Abstract
The canonical Wnt signaling pathway is critical for myogenesis and can induce muscle progenitors to switch from proliferation to differentiation; how Wnt signals integrate with muscle specific regulatory factors in this process is poorly understood. We previously demonstrated that the Barx2 homeobox protein promotes differentiation in cooperation with the muscle regulatory factor (MRF) MyoD. Pax7, another important muscle homeobox factor represses differentiation. We now identify Barx2, MyoD, and Pax7 as novel components of the Wnt effector complex, providing a new molecular pathway for regulation of muscle progenitor differentiation. Canonical Wnt signaling induces Barx2 expression in muscle progenitors and perturbation of Barx2 leads to misregulation of Wnt target genes. Barx2 activates two endogenous Wnt target promoters as well as the Wnt reporter gene TOPflash, the latter synergistically with MyoD. Moreover, Barx2 interacts with the core Wnt effectors β-catenin and TCF, is recruited to TCF/LEF sites, and promotes recruitment of β-catenin. In contrast, Pax7 represses the Wnt reporter gene and antagonizes the activating effect of Barx2. Pax7 also binds β-catenin suggesting that Barx2 and Pax7 may compete for interaction with the core Wnt effector complex. Overall, the data show for the first time that Barx2, Pax7, and MRFs can act as direct transcriptional effectors of Wnt signals in myoblasts and that Barx2 and Wnt signaling participate in a regulatory loop. We propose that antagonism between Barx2 and Pax7 in regulation of Wnt signaling may help mediate the switch from myoblast proliferation to differentiation.
Keywords: Muscle stem cells, differentiation, Wnt signaling, Homeobox
Maintenance and repair of skeletal muscle is mediated by stem cells called satellite cells [1–4]. Located between the basal lamina and the muscle fiber, satellite cells are quiescent in intact muscle; upon injury they are rapidly activated to form a pool of proliferative myoblasts that can differentiate to repair the injury. Pax7 is expressed in quiescent satellite cells and their proliferating myoblast progeny, but is downregulated during differentiation [5, 6]. Forced expression of Pax7 delays myoblast differentiation, in part by inhibition of MyoD function, suggesting a role for Pax7 in preventing precocious differentiation [5, 7–9]. We showed that the Barx2 homeobox protein is co-expressed with Pax7 in satellite cells and myoblasts and promotes differentiation [10–12]. Barx2 interacts directly with MyoD and regulates binding of MyoD to target genes [13]. Moreover, Barx2 exists in a regulatory loop with MRFs, whereby MRFs directly regulate Barx2 expression and Barx2 directly or indirectly regulates MRFs [14]. Barx2 null mice show impaired postnatal muscle growth, maintenance, and regeneration, consistent with the important role of Barx2 in satellite cell and myoblast function [10]. Currently the functional relationship between Pax7 and Barx2 is unknown.
Wnts have multiple roles in muscle development, acting as positional cues to specify the myotomes and inducing expression of MRFs [15–17]. Canonical Wnt signaling is activated during muscle regeneration in vivo [8] and canonical ligands Wnt3a and Wnt4 have been shown to promote myoblast differentiation in culture [18–20]. Recently Wnt signaling was shown to induce the switch from myoblast proliferation to differentiation [20, 21], which may involve regulation of MRFs and homeobox factors [22, 23].Canonical Wnt signaling is mediated by T-cell factor/lymphoid enhancer factor (TCF/LEF) proteins and β-catenin. In the absence of Wnt signals, these factors repress target genes by recruitment of TLE/Groucho family co-repressors. Activation of Wnt signalling stabilizes β-catenin, which then pairs with TCF/LEF, displaces co-repressors, and recruits co-activators to induce gene expression [24, 25]. How the activities of the ubiquitously expressed core Wnt effector proteins are transduced into tissue specific effects remains unclear.
Here we report for the first time physical and functional connections between Wnt/β-catenin signaling, Barx2, Pax7, and MRFs that begin to address the muscle-specific mechanisms of Wnt signaling. We show that Barx2 activates transcription via TCF/LEF sites in cooperation with MRFs,β-catenin and TCF proteins, while Pax7 antagonizes β-catenin and Barx2 function. We propose that functional antagonism between Barx2 and Pax7 with respect to Wnt signalling may be involved in the transition from myoblast proliferation to differentiation.
