Abstract
Genetically encoded fluorescent reporters of membrane potential promise to reveal aspects of neural function not detectable by other means. We present a palette of multi-colored brightly fluorescent genetically encoded voltage indicators with sensitivities from 8 – 13% ΔF/F per 100 mV, and half-maximal response times from 4 – 7 ms. A fluorescent protein is fused to an Archaerhodopsin-derived voltage sensor. Voltage-induced shifts in the absorption spectrum of the rhodopsin lead to voltage-dependent nonradiative quenching of the appended fluorescent protein. Through a library screen, we identify linkers and fluorescent protein combinations which report neuronal action potentials in cultured rat hippocampal neurons with a single-trial signal-to-noise ratio from 7 to 9 in a 1 kHz imaging bandwidth at modest illumination intensity. The freedom to choose a voltage indicator from an array of colors facilitates multicolor voltage imaging, as well as combination with other optical reporters and optogenetic actuators.
Introduction
Membrane voltage acts on all transmembrane proteins: the membrane electric field pulls on charged residues, shifting the free energy landscape for charge-displacing conformational transitions1. This bioelectric modulation is most famously observed in voltage-gated ion channels in neurons and cardiomyocytes, but voltage also affects the activity of G protein-coupled receptors (GPCRs) and some transmembrane enzymes2. Membrane voltage is dynamically regulated in bacteria3, fungi4, plants5, and many cell types and sub-cellular organelles in the human body, and is disregulated in states of neuronal, cardiac, and metabolic diseases. Thus there is a need for fast, sensitive, bright, and spectrally tunable reporters of membrane voltage.
Recent progress in genetically encoded voltage indicators (GEVIs) has led to several classes of proteins which robustly report action potentials (APs) in cultured neurons. The first GEVIs were based on fusion of fluorescent proteins to transmembrane voltage-sensing domains6. In some of these, voltage modulates the brightness of a single fluorescent moiety 7–10, while in others, voltage modulates the efficiency of Förster resonance energy transfer (FRET) between a pair of fluorescent moieties 11, 12. In our measurements of GFP-based GEVIs, described below, the most sensitive was ArcLight 8 (ΔF/F = −32% per 100 mV), but this reporter had half-maximal response times at room temperature of 90 ms (depolarizing step) and 104 ms (hyperpolarizing step). We measured the recently introduced ASAP1 9 reporter to have a sensitivity of ΔF/F = −29% per 100 mV, and a half-maximal response of 2 ms at room temperature. These numbers differ modestly from the original reports, likely due to differences in gene expression protocols and choices of filter sets.
A second class of GEVIs is based on microbial rhodopsin proton pumps3, 13, 14. In these, the transmembrane voltage modulates the endogenous near infrared fluorescence of the retinal chromophore. The most recently developed non-pumping mutants of Archaerhodopsin 3 (Arch), termed QuasArs, have voltage sensitivities between 30 and 90% per 100 mV (depending on the mutant), and half-maximal response times between 50 μs and 1.1 ms at room temperature13.
GFP-based and rhodopsin-based GEVIs have very different spectral properties. GFP-based GEVIs are excited by blue light (470 – 490 nm) and emit green (500 – 530 nm). QuasArs are excited by red light (594–640 nm) and emit in the near infrared (peak at 715 nm). GFP-based GEVIs are 30 to 80-fold brighter, and thus are typically imaged with excitation at ~10 W cm−2, while QuasArs are typically imaged at 300 – 800 W cm−2 (Ref. 13). In a comparison between ArcLight and QuasAr2 in cultured rat hippocampal neurons under their respective standard imaging conditions, QuasAr2 reported single APs with 4.7-fold higher signal-to-noise ratio (SNR) and was 15-fold more photostable13. In organotypic mouse hippocampal slice culture, QuasAr2 reported single APs with 4.5-fold higher SNR than ArcLight13. Nonetheless, the low brightness of Arch-based GEVIs presents a challenge for widespread use.
The availability of GEVIs spanning the visible spectrum is important when combining GEVIs with other optical reporters or optogenetic actuators. Having GEVIs of colors between GFP and Arch would facilitate multiplex voltage imaging, e.g. to distinguish activity of excitatory and inhibitory neurons in intact tissue. Furthermore, GEVIs spectrally distinct from GFP could be paired with other GFP-based reporters such as GCaMP (Ca2+) (Ref. 15), iGluSnFR (glutamate)16, Perceval (ATP)17, Clomeleon (Cl−)(Ref. 18), or Pyronic (pyruvate)19, or with other blue-excited optogenetic actuators. Thus there is strong motivation to develop a broad palette of GEVI colors.
We sought to combine the speed and sensitivity of Arch-based GEVIs with the brightness and spectral range of conventional fluorescent proteins. Traditionally, FRET is used to measure the physical distance between a donor and acceptor. However, we and others have also used FRET to measure changes in the absorption spectrum of a protein via its influence on nonradiative quenching of an attached fluorescent moiety20–22.
Here we use voltage-induced changes in the absorption spectrum of an Arch mutant to alter the degree of nonradiative quenching of a closely fused fluorescent protein. We call this mechanism electrochromic FRET (eFRET). We demonstrate fast and sensitive voltage sensing in fusions between QuasAr2 and a broad palette of fluorescent proteins: enhanced GFP (EGFP), yellow FP (Citrine), mOrange2, mRuby2. These probes have sensitivities from 8 – 13% ΔF/F per 100 mV, and half-maximal response times from 4 – 7 ms, which allows detection of action potentials in cultured rat hippocampal neurons with a single-trial signal-to-noise ratio from 7 to 9 in a 1 kHz imaging bandwidth at modest illumination intensity. A recent paper applied a similar strategy to make voltage sensors based on fusion of fluorescent proteins to a mutant of the L. maculans rhodopsin (Mac)23. While other GEVIs offer superior signal-to-noise ratio on the blue and far red ends of the spectrum, the present GEVIs fill in the visible spectrum with emission wavelengths between 530 – 640 nm. This spectral range facilitates multicolor voltage imaging with GEVIs, as well as combination with other optical reporters and optogenetic actuators.1
Results
Engineering eFRET GEVIs
Our electrochromic quencher is a mutant of Archaerhodopsin 3 (Arch), termed QuasAr2 (Fig. 1). The direct retinal fluorescence of QuasAr2 showed voltage sensitivity of 90% ΔF/F per 100 mV and a bi-exponential step response with time constants of 1.2 ms (68%) and 11.8 ms (32%) (Table 1) at room temperature13. It has not been feasible to measure directly the voltage-dependent absorption spectra of Arch or its mutants on account of the small quantity of protein found in the plasma membrane of a single cell. However, a mechanistic study of Arch showed that membrane voltage modulated fluorescence via a voltage-dependent shift in the acid-base equilibrium of the retinal Schiff base.24 Positive membrane voltage favored the protonated (and fluorescent) state. One can thus mimic the voltage-induced absorption changes by changing the pH in a solution of solubilized protein. At neutral pH, QuasAr2 had a broad absorption band peaked at 587 nm. At higher pH, this band dropped and a band at 411 nm appeared, corresponding to the deprotonated Schiff base (Supplementary Figure 1).
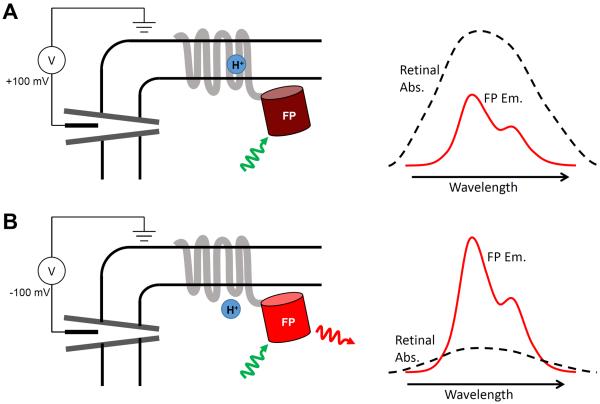
Figure 1. Proposed mechanism of voltage-dependent fluorescence in eFRET GEVIs.
