | 27.1 | 27 | 25.0 | 15 | 22.1 | 37 | 23.1 | 177 | 25.4 |
| Histology grouping | ||||||||||
| adenocarcinoma/BAC | 187 | 51.7 | 41 | 38.0 | 29 | 42.6 | 76 | 47.5 | 333 | 47.7 |
| squamous cell | 143 | 39.5 | 42 | 38.9 | 31 | 45.6 | 52 | 32.5 | 268 | 38.4 |
| other non-small cell | 31 | 8.6 | 18 | 16.7 | 8 | 11.8 | 21 | 13.1 | 78 | 11.2 |
| small cell carcinoma | 1 | 0.3 | 7 | 6.5 | 0 | 0.0 | 11 | 6.9 | 19 | 2.7 |
| Chemotherapy[1] | ||||||||||
| single or multiple agents | 97 | 89.8 | 146 | 91.2 | ||||||
| platinum and taxane | 72 | 66.7 | 111 | 69.4 | ||||||
| platinum only | 15 | 13.9 | 29 | 18.1 | ||||||
| taxane only | 5 | 4.6 | 3 | 1.9 | ||||||
| neither | 5 | 4.6 | 3 | 1.9 | ||||||
| chemotherapy, NOS | 11 | 10.2 | 14 | 8.8 | ||||||
| Surgical therapy | 360 | 99.4 | 102 | 94.4 | 62 | 91.2 | 103 | 64.4 | 627 | 89.8 |
| Radiation therapy | 44 | 12.2 | 53 | 49.1 | 19 | 27.9 | 98 | 61.3 | 214 | 30.7 |
First order (no chemotherapy vs. chemotherapy, NOS vs. single or multiple agent chemotherapy) classification based on NAACCR Item #1390 and second order classification (platinum and taxane vs. platinum only vs. taxane only vs. neither) classification based on chemotherapy exposures documented in NAACCR Item #2640 free text field. The second order classification disregards documented exposures to agents other than platinum or taxane.
We first used all N=698 patients to screen for: 1) chemotherapy/stage independence from SNP genotype, 2) association (without and with control for chemotherapy/stage) between genotype and survival, and 3) modification by chemotherapy/stage of association between genotype and survival. Results are presented in Supplemental Table 2. Chemotherapy (administered vs. not administered), stage (I-II vs. III-IV), and chemotherapy and stage jointly were independent of all evaluated ABCB1 and GSTP1 SNPs. One SNP in ABCB1 (rs2235013) showed a statistically significant association with overall survival (p=0.038, Wilcoxon-weighted log-rank test). This statistically significant association persisted in Cox regression controlled for stage (p=0.041). Notably, stage (p=0.001) and chemotherapy and stage jointly (p=0.025) significantly modified the association between rs2235013 genotype and survival.
Figure 1 explicates the modifying effect of chemotherapy/stage on association between rs2235013 and survival. Kaplan-Meier overall survival plots were qualitatively and statistically similar according to rs2235013 genotype in stage I-II patients not given chemotherapy, in stage I-II patients given chemotherapy, and in stage III-IV patients not given chemotherapy. However, in stage III-IV patients given chemotherapy, overall survival was relatively poor, in-between, and favorable in patients who inherited rs2235013 CC, CT, and TT, respectively (p=0.013). Among chemotherapy-treated stage III-IV patients who became disease-free after diagnosis, recurrence-free survival also differed according to rs2235013 genotype, with recurrence-free survival more favorable among 13 T-allele homozygotes than 60 heterozygotes and 20 C-allele homozygotes (p=0.034).
Figure 1.