Results
Canonical Wnt signaling promotes differentiation of primary myoblasts and expression of Barx2
We previously showed that Barx2 is upregulated at the onset of myoblast differentiation and promotes early differentiation events [13]. Conversely, Pax7 is down-regulated at the onset of differentiation and its forced expression delays differentiation [9]. We examined regulation of Barx2 and Pax7 expression in primary myoblasts by Wnt signaling. As various Wnts are reported to influence either proliferation or differentiation of myoblasts [18–20, 26], we first examined the effect of Wnt3a on myoblast cultures. Wnt3a increased the number of elongated myocytes and fused cells within 18 hours of treatment in differentiation media (Figure 1A); there was no increase in cell confluence attributable to Wnt3a during this period in either growth or differentiation media as assessed using the Incucyte (Essen) (Supplemental Figure S1).
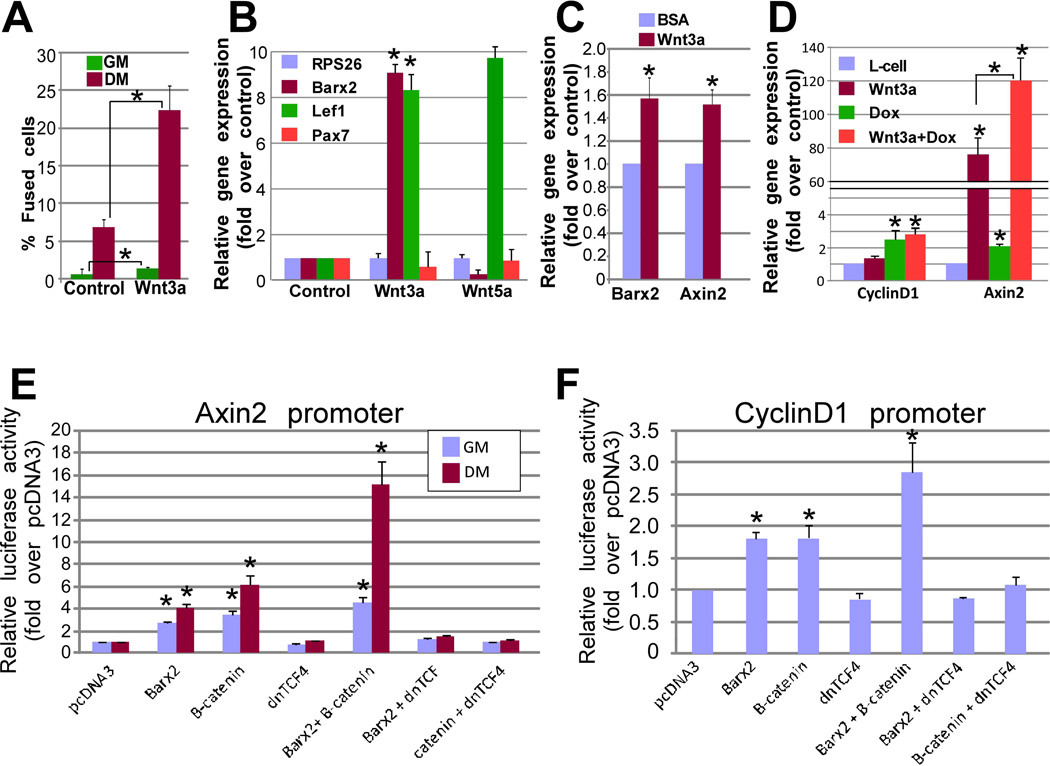
Figure 1.
Wnt3a induces differentiation and Barx2 expression in primary myoblasts, and Barx2 regulates Wnt-responsive target gene promoters. A. Primary myoblasts from TOPEGFP mice were treated with Wnt3a-conditioned media (Wnt3a-CM) or control media (L cell-CM) diluted in growth media (GM) or differentiation media (DM). The proportion of fused or elongated cells (myotubes and myocytes) was measured after 18 hours. B. Expression of Barx2, Pax7 and Lef1 were measured in primary myoblasts treated with Wnt3A-CM, Wnt5a-CM, or control L-cell-CM diluted in GM. Barx2 mRNA was increased by Wnt3a-CM but not Wnt5a-CM. Data was normalized to the housekeeping gene RPS26 and is the average of two experiments performed in duplicate. C. Expression of Barx2 and Axin2 mRNA in TA muscle from wildtype mice after induction of injury using cardiotoxin followed injection of either BSA (control) or 10mg/ml Wnt3a as described in the methods. D. Expression of Axin2 and cyclinD1 in Barx2-TET-ON C2C12 cells after induction with Doxycycline (to induce Barx2 expression) or Wnt3a; data is derived from two experiments performed in duplicate. E. Regulation of the Axin2 promoter by Barx2 and β-catenin in C2C12 cells; activity was assayed 48 hours post-transfection; the data is derived from at least 3 experiments performed in duplicate. F. Regulation of the cyclinD1 promoter by Barx2 and β-catenin in C2C12 cells; activity was assayed 48 hours post-transfection; the data is derived from at least 3 experiments performed in duplicate. For all data, asterisks indicate a significant difference from the control condition as assessed using Students T-Test with * P<0.05 and ** P<0.01.