Voltage controls the protonation, and thereby the absorption spectrum, of the Schiff base joining the retinal to the protein scaffold. A) At a depolarizing (positive) voltage, the microbial rhodopsin has a protonated retinal Schiff base, which absorbs strongly with a peak near 600 nm. The retinal absorption quenches the fluorescence of the appended fluorescent protein (FP). B) At hyperpolarizing (negative) voltages, the fluorescence is not quenched. Spectra shown are cartoons to illustrate mechanism.
Table 1.
Voltage sensing properties of GEVIs.
| HEK Cells | Neurons | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Fluorophore | ΔF/F per 100 mV(%) | Response to hyperpolarizing step | Response to depolarizing step | ΔF/F for single AP(%) | SNR for single AP | ||||||
| τfast (ms) | τslow (ms) | % fast | τ1/2 (ms) | τfast (ms) | τslow (ms) | % fast | τ1/2 (ms) | ||||
| ECFP | −0.8 | n.d. | n.d. | ||||||||
| EGFP | −7.7 | 4.3 | 27 | 45 | 7.6 | 3.0 | 26 | 61 | 3.9 | n.d. | |
| Citrine | −13.1 | 4.8 | 21 | 38 | 7.5 | 3.1 | 21 | 62 | 3.8 | 9.7 | 8.8 |
| mOrange2 | −10.0 | 4.3 | 26 | 45 | 7.5 | 3.9 | 27 | 60 | 5.1 | 9.9 | 9.0 |
| mRuby2 | −8.7 | 4.3 | 27 | 65 | 4.7 | 3.6 | 20 | 65 | 3.8 | 5.4 | 7.2 |
| mKate2 | −4.5 | 2.8 | 35 | 25 | 9.2 | 4.0 | 25 | 44 | 6.7 | n.d. | |
| ArcLight Q239 | −31.6 | 104 | 283 | 61 | 103 | 28 | 271 | 39 | 90 | 8.2 | 9.2 |
| ASAP1 | −28.8 | 3.0 | 29 | 67 | 3.5 | 2.3 | 39 | 88 | 1.9 | 17.8 | 13.1 |
| QuasAr2 | 90 | 1.0 | 16 | 80 | 0.9 | 1.2 | 12 | 68 | 1.4 | 48 | 41 |
Sensitivity, ΔF/F per 100 mV, was measured for a voltage step from −70 mV to +30 mV. Response times were characterized either by fast and slow time constants from a bi-exponential fit (τfast and τslow) or by τ1/2, the time to reach 50% of the steady state response to a step of membrane voltage of duration at least 2 τslow. Half-response times are shorter for steps that do not reach steady state. Hyperpolarizing step: +30 mV to −70 mV; depolarizing step: −70 mV to +30 mV. All measurements were performed at 23 °C and represent the average of 6 – 9 cells. Illumination intensities were 3 W cm−2 for all reporters except for QuasAr2. Illumination intensity was 200 W cm−2 for QuasAr2.23
Based on these spectra, we reasoned that QuasAr2 might mediate voltage-dependent quenching of an appended fluorescent protein. The emission spectrum of mOrange2 25 has a high degree of overlap with the absorption spectrum of Arch so we started with this fluorophore. To enable efficient FRET, it is critical to optimize the length of the linker, as rate of FRET is proportional to the reciprocal sixth power of the distance between the donor and acceptor chromophores. We examined the crystal structures of Arch-2 (a QuasAr2 homologue) (PDB ID 2EI4) 26 and mOrange (an mOrange2 homologue) (PDB ID 2H5O) 27, and found that the last 15 C-terminal residues of Arch-2 and 15 N-terminal residues of mOrange are largely unstructured (Fig. 2A). We reasoned that those unstructured residues could be removed without disrupting the function of either protein. We created a series of linker libraries by systematically truncating these resectable regions (Fig. 2B and Methods) and randomizing two residues at the junction.
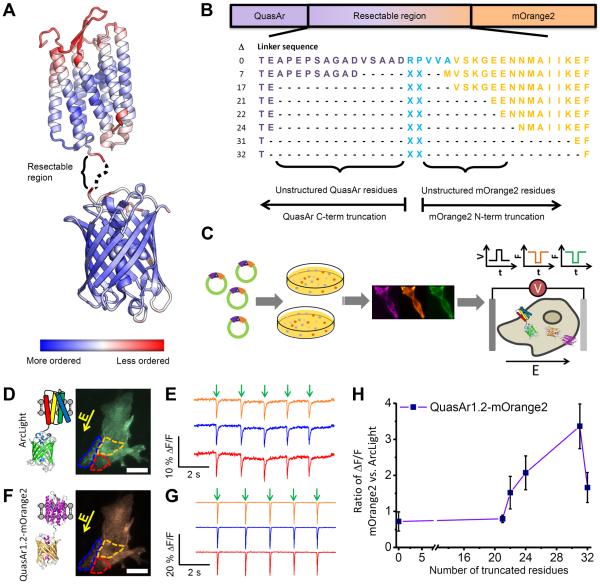
Figure 2. Directed evolution of an eFRET GEVI.
A) A model of QuasAr-mOrange2 constructs, represented by the crystal structures of Arch-2 (PDB ID 2EI4) 26 and mOrange (PDB ID 2H5O) 27. The color scale represents the degree of order in the crystal structure, as reported by the B-factor. Less ordered regions are presumed more dispensable. B) Design of linker truncation libraries. QuasAr1.2, the best available variant at the time of the screen, was used as the electrochromic quencher. C) Hierarchical screen of truncated linker libraries for eFRET GEVIs. Constructs were first screened in E. coli for mOrange2 brightness and then screened for membrane trafficking in HEK cells. Voltage-sensitivity was then tested via field stimulation in HeLa cells co-expressing the eFRET GEVI, ArcLight (as an internal control), and Kir2.1 (to lower the resting voltage to −60 mV 42). D) ArcLight fluorescence of three HeLa cells. Yellow arrow indicates direction of the applied electric field. Scale bar: 25 μm. E) Fluorescence intensity traces of the three regions shown in D). F) mOrange2 fluorescence from the same cells shown in D). G) Response of mOrange2 fluorescence during stimulation as in E). Fluorescence traces show single-trial measurement of cells imaged at 100 frames per second. H) Effect of linker length on voltage sensitivity in QuasAr1.2-mOrange2. For each linker length the most sensitive eFRET GEVI construct was compared to ArcLight measured in the same cells. Error bars represent standard deviation over n = 15–20 cells in the ratio ΔF/F.
We then developed a hierarchical screening strategy to identify variants with optimal mOrange2 brightness, membrane trafficking, and voltage response (Fig. 2C and Methods). We used a customized vector, termed pcDuEx1.0, for gene expression in both prokaryotic and eukaryotic cells. This vector circumvents the laborious cloning typically required to switch between a primary screen in bacteria and a secondary screen in mammalian cells (Methods). Colonies of E. coli were transformed with linker libraries in pcDuEx1.0. The colonies with the brightest mOrange2 fluorescence were picked for secondary screening in mammalian cells.
For the mammalian cell-based screen, a green GEVI, ArcLight Q239 7, was used as an internal reference for both membrane trafficking and voltage response (Fig. 2C–G and Methods). The sensitivity of eFRET variants generally increased as the linker was shortened. The most truncated variants (Δ31 and Δ32 libraries) had excellent voltage sensitivity, but substantially decreased brightness of mOrange2, likely due either to poor folding or to high basal quenching by the rhodopsin. In libraries with more than 20 residues truncated, the screen favored combinations of Arg, Leu, Ala, and Gly for the linker. The variant from the Δ24 library with linker sequence `Leu Arg' gave the best overall performance and was used henceforth. We added a trafficking sequence (TS) and endoplasmic reticulum export motif (ER2) to improve trafficking to the plasma membrane 28, although trafficking remained imperfect.