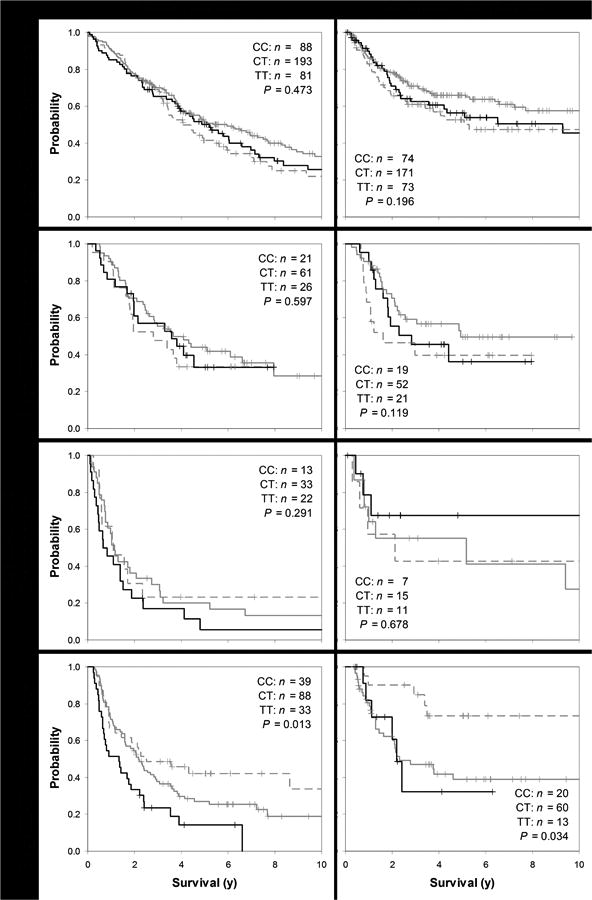
Overall (left hand plots) and recurrence-free survival (right hand plots; Kaplan-Meier plots, showing sample counts and statistical significance according to Wilcoxon-weighted log-rank test p-values) for each of 4 lung cancer subgroups, by stage and treatment, from top most to bottom most plots: stage I-II not given chemotherapy, stage I-II given chemotherapy, stage III-IV not given chemotherapy, and stage III-IV given chemotherapy, according to ABCB1 rs2235013 genotype (CC in black, CT in solid gray, and TT in dashed gray). Plus ("+") symbols signify censored observations.
Using Cox regression to control for year of diagnosis (1990-1999 / 2000-2008), sex (men / women), age at diagnosis (<55 / 55-64 / 65-69 / 70-74 / 75+), pack years (<35 / 35-49 / 50-74 / 75+), and stage (III / IV), stage III-IV patients given chemotherapy experienced a statistically significant (p=0.009) overall survival benefit in association with the ABCB1 rs2235013 T allele [hazard ratio (HR) (per T allele) 0.66, 95% confidence interval (CI) 0.49-0.90; Table 2]. As shown in Table 2, statistically significant (p<0.05) overall survival differences were also observed in association with five other ABCB1 SNPs, but not in association with any GSTP1 SNPs. These conclusions were unaffected by the addition of surgical therapy or radiation therapy to our multivariable models (data not shown). Supplemental Figure 1 shows an ABCB1 linkage disequilibrium (LD) plot for our stage III-IV lung cancer patients given chemotherapy. Five survival-associated SNPs (rs6949448, rs2235046, rs1128503, and rs10276036, in addition to rs2235013) were in high LD (r2>0.80) with each other.
Table 2.
Results from Cox proportional hazards regression models: SNP associations with overall and recurrence-free survival (additive genetic model; stage III-IV, chemotherapy-administered subgroup), adjusted for year of diagnosis (1990-1999 / 2000-2008), sex (men / women), age at diagnosis (<55 / 55-64 / 65-69 / 70-74 / 75+), pack years (<35 / 35-49 / 50-74 / 75+), and stage (III / IV)
| Overall survival | Recurrence-free survival[3] | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||||||
| Alleles[1] | All subjects | Standard treatment[2] | Disease-free post diagnosis | ||||||||||
|
| |||||||||||||
| SNP | N | HR | 95% CI | p-value | N | HR | 95% CI | p-value | N | HR | 95% CI | p-value | |
| ABCB1 | |||||||||||||
| rs1045642 | A/G | 160 | 0.77 | 0.59-0.99 | 0.045 | 106 | 0.74 | 0.50-1.09 | 0.126 | 93 | 0.62 | 0.38-1.00 | 0.050 |
| rs6949448 | C/T | 160 | 1.47 | 1.06-2.03 | 0.020 | 106 | 1.37 | 0.86-2.18 | 0.186 | 93 | 1.91 | 1.05-3.49 | 0.034 |
| rs2235067 | C/T | 160 | 0.75 | 0.48-1.10 | 0.141 | 106 | 0.67 | 0.38-1.10 | 0.116 | 93 | 0.54 | 0.21-1.11 | 0.098 |
| rs2235046 | C/T | 160 | 1.54 | 1.12-2.11 | 0.008 | 106 | 1.58 | 1.01-2.48 | 0.047 | 93 | 1.85 | 1.01-3.39 | 0.045 |
| rs2235013 | C/T | 160 | 0.66 | 0.49-0.90 | 0.009 | 106 | 0.67 | 0.44-1.02 | 0.062 | 93 | 0.55 | 0.31-0.95 | 0.032 |
| rs2235035 | G/A | 159 | 0.89 | 0.64-1.22 | 0.463 | 105 | 0.91 | 0.59-1.40 | 0.680 | 93 | 0.85 | 0.47-1.55 | 0.584 |