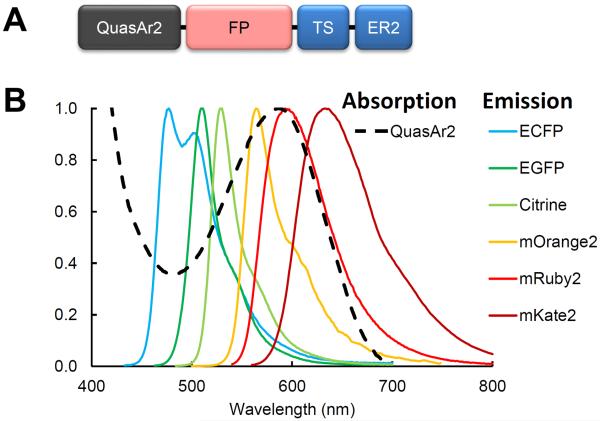
QuasAr2 absorbs strongly from 500 – 640 nm, so we reasoned that eFRET might work for fluorescent proteins with a range of emission wavelengths. We created a palette of eFRET constructs by replacing mOrange2 with: ECFP29, EGFP29, Citrine30, mRuby212, and mKate231. The emission spectra of these proteins all overlapped to some degree with the absorption spectrum of QuasAr2 (Fig. 3). Spectral properties of these fluorophores are given in Supplementary Table 1.
Figure 3. Spectral overlap of fluorescent proteins with QuasAr2 absorption.
A) eFRET GEVI domain structure. B) Overlay of the emission spectra of the fluorescent protein donors with the absorption spectrum of the QuasAr2 electrochromic quencher.
To test the hypothesized eFRET mechanism, we compared the fluorescence quantum yield of Citrine in the eFRET fusion and in a membrane-targeted construct lacking the QuasAr2 quencher, both expressed in HEK cells. We used two-photon fluorescence excitation and time-correlated single-photon counting to determine the electronic excited state lifetime (Methods). The native Citrine protein had a lifetime of τbare = 3.47 ns, while the eFRET construct had a lifetime of τFRET = 2.83 ns (Supplementary Figure 2). This difference in lifetime confirms that an additional nonradiative decay pathway exists in the eFRET construct compared to the bare Citrine protein, which we ascribe to FRET from Citrine to retinal. The FRET efficiency is given by EFRET = 1 - τFRET/τbare, from which EFRET = 0.18. Maximum voltage sensitivity occurs at EFRET = 0.5, implying that further optimization of the position and orientation of the FP may improve quenching efficiency and thereby voltage sensitivity.
Voltage responses in HEK cells
We characterized the eFRET GEVIs in HEK cells, imaged at 23 °C. All eFRET GEVIs trafficked efficiently to the plasma membrane (Fig. 4A) and exhibited bright fluorescence when excited at 3 W cm−2 with appropriate excitation and emission wavelengths (Supplementary Table 1). Membrane voltage was controlled via whole-cell voltage clamp. Fluorescence traces were extracted from images of cells either by manually defining a region of interest comprising the cell body, or by using an automated pixel-weighting algorithm14. Both approaches gave similar results. Fluorescence responses were normalized to fluorescence at a holding potential of −70 mV (after background subtraction) and are quoted as ΔF/F. Voltage sensitivity was calculated as [F(30 mV) – F(−70 mV)]/F(−70 mV). While rhodopsins require retinal as a cofactor, we found that the culture medium contained sufficient retinal that voltage sensitivity was not enhanced by addition of exogenous retinal (Supplementary Figure 3).
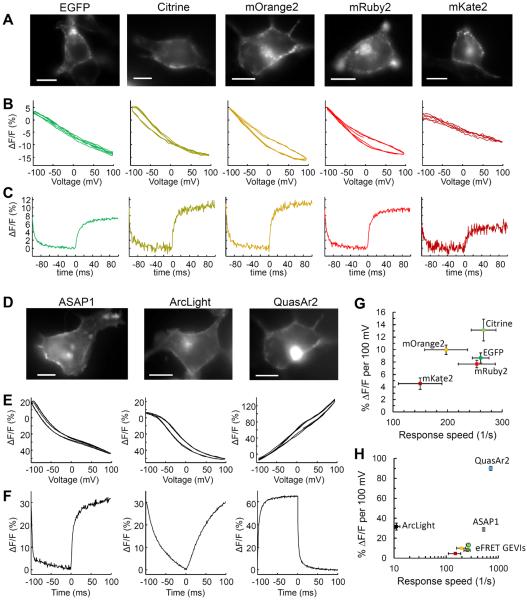
Figure 4. Voltage sensing with GEVIs in HEK293 cells.
Voltage response of eFRET GEVI reporters are shown in (A–C), and voltage responses of comparison GEVIs are shown in (D–F). A) and D) Fluorescence images of HEK cells expressing GEVI reporters. Scale bars 10 μm. B) and E) Fluorescence as a function of membrane voltage, normalized to fluorescence at V = −70 mV. Many eFRET-based GEVIs showed a small amount of hysteresis. Each trace is a single trial with 100 ms exposures. C) and F) Fluorescence response to a step in membrane voltage from −70 mV to +30 mV. Each trace is averaged over 20–30 trials, with 1 ms exposures. All data was acquired with laser illumination (3 W cm−2 for eFRET, ASAP1, and ArcLight GEVIs, 200 W cm−2 for QuasAr2). No temporal filtering was applied. Fluorescence traces were corrected for photobleaching. G) Quantification of eFRET GEVI sensitivity and speed. Voltage sensitivity of eFRET GEVIs approximately corresponds to the degree of spectral overlap between the emission of the GEVI and the absorption of QuasAr2. H) eFRET GEVI sensitivity and speed in the context of other GEVIs. Response speed was defined as the inverse of the time to reach 50% of steady-state response to a voltage step from −70 mV to +30 mV. Data in G) and H) represents an average of n = 6 to 9 cells for each construct. Error bars represent s.e.m.
Fluorescence of all eFRET GEVIs showed approximately linear dependence on applied voltage (Fig. 4B). The most sensitive construct was the Citrine fusion with ΔF/F = −13.1 ± 1.8% per 100 mV (n = 6 cells, all statistics are mean ± s.e.m.). The EGFP, mOrange2, and mRuby2 constructs showed sensitivities between 7.7 and 10.0% per 100 mV (Table 1). The bluest and reddest constructs, containing ECFP and mKate2, exhibited poor voltage sensitivity, which we attribute to poor spectral overlap with QuasAr2 absorption. The negative slope of dF/dV was consistent with greater spectral overlap of mOrange2 emission and QuasAr2 absorption at depolarizing voltages.
Fast fluorescence response to a change in voltage is essential for detecting neuronal APs. We applied a square wave of voltage (−70 mV to +30 mV, 5 Hz) and recorded the fluorescence at a frame rate of 1,000 frames s−1 (Fig. 4C). Fluorescence responses were approximately fit by a sum of two exponentials, with a dominant fast and a smaller slow time constant. The fastest construct was the mRuby2 GEVI, with τfast = 3.6 ± 0.3 ms (65%) and τslow = 20 ± 2 ms (35%) for depolarizing membrane potential step (n = 8 cells). The GFP, Citrine, and mOrange2 had half-response times between 3.9 and 7.6 ms (Table 1). Response times were similar on rising and falling edges. Response amplitudes to a 5 ms step from −70 to +30 mV are given in Supplementary Table 2.
We made side-by-side comparisons of the performance of the eFRET GEVIs to two GFP-based GEVIs, ArcLight Q239 (Ref. 7), and ASAP1 (Ref. 9), and to direct fluorescence of QuasAr2 (Fig. 4D–F). Cell culture, gene expression, electrophysiology, imaging, and analysis protocols were identical across constructs (except for changes in illumination wavelength and intensity as noted). ArcLight showed voltage sensitivity of −31.6 ± 3.5% ΔF/F per 100 mV (n = 7 cells, Fig. 4E), consistent with previous reports. The step response of ArcLight followed a bi-exponential decay with time constants of 28 ± 8 ms (39%) and 271 ± 6 ms (61%) (for a depolarizing voltage step from −70 mV to +30 mV) and a bi-exponential fluorescence rise with time constants of 104 ± 21 ms (61%) and 283 ± 40 ms (39%) (for voltage step from +30 mV to −70 mV) (n = 7 cells, Supplementary Figure 4). These numbers are slower than in the original report on ArcLight7, due to the difference in measurement temperature: 23 °C in our measurements vs. 33–35 °C in the prior report. ASAP1 showed voltage sensitivity of 28.8 ± 1.7% per 100 mV and depolarizing volrage step response time constants of 2.3 ± 0.1 ms (88%) and 39 ± 3 ms (12%) (n = 6 cells, Fig. 4E–F and Table 1).
The voltage response of direct retinal fluorescence of QuasAr2 was measured in the QuasAr2-mOrange eFRET GEVI. To achieve comparable levels of QuasAr2 and mOrange2 fluorescence required 70-fold greater illumination intensity for QuasAr2 (640 nm, 200 W cm−2 vs. 532 nm, 3 W cm−2). Compared to the mOrange2 fluorescence, QuasAr2 fluorescence was more sensitive to voltage (ΔF/F = 90 ± 2% per 100 mV, n = 6 cells, Fig. 4E–H) and had a somewhat faster response time (double-exponential fit yielding: τ1 = 1.2 ms, 68%, τ2 = 12 ms, 32%, Fig. 4E–H). The discrepancy in response time between the QuasAr2 and mOrange2 may arise because, in addition to the fast fluorescence-determining transition, QuasAr2 may also undergo slower voltage-dependent transitions that change its absorption spectrum but not its fluorescence. Alternatively, subtle voltage-induced conformational shifts could change the relative position and orientation of the mOrange2 and the retinal, and thereby change the efficiency of nonradiative quenching.
Characterization in neurons
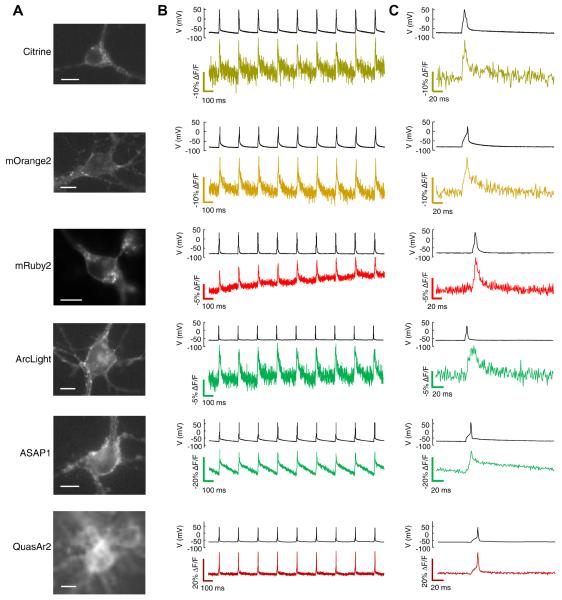
The Citrine, mOrange2, and mRuby2 eFRET GEVIs showed sufficient sensitivity and speed to merit testing in neurons. We expressed these via transient transfection in cultured rat hippocampal neurons. Constructs exhibited good trafficking to the plasma membrane (Fig. 5). Injection of current pulses via a patch pipette (500 – 600 pA, 5 – 10 ms, 5 Hz) induced trains of APs, which induced downward fluorescence transients of 9.7 ± 1.7% (Citrine), 9.9 ± 1.3% (mOrange2), and 5.4 ± 0.4 % (mRuby2) (n = 7–11 cells for each, Table 1). Our eFRET constructs also reported spontaneous activity in cultured neurons (Fig. 6).
Figure 5. Single-trial recording of neuronal APs with GEVIs.
Each GEVI construct was expressed in primary rat hippocampal neurons under a CamKIIα promoter. Action potentials were induced by current injections through a patch pipette. A) Images of neurons expressing the indicated GEVI. Scale bars 10 μm. B) Simultaneous patch clamp and fluorescence recordings of AP waveforms. All recordings were acquired with laser illumination (3 W cm−2 for eFRET, ASAP1, and ArcLight GEVIs, 200 W cm−2 for QuasAr2) at an exposure time of 1 ms. Fluorescence traces are presented without temporal filtering nor correction for photobleaching. All but the QuasAr2 recording have been inverted (hence photobleaching appears as an upward trend in the mRuby2 trace). Recordings are representative traces from 5 – 11 cells recorded for each construct. C) Close-up showing single-trial, unfiltered, electrically and optically reported AP waveforms.
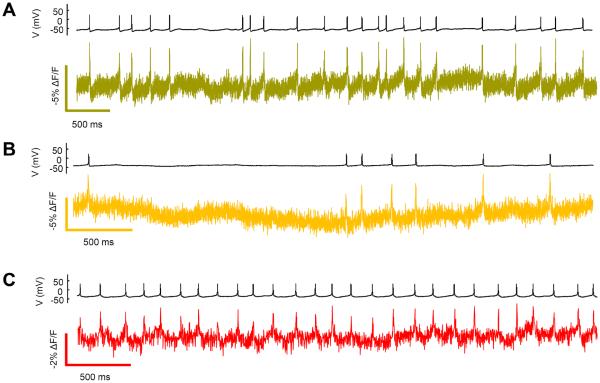
Figure 6. Optical recording of spontaneous neuronal firing with eFRET GEVIs.
In neurons synaptically coupled to a spontaneously active circuit, the (A) Citrine, (B) mOrange, and (C) mRuby2 eFRET GEVIs clearly revealed single APs. Patch clamp voltage recordings are shown above optical recording traces. All recordings were acquired with laser illumination (3 W cm−2) at an exposure time of 2 ms. Fluorescence traces are presented without temporal filtering. The optical recordings have been corrected for photobleaching with a linear fit.
Arclight also reported single-trial action potentials when used at 23 °C, with a downward transient of 8.2 ± 0.7% (n = 7 cells), though this response appeared to be dominated by the response to the slow sub-threshold depolarization as opposed to the fast spike. Thus, despite the greater steady-state voltage sensitivity of ArcLight than the eFRET GEVIs, the slow response of ArcLight acted as a low-pass filter on millisecond-timescale APs, decreasing the fluorescence response 7. ASAP1 reported single-trial neuronal APs with downward fluorescence transients of 17.8 ± 3.4% (n = 5 cells). On account of its greater sensitivity at negative voltages, ASAP1 magnified the sub-threshold signal (55% of fluorescence response) relative to the AP spike (45% of fluorescence response). In a similar assay, Mac-based eFRET GEVIs were reported to resolve single APs with downward transients of 7.2% for MacQ-mOrange2 and 4.8% for MacQ-mCitrine 23.
The SNR of fluorescence detection, defined as the ratio of the peak amplitude to standard deviation of fluorescence at the baseline, was 8.8 ± 1.0 (Citrine), 9.0 ± 1.2 (mOrange2), and 7.2 ± 0.6 (mRuby2) (1 kHz frame rate, illumination at 3 W cm−2, 23 °C for all eFRET GEVIs) (n = 7 – 11 cells for each, Table 1). Parallel measurements on ArcLight had a single-trial SNR of 9.2 ± 1.5 and substantially broadened the AP waveform (1 kHz frame rate, illumination at 3 W cm−2, 23 °C), while ASAP1 had a single-trial SNR of 13.1 ± 2.9. Previous measurements with QuasAr2 reported a single-trial SNR for AP detection of 41 (300 W cm−2) and 70 (800 W cm−2) 13.
Discussion
With the proliferation of GEVI constructs, it can be difficult to identify the best construct for a particular experiment. Speed, sensitivity, and SNR for single APs for recently reported GEVIs are summarized in Table 1. SNR depends on both voltage sensitivity (ΔF/F) and overall fluorescence counts (F). In the shot noise-limited regime, SNR for AP detection scales as , where ΔF is the change in fluorescence counts for a single AP. Baseline fluorescence counts, F, can vary considerably between experiments, depending upon illumination intensity and wavelength, protein expression level, sources of background autofluorescence, collection efficiency of the objective, choice of emission filter, quantum efficiency of the detector, and selection of the region of interest. The SNR values reported here provide a comparative guide for measurements under matched conditions, but will differ for measurements performed under different conditions.