| rs1128503 | G/A | 160 | 1.53 | 1.11-2.09 | 0.009 | 106 | 1.51 | 0.96-2.37 | 0.075 | 93 | 2.04 | 1.11-3.77 | 0.021 |
| rs10276036 | T/C | 160 | 1.53 | 1.11-2.09 | 0.009 | 106 | 1.51 | 0.96-2.37 | 0.075 | 93 | 2.04 | 1.11-3.77 | 0.021 |
| rs1922240 | T/C | 160 | 0.87 | 0.62-1.20 | 0.383 | 106 | 0.87 | 0.57-1.34 | 0.542 | 93 | 0.85 | 0.47-1.55 | 0.584 |
| rs1202179 | T/C | 160 | 0.79 | 0.58-1.06 | 0.117 | 106 | 0.85 | 0.57-1.25 | 0.413 | 93 | 0.69 | 0.38-1.18 | 0.181 |
| rs4728709 | G/A | 160 | 0.64 | 0.32-1.15 | 0.144 | 106 | 0.71 | 0.27-1.53 | 0.408 | 93 | 0.78 | 0.28-1.76 | 0.582 |
|
| |||||||||||||
| GSTP1 | |||||||||||||
| rs6591256 | A/G | 160 | 0.95 | 0.73-1.24 | 0.725 | 106 | 1.10 | 0.76-1.57 | 0.615 | 93 | 0.79 | 0.50-1.24 | 0.303 |
| rs17593068 | G/T | 160 | 0.95 | 0.72-1.25 | 0.726 | 106 | 1.13 | 0.78-1.63 | 0.507 | 93 | 0.83 | 0.51-1.32 | 0.429 |
| rs4147581 | G/C | 160 | 1.10 | 0.84-1.44 | 0.474 | 106 | 1.41 | 0.98-2.03 | 0.067 | 93 | 0.90 | 0.57-1.44 | 0.673 |
| rs762803 | C/A | 159 | 0.95 | 0.72-1.24 | 0.699 | 106 | 1.13 | 0.78-1.63 | 0.507 | 92 | 0.87 | 0.53-1.40 | 0.568 |
| rs1695 | A/G | 160 | 0.91 | 0.68-1.22 | 0.535 | 106 | 1.10 | 0.73-1.66 | 0.639 | 93 | 0.66 | 0.38-1.12 | 0.126 |
| rs1138272 | C/T | 160 | 1.06 | 0.62-1.75 | 0.819 | 106 | 1.32 | 0.60-2.66 | 0.474 | 93 | 0.42 | 0.12-1.09 | 0.078 |
| rs4891 | T/C | 160 | 0.99 | 0.68-1.46 | 0.965 | 106 | 1.24 | 0.70-2.19 | 0.461 | 93 | 0.73 | 0.34-1.54 | 0.418 |
| rs947895 | C/A | 159 | 0.87 | 0.65-1.17 | 0.374 | 105 | 1.08 | 0.71-1.64 | 0.720 | 93 | 0.76 | 0.44-1.29 | 0.317 |
Abbreviations: HR hazard ratio, CI confidence interval, P statistical significance (p-value)
Common/minor allele
Restricted to stage III-IV non-small cell lung cancer cases who received planned multi-agent chemotherapy first course of treatment with exposure to platinum (carboplatin or cisplatin) and taxane documented
Fifty two (52) patients remaining disease free over a median post-diagnosis 4.0-year follow-up period (interquartile range 1.4-6.3 years)
We examined SNP associations with overall survival in a subgroup (N=106) of advanced stage non-small cell lung cancer patients who received standard treatment, defined to include multi-agent chemotherapy (NAACCR Item #1390 code 03) that included both platinum (carboplatin or cisplatin), taxane, and possibly other agents. Compared with results observed in all stage III-IV patients given chemotherapy, results observed in stage III-IV non-small cell lung cancer patients documented to have received standard treatment were similar both in direction and magnitude. In this subgroup, one SNP retained its statistically significant association with overall survival (rs2235046, adjusted HR 1.58, 95% CI 1.01-2.48, p=0.047; Table 2). ABCB1 differences observed with respect to overall survival among all chemotherapy-treated stage III-IV patients were also observed with respect to recurrence-free survival among chemotherapy-treated stage III-IV patients who became disease-free after diagnosis (Table 2).