For multi-color voltage imaging, the choice of emission filter is critical. We used the noise scaling to estimate the SNR for each eFRET GEVI for 30 nm-wide detection bands between 510 – 690 nm. We then determined which eFRET GEVI gave the maximum SNR in each band (Supplementary Figure 5). This calculation suggests that the eFRET GEVIs enable detection of APs with SNR > 5 for any 30 nm emission band between 510 – 660 nm. In the GFP spectral band, ASAP1 outperforms the eFRET GEVIs and ArcLight in terms of sensitivity, speed, and SNR. In the yellow and orange emission bands, the MacQ-based eFRET GEVIs 23 are somewhat more sensitive, while the QuasAr2-based eFRET GEVIs are somewhat faster. For detection of APs in cultured neurons, both types of constructs yield similar SNR. At the far red end of the spectrum, QuasAr2 outperforms both ASAP1 and the eFRET GEVIs in terms of sensitivity, speed, SNR, and photostability; but the requirement for intense laser illumination is still a limitation.
The excitation and emission wavelengths of a GEVI or other optical reporter influence performance when imaging in intact tissue. The excitation and emission spectra of flavins, a major contributor to brain autofluorescence, overlap strongly with those of GFP 32, 33. Consequently, a rule of thumb is that the signal-to-background ratio for GFP-based reporters is 10-fold lower in tissue than in cell culture. Redder reporters have significantly less background in brain tissue. Light scattering in tissue scales with wavelength λ approximately as λ−2.33, so redder reporters can be imaged deeper34. Illumination at 640 nm propagates nearly 1.9-fold further into tissue than does illumination at 488 nm. While two-photon excitation of GFP-based reporters enables even greater depth penetration, this comes at the cost of requiring serial scanning, and a complex optical setup. Absolute brightness is also important when imaging in tissue, due to the need to overcome background autofluorescence and to minimize optical power dissipation in the tissue. However, in a comparison between ArcLight and QuasAr2 in rat brain slice, the far dimmer but redder QuasAr2 reported action potentials with 4.5-fold higher SNR than ArcLight, albeit at 24-fold higher illumination intensity.13 Due to the complex combination of factors that determine GEVI performance in vivo, an empirical test is far preferable to extrapolations from measurements in vitro.
Trafficking remains a key limitation of all GEVIs, despite adoption of trafficking motifs in most. The efficiency of trafficking to the plasma membrane often varies dramatically between preparations, between cells of the same type in vitro vs. in vivo, and between different cell types from the same organism. For now, an empirical approach is warranted to identify the construct best suited to a particular experiment.
The varied F vs. V curves for available GEVIs accentuate different aspects of cellular electrophysiology. The more linear GEVIs (eFRET and QuasArs) preserve the waveform of the underlying voltage dynamics, a useful feature for quantification of action potential waveforms. The more sigmoidal GEVIs (ASAP1 and ArcLight) emphasize voltage changes in the region of maximum slope. For ASAP1 and ArcLight this feature would emphasize sub-threshold dynamics over action potentials in population-level recordings averaged over many cells. Due to their slightly different voltages of maximum slope, ASAP1 would show greatest sensitivity for hyperpolarizing potentials while ArcLight would show greatest sensitivity for depolarizing potentials (Fig. 4E). ArcLight variants have been demonstrated with maximum sensitivity over a range of voltages7.
Several challenges remain in genetically encoded voltage imaging. Due to uncontrolled variation in expression level, none of the GEVIs discussed here reports absolute membrane voltage. To achieve absolute accuracy requires a more complex imaging scheme, either ratiometric35 or time-resolved 36. Two-photon voltage imaging, which would facilitate imaging in tissue, also remains a challenge. This goal has been achieved with voltage-sensitive dyes 37, but two-photon GEVI imaging has not yet clearly resolved single action potentials, though some recent work succeeded in detecting activity-evoked signals38.
Further improvements in the eFRET GEVIs are likely to come from optimization of the rhodopsin-based quencher and from improvements in the attachment of the fluorescent protein to the rhodopsin. A rhodopsin need not show endogenous fluorescence to serve as an efficient eFRET quencher, and thus many rhodopsins that are not suitable as direct voltage indicators may still be useful for eFRET. Thousands of microbial rhodopsin genes have been identified in the wild39, 40, and among these improved quenchers are likely to exist. The estimated FRET efficiency of 18% in the Citrine construct implies that increases in voltage sensitivity could be attained by improving energy transfer from the fluorescent protein to the rhodopsin. This increase in efficiency could come from greater intimacy of contact between the fluorescent protein and the rhodopsin, or from optimizing the relative orientation of the donor and acceptor fluorophores. To achieve these goals, one may insert circularly permuted fluorescent proteins into intracellular loops on the rhodopsin.
With simultaneous multicolor voltage imaging one could apply genetic targeting to record voltage simultaneously from distinct structures that might otherwise be difficult to distinguish. For instance-wide-field images that lack single-cell resolution could still distinguish population-average activity levels in intermingled excitatory and inhibitory neurons. The broad palette of eFRET GEVIs also facilitates combination with other GFP-based fluorescent reporters or a wide variety of optogenetic actuators.
Methods
Molecular biology
Synthetic DNA oligonucleotides were purchased from Integrated DNA Technologies. PfuUltraII polymerase (Agilent Technologies) or Phusion Polymerase (New England Biolabs) were used for high fidelity PCR amplifications in the buffer supplied by the respective manufacturer. PCR products and products of restriction digests were purified either with PCR clean up kit (QIAGEN) or using preparative agarose gel electrophoresis followed by DNA isolation using the Zymoclean gel DNA recovery kit (Zymo Research). Restriction endonucleases were purchased from New England Biolabs and used according to the manufacturer's instructions. Ligations were performed using T4 DNA ligase (Invitrogen) or Gibson Assembly (New England Biolabs). Small-scale isolation of plasmid DNA was performed by plasmid miniprep kit (QIAGEN). The cDNA sequences for all fusion constructs were confirmed by sequencing (Genewiz). Site-directed mutagenesis was performed with QuikChange kit (Agilent Technologies).
Construction of linker libraries
A vector for expression in prokaryotic and eukaryotic systems was constructed based on mammalian expression vector pcDNA3.1 (+). To facilitate prokaryotic expression, a customized constitutive promoter was introduced (ttgctttgtgagcggataacaattataatagattca) based on the phage early T5 promoter for prokaryotic transcription 41. An E. coli ribosome binding site (aggaggaa) for prokaryotic translation was also introduced via QuikChange reactions. The resultant vector, designated pcDuEx1.0, exhibits moderate expression of Arch-mOrange2 fusions in E. coli cells and very similar expression levels in HeLa cells compared to its predecessor. We used pcDuEx1.0 as the vector for screening of linker libraries.
We focused on mOrange2 fusions for determining optimized linker length for FRET efficiency. We chose QuasAr1.2 (Arch D95N/D106H, the best available at the time of the screen) as the Arch body. For construction of QuasAr1.2-mOrange2 Δ0, we fused QuasAr1.2 to the N-terminus of mOrange2 fusion via a 5 residue linker (RPVVA) using overlap PCR. The DNA was PCR amplified with the flanking restriction sites BamHI and XbaI, followed by double digestion and ligation into pcDuEx1.0 linearized by digestion with the same two enzymes.
From the template of QuasAr1.2-mOrange2 Δ0, we constructed linker libraries by systematic truncation of the connecting region between QuasAr1.2 and mOrange2 using Quikchange reactions (Fig. 2). For each library, 2 randomized amino acid residues (nucleotide sequence NNKNNK, where N = A, G, C, T and K = G, T) were placed between the C-terminus of Arch and the N-terminus of mOrange2, to generate 400 amino acid combinations (1024 nucleotide combinations). The libraries were cloned into pcDuEx1.0 via standard procedure of restriction enzyme digestion and ligation.
Hierarchical screen of linker libraries
E. coli (DH10B ™, Invitrogen) colonies expressing the linker library exhibited varied intensities of mOrange2 fluorescence. For each library, the 24 variants with brightest mOrange2 fluorescence were picked and grown in overnight culture at 37 °C. The plasmid DNA of each variant was prepared from overnight culture using standard mini-prep procedure. For each variant, the plasma membrane trafficking was examined in HEK cells (CRL-1573™, ATCC) co-expressing ArcLight, which served as internal reference. The 5 variants with the best membrane trafficking were then expressed in HeLa cells for testing of voltage sensitivity.
HeLa cells (CCL2 ™, ATCC) were grown to 40–60% confluence on home-made 35 mm glass bottom dishes or 24-well glass bottom plates, and transfected with 1 μg of plasmid DNA and 2 μL Turbofect (Thermo Scientific) according to the manufacturer's instructions. HeLa cells were transfected with plasmids encoding the QuasAr1.2-mOrange2 variant, ArcLight Q239 (Addgene: 36856) and Kir2.1 (Addgene: 32641) in equal weight ratio. Expression of Kir2.1 in HeLa cells maintained the resting potential close to −60 mV, appropriate for a neuronal voltage indicator 42. After 3 h incubation, the media was exchanged to DMEM with 10% fetal bovine serum and the cells were incubated for an additional 24 h at 37 °C in a CO2 incubator. Immediately prior to imaging, cells were washed twice with Hanks balanced salt solution (HBSS) and then 1 mL of 20 mM HEPES buffered HBSS was added. No retinal was added to the buffer--loading of retinal into the rhodopsin was presumed to occur from endogenous retinal.
Cell imaging was performed with an inverted Eclipse Ti-E (Nikon) equipped with a Photometrics QuantEM 512SC camera and a 150 W mercury-xenon lamp (Hamamatsu). A home-made parallel platinum electrode pair with a separation distance of 0.5 cm was mounted in a custom plastic support and was placed in the imaging dish or well. The waveforms of voltage pulses were generated by a pulse generator PG 58A (Gould Advance Ltd.) and amplified by an Agilent 6824A 40V/25A DC Power Supply (Hewlett Packard). The typical waveform had square wave pulses lasting 20 ms with pulse field strength ranging from 50 – 60 V cm–1. The mOrange2 fluorescence was imaged at 100 Hz frame rate in 4×4 binning mode for 10 s using the following filter set: 545/30 nm (excitation), 620/60 nm (emission), and 565 nm (dichroic). For imaging ArcLight, the filter set was: 480/40 nm (excitation), 535/40 nm (emission), and 505 nm (dichroic).
The raw fluorescence traces of both mOrange2 and ArcLight were extracted from identical regions of interest in cells expressing both constructs, and exported into a Microsoft Excel spreadsheet using the microscope software NIS-Elements Advanced Research (Nikon). Background subtraction, photobleaching corrections, calculations of average ΔF/F, and calculation of signal-to-noise ratios (SNR) were performed automatically in Excel. The average ΔF/F and SNR of mOrange2 signals was compared to those of ArcLight signals from the same cells, and the ratios of ΔF/F of mOrange2 vs. ArcLight were reported. At least 20 cells co-expressing mOrange2 fusion and ArcLight were analyzed for each variant. The best variant with maximum mean ratio in each library was determined and sequenced.
pH-dependent absorption spectrum of QuasAr2
E. coli cells transformed with a pBAD vector encoding QuasAr2 were grown in 12 mL liquid LB medium with 200 μg/mL ampicillin overnight. The next day, 12 mL of liquid LB medium containing 50 μM retinal, 200 μg/mL ampicillin and 0.2% L-arabinose was added into the culture to induce expression of QuasAr2, followed by additional incubation at 37 °C for 3.5 hours. The cell pellets were collected by centrifugation, washed with Tris buffered saline (TBS, 30 mM Tris, 150 mM NaCl, pH 7.4), and lysed with a tip sonicator for 10 min. The cytoplasmic fraction was discarded after centrifugation and the colored insoluble fraction was resuspended in a series of modified TBS buffers (30 mM Tris, 150 mM NaCl, adjusted to the pH value of the spectral measurement) containing 1% n-dodecyl-β-D-maltopyranoside (Affymetrix, Inc.). The suspension was then centrifuged (17,000 g for 15 min, 4 °C). Absorption spectra of QuasAr2 in the supernatant in different pH were recorded on a DU-800 UV-visible spectrophotometer (Beckman).
Expression vectors for HEK cells and neurons
We chose lentivirus vector FCK-Arch-EGFP (Addgene: 22217) as the backbone for all eFRET constructs. This vector features a CaMKIIα promoter and a Woodchuck Hepatitis Virus Posttranscriptional Regulatory Element (WPRE) after the 3' end of the open reading frame. To enhance membrane trafficking of fusion proteins, we added a trafficking signal (TS) and ER export signal peptide sequence (FCYENEV), derived from the inward rectifier potassium channel Kir2.1, as previously described 28. QuasAr2-FP fusion constructs were made by Gibson Assembly: the vector was linearized by double digestion with BamHI and BsrGI, and QuasAr2 and fluorescent protein cDNA segments were generated by PCR amplification.
The linker configuration of all eFRET fusion proteins was constructed based on the optimized linker sequence found by the linker screening. In all eFRET fusion proteins, the N-terminal 2 amino acids of QuasAr2 were changed from DP to VS to facilitate expression, and the C-terminal 14 amino acids of QuasAr2 (APEPSAGADVSAAD) were replaced with a 2-amino acid linker, Leu-Arg (based on the optimized linker sequence found in linker library Δ24), to shorten the distance between QuasAr2 and fluorescent protein. The N-terminal amino acids in the fluorescent proteins were also truncated (Supplementary Table 3). The nucleotide sequences of all constructs are in the Supplementary Note 1.
Two-photon lifetime measurements to determine FRET efficiency
We measured fluorescence lifetime on a homebuilt beam-scanning two-photon microscope with an 80 MHz, 100 fs tunable pulsed laser (SpectraPhysics Insight DeepSee). Pulse compression in the sample plane was performed through maximizing the fluorescence intensity of a bead-sample, using the motorized prism compressor built into the laser. The citrine measurements were performed at 1040 nm excitation wavelength with a time-averaged excitation power of 30 mW, or 0.4 nJ per pulse, focused down to a ~500 nm spot with a 1.2 NA water immersion objective (Olympus UplanSapo) for a time-averaged intensity of 15 MW cm−2; measurements were performed for linear speeds of the scanned spot varying between 0 and 30 cm s−1 without any measurable effect on fluorescence lifetime. Excitation light and fluorescence were separated using a FF775-Di01 dichroic mirror and FF01-790/SP-25 shortpass filter (Both Semrock); fluorescence was detected using a Hamamatsu R943-02 photomultiplier tube in photon counting mode, cooled to −20 °C. THe PMT signal was amplified through an SRS PR325 amplifier and discretized with a Hamamatsu Photon counting unit C9744, before being fed into a Picoharp 300 TCSPC module (Picoquant). The setup was controlled by Labview software written in-house.
Simultaneous electrophysiology and fluorescence in HEK cells
HEK293t/17 cells (ATCC CRL-11268)were cultured and transfected following standard protocols. Briefly, cells were grown at 37 °C, 5% CO2, in DMEM supplemented with 10% FBS and penicillin-streptomycin. Plasmids were transfected using TransIT-293 reagent (Mirus Bio LLC) following the manufacturer's instructions. 24 hours post-transfection, cells were re-plated onto glass-bottom dishes (MatTek) at a density of ~10,000 cells cm−2. Cells were assayed 40–60 hours post-transfection.
Despite the presence of saturating retinal in the culture medium, cells were supplemented with retinal prior to measurement to remove a possible source of variability. Cells were supplemented with retinal by pre-incubating with 5 μM retinal in growth medium (diluted from 40 mM stock solution in DMSO) in the incubator for 0.5 – 1 hour immediately prior to imaging. All imaging and electrophysiology were performed in Tyrode's buffer (containing 125 mM NaCl, 2.5 mM KCl, 3 mM CaCl2, 1 mM MgCl2, 10 mM HEPES, 30 mM glucose, at pH 7.3, and adjusted to 305–310 mOsm with sucrose). A gap junction blocker, 2-aminoethoxydiphenyl borate (50 μM, Sigma), was added to eliminate electrical coupling between cells.