Pharmacogenomic studies have frequently considered haplotypes formed from three ABCB1 SNPs, rs1045642 (3435C>T, Ile1145Ile), rs2032582 (2677G>TA, Ala893Ser/Thr), and rs1128503 (1236C>T, Gly412Gly) [10]. Our SNP panel included the first and third SNP, two survival-associated SNPs among stage III-IV cases given chemotherapy (Table 2). Limitations of the Illumina GoldenGate technology did not allow us to genotype rs2032582. However, we genotyped rs2235046, a survival-associated (Table 2) intronic SNP in high LD with rs2032582 (r2=0.94, HapMap CEU population). Substituting rs2235046 for rs2032582, we estimated rs1045642-rs2235046-rs1128503 haplotype probability weights and entered these weights jointly into Cox regression models. Referenced against the common GCG haplotype, haplotypes containing T at rs2235046 and A at rs1128503 signified poorer overall survival (per ATA allele: adjusted HR 1.46, 95% CI 1.05-2.03; per GTA allele: adjusted HR 2.19, 95% CI 1.06-4.52; Table 3). Statistically significant survival differences were not observed for the haplotype containing A at rs1045642 (per ACG allele: adjusted HR 1.18, 95% CI 0.77-1.82).
Table 3.
Results from Cox proportional hazards regression models: joint rs1045642-rs2235046-rs1128503 haplotype (additive genetic model) association with overall survival (white race, stage III-IV, chemotherapy administered, N=160)
| Haplotype | Freq | Unadjusted | Adjusted[1] | ||||
|---|---|---|---|---|---|---|---|
|
|
|
||||||
| HR | 95% CI | p-value | HR | 95% CI | p-value | ||
| GCG | 0.46 | REF | REF | ||||
| ATA | 0.39 | 1.54 | 1.14-2.09 | 0.006 | 1.46 | 1.05-2.03 | 0.023 |
| ACG | 0.11 | 1.22 | 0.80-1.86 | 0.353 | 1.18 | 0.77-1.82 | 0.446 |
| GTA | 0.04 | 2.42 | 1.21-4.83 | 0.012 | 2.19 | 1.06-4.52 | 0.035 |
Abbreviations: Freq estimated haplotype frequency, HR hazard ratio (per allele), CI confidence interval
Adjusted for year of diagnosis (1990-1999 / 2000-2008), sex (men / women), age at diagnosis (<55 / 55-64 / 65-69 / 70-74 / 75+), pack years (<35 / 35-49 / 50-74 / 75+), and stage (III / IV)
Discussion
We systematically evaluated 11 ABCB1 and 8 GSTP1 SNPs for survival differences in 698 white lung cancer patients. Five ABCB1 SNPs in high LD (rs6949448, rs2235046, rs2235013, rs1128503, and rs10276036) and a sixth independent ABCB1 SNP (rs1045642) showed statistically significant and specific associations with survival outcomes in patients with advanced lung cancer managed with chemotherapy. We did not observe statistically significant survival differences according to inherited variation in GSTP1.
Glutathione S-transferase pi 1 (GSTP1) codes for a detoxifying phase II metabolic enzyme that conjugates reduced glutathione to many xenobiotic compounds, including platinum-containing cancer drugs [4]. ATP-binding cassette, sub-family B (MDR/TAP), member 1 (ABCB1) codes for the multi-drug-resistance protein, P-glycoprotein 1, a membrane-associated protein that transports many xenobiotic compounds from the cytoplasm to the extracellular space. ABCB1 and three other ATP-binding cassette genes (ABCG2, ABCC1, and ABCC2) participate in the intra- to extra-cellular transport of taxane chemotherapeutics [5]. Inherited variation in these genes may affect treatment response and survival. By 1990, the start of enrollment for our study, multimodality treatments that included platinum were known to improve the survival of patients with locally advanced (stage III) [11] and advanced (stage IV) [12] non-small cell lung cancer. Subsequent developments soon established multi-agent chemotherapy, combining platinum with second agents, frequently taxanes, as standard primary treatments for advanced non-small cell lung cancer [11,12]. Our study sample included 11 and 217 advanced stage small and non-small cell lung cancer cases, respectively (Table 1). All 11 small cell and 149 non-small cell cases received chemotherapy as part of primary treatment, with exposure to both platinum and taxane documented in 111 (49% of 228 advanced stage cases). These results not only establish taxane, in our sample, as the preferred second agent to combine with platinum for treatment of advanced non-small cell lung cancer, but also create opportunity to observe variable outcomes in relation to an inherited factor that affects cellular delivery of taxane chemotherapeutics. In this context, survival associations with the ABCB1 SNPs of interest were preserved, though at reduced levels of statistical significance, in the smaller set of advanced non-small cell lung cancer patients with documented exposures to both platinum and taxane.
A literature search for studies of lung cancer outcomes in relation to inherited variability in ABCB1 produced 10 results [13-22], many included in a 2011 meta-analysis [23]. The nine earliest studies [13-20,22], from the U.S. [16], Europe [13,19,22], and Asia [14-18,20], examined no more than three ABCB1 variants, most often rs1045642 (3435C>T, Ile1145Ile). The limited evidence provided by these small studies, each containing between 54 [17] and 107 [14] cases of advanced (stage IIIB-IV) non-small cell lung cancer, suggest, consistent with our results, better tumor response in patients homozygous for the ABCB1 3435C allele, which corresponds to the rs1045642 G allele in our study. No SNP in ABCB1 or GSTP1 passed initial screens of genome-wide association with advanced lung cancer survival after chemotherapy in Japanese (N=105, [24]), U.S. Caucasian (N=327 [25]), or Han Chinese (N=528 [26]) populations.
Genotyping 206 and 171 Caucasian Germans with stage II-IV non-small cell and small cell lung cancer (75% platinum treated), respectively, Campa et al. [21] recently published results from a systematic study of tumor response, progression-free survival, and overall survival in relation to 25 ABCB1, 12 ABCC2, and 16 ABCG2 haplotype tagging SNPs. The most statistically significant result appeared in small cell lung cancer patients treated with platinum (N=126), where ABCC2 rs717620 minor allele containing genotypes portended poorer progression-free survival (sex-, age-, and stage-adjusted HR 1.95, 95% CI 1.29-2.94, p=0.0015). Campa et al. also called attention to statistically significant associations between inherited variations in ABCB1 rs6979885 and survival in patients with small cell lung cancer. Using HapMap CEU data, rs6979885 did not link with any of the 11 SNPs in our ABCB1 panel. Two SNPs in our panel, rs1045642 and rs2235013, were also evaluated by Campa et al. In our study population, the rs1045642 and rs2235013 minor alleles were associated with better survival. In line with our results, Campa et al. reported better survival (p < 0.05) in small cell lung cancer patients who inherited the rs1045642 G allele and in both small cell and non-small cell patients who inherited the rs2235013 T allele.
Our result showing association between inherited variation in ABCB1 and survival in lung cancer patients, the majority having received taxane (Table 2), is consistent with the observation that ABCB1 transports taxane out of cells [5]. Since our patients almost always received taxane in combination with platinum, we could not separately determine the association between ABCB1 and survival in patients who received only platinum or only taxane. Cisplatin and carboplatin are not substrates for the ABCB1 transporter [27,28], which argues against a contribution from these agents to the survival effects we observed. However, a recent study identified a novel ABCB1 transporter function, caspase-3 blockade, a mechanism whereby variation in ABCB1 might explain cisplatin resistance [29]. Another report connected down regulation of ABCB1 with reversal of cisplatin resistance in cervical cancer cells [30].