Filamented glass micropipettes (WPI) were pulled to a tip resistance of 5–10 MΩ, and filled with internal solution containing 125 mM potassium gluconate, 8 mM NaCl, 0.6 mM MgCl2, 0.1 mM CaCl2, 1 mM EGTA, 10 mM HEPES, 4 mM Mg-ATP, 0.4 mM Na-GTP (pH 7.3); adjusted to 295 mOsm with sucrose. Pipettes were positioned with a Sutter MP285 manipulator. Whole-cell, voltage and current clamp recordings were acquired using a patch clamp amplifier (A-M Systems, Model 2400), filtered at 5 kHz with the internal filter and digitized with a National Instruments PCIE-6323 acquisition board at 10 kHz.
Simultaneous whole-cell patch clamp recordings and fluorescence recordings were acquired on a home-built, inverted epifluorescence microscope, described below in the section “Imaging Apparatus”.
Neuronal culture and electrophysiology
All procedures involving animals were in accordance with the National Institutes of Health Guide for the care and use of laboratory animals and were approved by the Institutional Animal Care and Use Committee (IACUC) at the institution at which they were carried out.
Hippocampal neurons from P0 rat pups were dissected and cultured in neurobasal-based medium (NBActiv4, Brainbits llc.) at a density of 40,000 cm−2 on glass-bottom dishes (MatTek) pre-coated with poly-d-lysine (Sigma P7205) and matrigel (BD biosciences 356234). At 3 days in vitro (DIV), cytarabine was added to the neuronal culture medium at a final concentration of 2 μM to inhibit glial growth 43.
Neurons were transfected on DIV 7 with the eFRET plasmid using Lipofectamine 2000 transfection reagent (Life Technologies). Procedures followed manufacturer's instructions but reduced the amount of reagent by 50–80% to avoid toxicity.
Measurements were performed on primary cultures at DIV 10 – 20. Experiments were conducted in Tyrode's solution containing 125 mM NaCl, 2.5 mM KCl, 3 mM CaCl2, 1 mM MgCl2, 10 mM HEPES, 30 mM glucose (pH 7.3) and adjusted to 305–310 mOsm with sucrose. Immediately prior to imaging, neurons were incubated with 5 μM all-trans retinal in the culture medium for 30 minutes and then washed with Tyrode's solution. Experiments were performed at 23 °C under ambient atmosphere.
Imaging apparatus
Experiments were conducted on a home-built inverted fluorescence microscope equipped with 488nm, 532nm, 561nm, 594nm, and 640nm laser lines and a scientific CMOS camera (Hamamatsu ORCA-Flash 4.0). The power and manufacturer of laser lines are summarized in Supplementary Table 4. Illumination from lasers were combined using dichroic mirrors, sent through an acousto-optic tunable filter (AOTF; Gooch and Housego 48058-2.5-.55-5W) for intensity modulation, and then expanded and focused onto the back-focal plane of a 60x water immersion objective, numerical aperture 1.20 (Olympus UIS2 UPlanSApo 60x/1.20 W). Imaging of fluorescent proteins was performed at illumination intensities of 2 – 4 W cm–2. Imaging of QuasAr2 direct fluorescence was performed at an illumination intensity of 200–400 W cm−2. Supplementary Table 5 summarizes the laser lines, dichroic mirrors and emission filters used for fluorescence imaging. For fast data acquisition, a small field of view around the cell of interest was chosen at the center of the camera to achieve a frame rate of 1,000 frames per second.
Data analysis
Data was analyzed with homemade software written in MATLAB. Fluorescence intensities from raw movies were extracted using a maximum likelihood pixel weighting algorithm described in Ref. 14. Briefly, the fluorescence at each pixel was correlated with the mean fluorescence. Pixels that showed stronger correlation to the mean were preferentially weighted. This algorithm automatically found the pixels carrying the most information, and de-emphasized background pixels. Alternatively, a region of interest (ROI) comprising the cell body was defined by the user, and fluorescence intensity was calculated from the unweighted mean of pixel values within the ROI. With the improved trafficking of the QuasAr2 mutants resulting from the TS and ER2 motifs, the ROI approach gave similar results as the maximum likelihood pixel weighting algorithm.
For eFRET speed analysis, the time constants for the step response were calculated by fitting a double exponential to the rising and decaying portions of the fluorescence traces. All error ranges represent standard error of the mean.
Supplementary Material
Acknowledgments
We thank V. Nathan and N. Smedemark-Margulies for technical assistance, and V. Venkatachalam, S. Farhi, and J. Kralj for helpful discussions. We thank A. Y. Ting, F. St. Pierre, and M. Z. Lin for plasmids and spectral information. PZ, ADD, DRH, DB, CAW, and AEC were supported by the Howard Hughes Medical Institute, the Harvard Center for Brain Science, PECASE award N00014-11-1-0549, US National Institutes of Health grants 1-R01-EB012498-01 and New Innovator grant 1-DP2-OD007428. DRH was supported by an NSF Graduate Fellowship. DB was supported by an NWO-Rubicon Fellowship. REC was supported by the Natural Sciences and Engineering Research Council of Canada (Discovery Grants) and the Canadian Institutes of Health Research. YZ was supported by a graduate scholarships from the University of Alberta and Alberta Innovates. R.E.C. holds a Tier II Canada Research Chair.
Footnotes
Author contributions PZ, YZ, DRH, ADD, CAW, DB conducted experiments and analyzed data. REC supervised the screen for linker length optimization. AEC proposed eFRET for voltage sensing. PZ, YZ, REC, and AEC wrote the paper.
Competing financial interest statement AEC is a founder of Q-State Biosciences.