Our white only patient sample enabled study of associations between inherited differences and survival, specific to stage (stage I-II vs. stage III-IV) and chemotherapy as part of first course of treatment. Notwithstanding absent information about drug dosages and number of completed cycles, our study was not able, as noted above, to differentiate SNP effects specific to type of chemotherapy, taxane vs. platinum. SNPs were primarily selected based on biological plausibility and not all common variation in ABCB1 and GSTP1 was evaluated. It is possible that important associations and/or interactions were not identified due to the limited number of SNPs investigated. Nominal p-values shown in Table 2 and elsewhere do not correct for multiple comparisons. In addition, sample size prohibited study of rare gene variants. We determined recurrence-free survival from clinical sources where monitoring for recurrence did not occur according to a fixed protocol. Lacking information about gene expression or function in tumor or host tissues, we can only speculate about the biological significance of our observations. An influential study, published in 2000 [31], attributed functional significance to rs1045642, a synonymous polymorphism in ABCB1 exon 26. More recent critical reviews find only inconsistencies in the literature with respect to the effects of ABCB1 genotypes on MDR protein expression or function [10,32]. The absence of reproducible findings regarding the clinical significance of individual ABCB1 SNPs motivates study of ABCB1 haplotypes [32]. Our analysis of 3-SNP haplotypes formed with commonly studied variants (Table 3) concurs with results from single SNP analysis (Table 2). In this context, a recent study observed strong association between similarly constructed three-variant haplotypes and imatinib response in chronic myeloid leukemia [33].
In conclusion, partially validating findings in an independent patient population [21], we observed statistically significant associations between inherited variation in ABCB1 and clinical outcomes in advanced stage lung cancer patients treated with chemotherapy.
Supplementary Material
Acknowledgments
This work was supported by National Cancer Institute grant 5P50 CA090440 to J.M.S. and Cancer Center Core Grant 2P30 CA047904 to the University of Pittsburgh Cancer Institute. This project used the UPCI Cancer Biomarkers Facility Illumina Platform that is supported in part by award P30CA047904.
Abbreviations
- ABCB1
ATP-binding cassette, sub-family B, member 1
- AJCC
American Joint Committee on Cancer
- BAC
bronchioloalveolar carcinoma
- CI
confidence interval
- GSTP1
glutathione S-transferase pi 1
- HR
hazard ratio
- LD
linkage disequilibrium
- MAF
minor allele frequency
- MDR
multidrug resistance
- NAACCR
North American Association of Central Cancer Registries
- SNP
single nucleotide polymorphism
Footnotes
References
- 1.Surveillance E, and End Results (SEER) Program (www.seer.cancer.gov) SEER*Stat Database: Incidence - SEER 18 Regs Research Data + Hurricane Katrina Impacted Louisiana Cases, Nov 2012 Sub (1973-2010 varying) - Linked To County Attributes - Total U.S., 1969-2011 Counties, National Cancer Institute, DCCPS, Surveillance Research Program, Surveillance Systems Branch, released, April 2013, based on the November 2012 submission.
- 2.Chemotherapy in non-small cell lung cancer: A meta-analysis using updated data on individual patients from 52 randomised clinical trials. Non-small Cell Lung Cancer Collaborative Group. BMJ. 1995;311:899–909. [PMC free article] [PubMed] [Google Scholar]
- 3.Buch SC, Diergaarde B, Nukui T, et al. Genetic variability in DNA repair and cell cycle control pathway genes and risk of smoking-related lung cancer. Mol Carcinog. 2012;51(Suppl 1):E11–20. doi: 10.1002/mc.20858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Marsh S, McLeod H, Dolan E, et al. Platinum pathway. Pharmacogenet Genomics. 2009;19:563–4. doi: 10.1097/FPC.0b013e32832e0ed7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Oshiro C, Marsh S, McLeod H, Carrillo M, Klein T, Altman R. Taxane Pathway. Pharmacogenet Genomics. 2009;19:979–83. doi: 10.1097/FPC.0b013e3283335277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fritz A, Percy C, Jack A, et al., editors. ICD-O: International Classification of Diseases for Oncology. 3rd. Geneva: World Health Organization; 2000. [Google Scholar]
- 7.Greene FL, Page DL, Fleming ID, et al., editors. AJCC Cancer Staging Manual. 6th. New York: Springer; 2002. [Google Scholar]
- 8.Facility Oncology Registry Data Standards: Revised for 2011. American College of Surgeons Commission on Cancer; 2011. [Google Scholar]
- 9.Thornton M, O'Connor L, editors. Standards for Cancer Registries Volume II: Data Standards and Data Dictionary, Record Layout Version 12. 14th. Springfield, Ill: North American Association of Central Cancer Registries; Feb, 2009. rev. August 2009. [Google Scholar]
- 10.Leschziner GD, Andrew T, Pirmohamed M, Johnson MR. ABCB1 genotype and PGP expression, function and therapeutic drug response: A critical review and recommendations for future research. Pharmacogenomics J. 2007;7:154–79. doi: 10.1038/sj.tpj.6500413. [DOI] [PubMed] [Google Scholar]
- 11.Choy H, Pyo H, Kim JS, MacRae R. Role of taxanes in the combined modality treatment of patients with locally advanced non-small cell lung cancer. Expert Opin Pharmacother. 2001;2:963–74. doi: 10.1517/14656566.2.6.963. [DOI] [PubMed] [Google Scholar]
- 12.Ramalingam S, Belani CP. Taxanes for advanced non-small cell lung cancer. Expert Opin Pharmacother. 2002;3:1693–709. doi: 10.1517/14656566.3.12.1693. [DOI] [PubMed] [Google Scholar]
- 13.Isla D, Sarries C, Rosell R, et al. Single nucleotide polymorphisms and outcome in docetaxel-cisplatin-treated advanced non-small-cell lung cancer. Ann Oncol. 2004;15:1194–203. doi: 10.1093/annonc/mdh319. [DOI] [PubMed] [Google Scholar]
- 14.Han JY, Lim HS, Yoo YK, et al. Associations of ABCB1, ABCC2, and ABCG2 polymorphisms with irinotecan-pharmacokinetics and clinical outcome in patients with advanced non-small cell lung cancer. Cancer. 2007;110:138–47. doi: 10.1002/cncr.22760. Erratum appears in Cancer. 2010 Aug 1;116(15):3749. [DOI] [PubMed] [Google Scholar]
- 15.Pan JH, Han JX, Wu JM, Sheng LJ, Huang HN, Yu QZ. MDR1 single nucleotide polymorphisms predict response to vinorelbine-based chemotherapy in patients with non-small cell lung cancer. Respiration. 2008;75:380–5. doi: 10.1159/000108407. [DOI] [PubMed] [Google Scholar]
- 16.Gandara DR, Kawaguchi T, Crowley J, et al. Japanese-US common-arm analysis of paclitaxel plus carboplatin in advanced non-small-cell lung cancer: A model for assessing population-related pharmacogenomics. J Clin Oncol. 2009;27:3540–6. doi: 10.1200/JCO.2008.20.8793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pan JH, Han JX, Wu JM, Huang HN, Yu QZ, Sheng LJ. MDR1 single nucleotide polymorphism G2677T/A and haplotype are correlated with response to docetaxel-cisplatin chemotherapy in patients with non-small-cell lung cancer. Respiration. 2009;78:49–55. doi: 10.1159/000158454. [DOI] [PubMed] [Google Scholar]
- 18.Chen S, Huo X, Lin Y, et al. Association of MDR1 and ERCC1 polymorphisms with response and toxicity to cisplatin-based chemotherapy in non-small-cell lung cancer patients. Int J Hyg Environ Health. 2010;213:140–5. doi: 10.1016/j.ijheh.2010.01.004. [DOI] [PubMed] [Google Scholar]