References
- 1.Cohen AE, Venkatachalam V. Bringing bioelectricity to light. Annu. Rev. Biophys. 2014;43:11.1–11.22. doi: 10.1146/annurev-biophys-051013-022717. [DOI] [PubMed] [Google Scholar]
- 2.Bezanilla F. How membrane proteins sense voltage. Nat. Rev. Mol. Cell Biol. 2008;9:323–332. doi: 10.1038/nrm2376. [DOI] [PubMed] [Google Scholar]
- 3.Kralj JM, Hochbaum DR, Douglass AD, Cohen AE. Electrical spiking in Escherichia coli probed with a fluorescent voltage indicating protein. Science. 2011;333:345–348. doi: 10.1126/science.1204763. [DOI] [PubMed] [Google Scholar]
- 4.Slayman CL, Slayman CW. Depolarization of the plasma membrane of Neurospora during active transport of glucose: evidence for a proton-dependent cotransport system. Proc. Nat. Acad. Sci. U. S. A. 1974;71:1935–1939. doi: 10.1073/pnas.71.5.1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mousavi SA, Chauvin A, Pascaud F, Kellenberger S, Farmer EE. GLUTAMATE RECEPTOR-LIKE genes mediate leaf-to-leaf wound signalling. Nature. 2013;500:422–426. doi: 10.1038/nature12478. [DOI] [PubMed] [Google Scholar]
- 6.Siegel MS, Isacoff EY. A Genetically Encoded Optical Probe of Membrane Voltage. Neuron. 1997;19:735–741. doi: 10.1016/s0896-6273(00)80955-1. [DOI] [PubMed] [Google Scholar]
- 7.Jin L, et al. Single action potentials and subthreshold electrical events imaged in neurons with a fluorescent protein voltage probe. Neuron. 2012;75:779–785. doi: 10.1016/j.neuron.2012.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cao G, et al. Genetically Targeted Optical Electrophysiology in Intact Neural Circuits. Cell. 2013;154:904–913. doi: 10.1016/j.cell.2013.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.St-Pierre F, et al. High-fidelity optical reporting of neuronal electrical activity with an ultrafast fluorescent voltage sensor. Nat. Neurosci. 2014;17:884–889. doi: 10.1038/nn.3709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Baker BJ, et al. Genetically encoded fluorescent voltage sensors using the voltage-sensing domain of Nematostella and Danio phosphatases exhibit fast kinetics. J. Neurosci. Methods. 2012;208:190–196. doi: 10.1016/j.jneumeth.2012.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mutoh H, Akemann W, Knöpfel T. Genetically engineered fluorescent voltage reporters. ACS Chem. Neuro. 2012;3:585–592. doi: 10.1021/cn300041b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lam AJ, et al. Improving FRET dynamic range with bright green and red fluorescent proteins. Nat. Methods. 2012;9:1005–1012. doi: 10.1038/nmeth.2171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hochbaum DR, et al. All-optical electrophysiology in mammalian neurons using engineered microbial rhodopsins. Nat. Methods. 2014 doi: 10.1038/nmeth.3000. in press, doi:10.1038/nmeth.3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kralj JM, Douglass AD, Hochbaum DR, Maclaurin D, Cohen AE. Optical recording of action potentials in mammalian neurons using a microbial rhodopsin. Nat. Methods. 2012;9:90–95. doi: 10.1038/nmeth.1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen T, et al. Ultrasensitive fluorescent proteins for imaging neuronal activity. Nature. 2013;499:295–300. doi: 10.1038/nature12354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marvin JS, et al. An optimized fluorescent probe for visualizing glutamate neurotransmission. Nat. Methods. 2013;10:162–170. doi: 10.1038/nmeth.2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tantama M, Martínez-François JR, Mongeon R, Yellen G. Imaging energy status in live cells with a fluorescent biosensor of the intracellular ATP-to-ADP ratio. Nat. Commun. 2013;4:e2550. doi: 10.1038/ncomms3550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kuner T, Augustine GJ. A genetically encoded ratiometric indicator for chloride: capturing chloride transients in cultured hippocampal neurons. Neuron. 2000;27:447–459. doi: 10.1016/s0896-6273(00)00056-8. [DOI] [PubMed] [Google Scholar]
- 19.San Martín A, et al. Imaging mitochondrial flux in single cells with a FRET sensor for pyruvate. PLoS One. 2014;9:e85780. doi: 10.1371/journal.pone.0085780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bayraktar H, et al. Ultrasensitive measurements of microbial rhodopsin photocycles using photochromic FRET. Photochem. Photobiol. 2011;88:90–97. doi: 10.1111/j.1751-1097.2011.01011.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Barker SL, Kopelman R, Meyer TE, Cusanovich MA. Fiber-optic nitric oxide-selective biosensors and nanosensors. Anal. Chem. 1998;70:971–976. doi: 10.1021/ac970706k. [DOI] [PubMed] [Google Scholar]
- 22.Kuznetsova S, et al. The enzyme mechanism of nitrite reductase studied at single-molecule level. Proc. Natl. Acad. Sci. U. S. A. 2008;105:3250–3255. doi: 10.1073/pnas.0707736105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gong Y, Wagner MJ, Li JZ, Schnitzer MJ. Imaging neural spiking in brain tissue using FRET-opsin protein voltage sensors. Nat. Commun. 2014;5:3674. doi: 10.1038/ncomms4674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maclaurin D, Venkatachalam V, Lee H, Cohen AE. Mechanism of voltage-sensitive fluorescence in a microbial rhodopsin. Proc. Natl. Acad. Sci. USA. 2013;110:5939–5944. doi: 10.1073/pnas.1215595110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shaner NC, et al. Improving the photostability of bright monomeric orange and red fluorescent proteins. Nat. Methods. 2008;5:545–551. doi: 10.1038/nmeth.1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Enami N, et al. Crystal structures of archaerhodopsin-1 and-2: Common structural motif in archaeal light-driven proton pumps. J. Mol. Biol. 2006;358:675–685. doi: 10.1016/j.jmb.2006.02.032. [DOI] [PubMed] [Google Scholar]
- 27.Shu X, Shaner NC, Yarbrough CA, Tsien RY, Remington SJ. Novel chromophores and buried charges control color in mFruits. Biochemistry (N. Y.) 2006;45:9639–9647. doi: 10.1021/bi060773l. [DOI] [PubMed] [Google Scholar]
- 28.Gradinaru V, et al. Molecular and Cellular Approaches for Diversifying and Extending Optogenetics. Cell. 2010;141:154–165. doi: 10.1016/j.cell.2010.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Heim R, Tsien RY. Engineering green fluorescent protein for improved brightness, longer wavelengths and fluorescence resonance energy transfer. Curr. Biol. 1996;6:178–182. doi: 10.1016/s0960-9822(02)00450-5. [DOI] [PubMed] [Google Scholar]
- 30.Griesbeck O, Baird GS, Campbell RE, Zacharias DA, Tsien RY. Reducing the environmental sensitivity of yellow fluorescent protein. Mechanism and applications. J. Biol. Chem. 2001;276:29188–29194. doi: 10.1074/jbc.M102815200. [DOI] [PubMed] [Google Scholar]
- 31.Shcherbo D, et al. Far-red fluorescent tags for protein imaging in living tissues. Biochem. J. 2009;418:567–574. doi: 10.1042/BJ20081949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chung YG, Schwartz JA, Sawaya RE. Diagnostic potential of laser-induced autofluorescence emission in brain tissue. J. Korean Med. Sci. 1997;12.2:135–142. doi: 10.3346/jkms.1997.12.2.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lin W, Toms SA, Motamedi M, Jansen ED, Mahadevan-Jansen A. Brain tumor demarcation using optical spectroscopy; an in vitro study. J. Biomed. Opt. 2000;5:214–220. doi: 10.1117/1.429989. [DOI] [PubMed] [Google Scholar]
- 34.Flock ST, Jacques SL, Wilson BC, Star WM, van Gemert MJ. Optical properties of Intralipid: a phantom medium for light propagation studies. Lasers Surg. Med. 1992;12:510–519. doi: 10.1002/lsm.1900120510. [DOI] [PubMed] [Google Scholar]
- 35.Zhang J, Davidson RM, Wei M, Loew LM. Membrane electric properties by combined patch clamp and fluorescence ratio imaging in single neurons. Biophys. J. 1998;74:48–53. doi: 10.1016/S0006-3495(98)77765-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hou JH, Venkatachalam V, Cohen AE. Temporal Dynamics of Microbial Rhodopsin Fluorescence Reports Absolute Membrane Voltage. Biophys. J. 2014;106:639–648. doi: 10.1016/j.bpj.2013.11.4493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fisher JA, et al. Two-photon excitation of potentiometric probes enables optical recording of action potentials from mammalian nerve terminals in situ. J. Neurophysiol. 2008;99:1545–1553. doi: 10.1152/jn.00929.2007. [DOI] [PubMed] [Google Scholar]
- 38.Akemann W, et al. Two-photon voltage imaging using a genetically encoded voltage indicator. Sci. Rep. 2013;3:2231. doi: 10.1038/srep02231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Beja O, Spudich EN, Spudich JL, Leclerc M, Delong EF. Proteorhodopsin phototrophy in the ocean. Nature. 2001;411:786–789. doi: 10.1038/35081051. [DOI] [PubMed] [Google Scholar]
- 40.Spudich JL. The multitalented microbial sensory rhodopsins. Trends Microbiol. 2006;14:480–487. doi: 10.1016/j.tim.2006.09.005. [DOI] [PubMed] [Google Scholar]
- 41.Bujard H, et al. A T5 promoter-based transcription-translation system for the analysis of proteins in vitro and in vivo. Meth. Enzymol. 1987;155:416–433. doi: 10.1016/0076-6879(87)55028-5. [DOI] [PubMed] [Google Scholar]
- 42.Hille B. Ion channels of excitable membranes. Sinauer; Sunderland, MA: 2001. [Google Scholar]
- 43.Chen G, Harata NC, Tsien RW. Paired-pulse depression of unitary quantal amplitude at single hippocampal synapses. Proc. Nat. Acad. Sci. U. S. A. 2004;101:1063–1068. doi: 10.1073/pnas.0307149101. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.