- 19.Viñolas N, Provencio M, Reguart N, et al. Single nucleotide polymorphisms in MDR1 gene correlates with outcome in advanced non-small-cell lung cancer patients treated with cisplatin plus vinorelbine. Lung cancer. 2011;71:191–8. doi: 10.1016/j.lungcan.2010.05.005. [DOI] [PubMed] [Google Scholar]
- 20.Yan PW, Huang XE, Yan F, Xu L, Jiang Y. Influence of MDR1 gene codon 3435 polymorphisms on outcome of platinum-based chemotherapy for advanced non small cell lung cancer. Asian Pac J Cancer Prev. 2011;12:2291–4. [PubMed] [Google Scholar]
- 21.Campa D, Muller P, Edler L, et al. A comprehensive study of polymorphisms in ABCB1, ABCC2 and ABCG2 and lung cancer chemotherapy response and prognosis. Int J Cancer. 2012;131:2920–8. doi: 10.1002/ijc.27567. [DOI] [PubMed] [Google Scholar]
- 22.Dogu GG, Kargi A, Turgut S, et al. MDR1 single nucleotide polymorphism C3435T in Turkish patients with non-small-cell lung cancer. Gene. 2012;506:404–7. doi: 10.1016/j.gene.2012.06.057. [DOI] [PubMed] [Google Scholar]
- 23.Wei HB, Lu XS, Shang LH, et al. Polymorphisms of ERCC1 C118T/C8092A and MDR1 C3435T predict outcome of platinum-based chemotherapies in advanced non-small cell lung cancer: A meta-analysis. Arch Med Res. 2011;42:412–20. doi: 10.1016/j.arcmed.2011.07.008. [DOI] [PubMed] [Google Scholar]
- 24.Sato Y, Yamamoto N, Kunitoh H, et al. Genome-wide association study on overall survival of advanced non-small cell lung cancer patients treated with carboplatin and paclitaxel. J Thorac Oncol. 2011;6:132–8. doi: 10.1097/JTO.0b013e318200f415. [DOI] [PubMed] [Google Scholar]
- 25.Wu X, Ye Y, Rosell R, et al. Genome-wide association study of survival in non-small cell lung cancer patients receiving platinum-based chemotherapy. J Natl Cancer Inst. 2011;103:817–25. doi: 10.1093/jnci/djr075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hu L, Wu C, Zhao X, et al. Genome-wide association study of prognosis in advanced non-small cell lung cancer patients receiving platinum-based chemotherapy. Clin Cancer Res. 2012;18:5507–14. doi: 10.1158/1078-0432.CCR-12-1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Herraez E, Gonzalez-Sanchez E, Vaquero J, et al. Cisplatin-induced chemoresistance in colon cancer cells involves FXR-dependent and FXR-independent up-regulation of ABC proteins. Mol Pharm. 2012;9:2565–76. doi: 10.1021/mp300178a. [DOI] [PubMed] [Google Scholar]
- 28.Zhang DM, Shu C, Chen JJ, et al. BBA, a derivative of 23-hydroxybetulinic acid, potently reverses ABCB1-mediated drug resistance in vitro and in vivo. Mol Pharm. 2012;9:3147–59. doi: 10.1021/mp300249s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gibalova L, Seres M, Rusnak A, et al. P-glycoprotein depresses cisplatin sensitivity in L1210 cells by inhibiting cisplatin-induced caspase-3 activation. Toxicol In Vitro. 2012;26:435–44. doi: 10.1016/j.tiv.2012.01.014. [DOI] [PubMed] [Google Scholar]
- 30.Chang A. Chemotherapy, chemoresistance and the changing treatment landscape for NSCLC. Lung cancer. 2011;71:3–10. doi: 10.1016/j.lungcan.2010.08.022. [DOI] [PubMed] [Google Scholar]
- 31.Hoffmeyer S, Burk O, von Richter O, et al. Functional polymorphisms of the human multidrug-resistance gene: multiple sequence variations and correlation of one allele with P-glycoprotein expression and activity in vivo. Proc Natl Acad Sci U S A. 2000;97:3473–8. doi: 10.1073/pnas.050585397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fung KL, Gottesman MM. A synonymous polymorphism in a common MDR1 (ABCB1) haplotype shapes protein function. Biochim Biophys Acta. 2009;1794:860–71. doi: 10.1016/j.bbapap.2009.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vivona D, Bueno CT, Lima LT, et al. ABCB1 haplotype is associated with major molecular response in chronic myeloid leukemia patients treated with standard-dose of imatinib. Blood Cells Mol Dis. 2012;48:132–6. doi: 10.1016/j.bcmd.2011.11.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