The level of Barx2 mRNA in these primary myoblasts was increased approximately 8-fold by Wnt3a relative to control treatment, but not by non-canonical ligand Wnt5a. Both ligands induced expression of other known Wnt target genes, but not expression of Pax7. We also examined induction of Barx2 expression in vivo by injecting Wnt3a into tibialis anterior (TA) muscle after cardiotoxin-induced injury (Figure 1C). There were very modest increases in Barx2 and Axin2 mRNA (~1.6 fold) at 3 days post Wnt3a injection. Low level induction may relate to poor retention of the ligand at the injection site. However, taken together our data suggest that Wnt3a can induce Barx2 in myoblasts both in vivo and ex-vivo. We used a series of Barx2 promoter constructs spanning 3kb upstream of the transcription start site [14] to explore the mechanism of induction; however the promoters showed no significant response to Wnt3a (data not shown), suggesting that induction requires elements outside of this region and/or involves post-transcriptional mechanisms.
Barx2 regulates known Wnt target genes
We profiled gene expression in Barx2+/+ and Barx2−/− primary myoblasts using the Wnt Targets and Wnt Signaling Pathway PCR Arrays (Qiagen). 22 genes were downregulated and 11 genes were upregulated more than 1.5-fold in Barx2−/− myoblasts relative to Barx2+/+ myoblasts (Table 1). We also performed RNAseq analysis of Barx2+/+ and Barx2−/− primary myoblasts; this data was highly concordant with the PCR arrays (Supplemental Table S1). Next we assessed a subset of Wnt targets in vivo by RTPCR analysis of whole TA muscles from postnatal day (P)21 Barx2−/− and Barx2+/+ mice (Supplemental Figure S2). CyclinD1 and Wif1 were lower in null muscle, while MMP9 was higher and Axin2 was unchanged; overall there was much greater inter-individual variation in whole muscles than in myoblast cultures. To assess regulation of Wnt targets in a system with lower inherent variability, we used a tetracycline inducible expression system in C2C12 cells (Barx2-TET-ON) (Supplemental Table S2). Several genes that were downregulated in Barx2−/− myoblasts were upregulated by induction of Barx2 expression in C2C12 cells, including Axin2 and cyclin D1 (Figure 1D). Interestingly, the combination of Barx2 and Wnt3a synergistically induced Axin2.
The Axin2 and cyclinD1 promoters are regulated by Barx2
To better understand regulation by Barx2, we used cyclinD1 and Axin2 promoter constructs [27, 28]. Barx2 induced the Axin2 promoter/intron construct ~2-fold in growth conditions and ~4-fold in differentiation conditions. β-catenin activated the promoter 3- and 6-fold in growth and differentiation conditions respectively (Figure 1E). Barx2 and β-catenin synergistically induced the promoter in differentiation conditions but not in growth conditions. Dominant negative (dn)TCF4 blocked activation by both Barx2 and β-catenin (Figure 1E). Both Barx2 and β-catenin induced the cyclinD1 promoter nearly 2-fold and dnTCF4 blocked activation of the cyclinD1 promoter by both factors (Figure 1F). The level of activation of the promoter by β-catenin is similar to that seen previously [27]. In contrast to the Axin2 promoter, fold changes in the cyclinD1 promoter were identical in growth and differentiation conditions and there was no synergy between Barx2 and β-catenin (not shown).
Barx2 activates the Wnt reporter gene TOPflash
To better understand how Barx2 regulates Wnt target promoters, we used the TOPflash reporter gene that contains eight TCF/LEF binding sites [29], reasoning that this would reduce confounding effects due to other regulatory elements. Expression of full length Barx2 (FL Barx2) increased TOPflash activity 4-5 fold in C2C12 myoblasts (see Figure 2A). To delineate the Barx2 domains involved in activation, we prepared deletion constructs (Figure 2A, B). Barx2 contains a centrally located homeodomain (HD) and adjacent 17 amino acid Barx basic region (BBR) that together mediate DNA-binding, flanked by N- and C-terminal domains [30]. Previous work showed that the C-terminal domain mediates transactivation and the N-terminal domain can mediate repression [30, 31]. Barx2 HDBBRC and NHDBBR constructs both activated TOPflash, although the HDBBRC fragment was more potent. Constructs with progressively truncated C-termini showed proportionally reduced activation, suggesting that the activation function may be distributed throughout this region (Figure 2B)
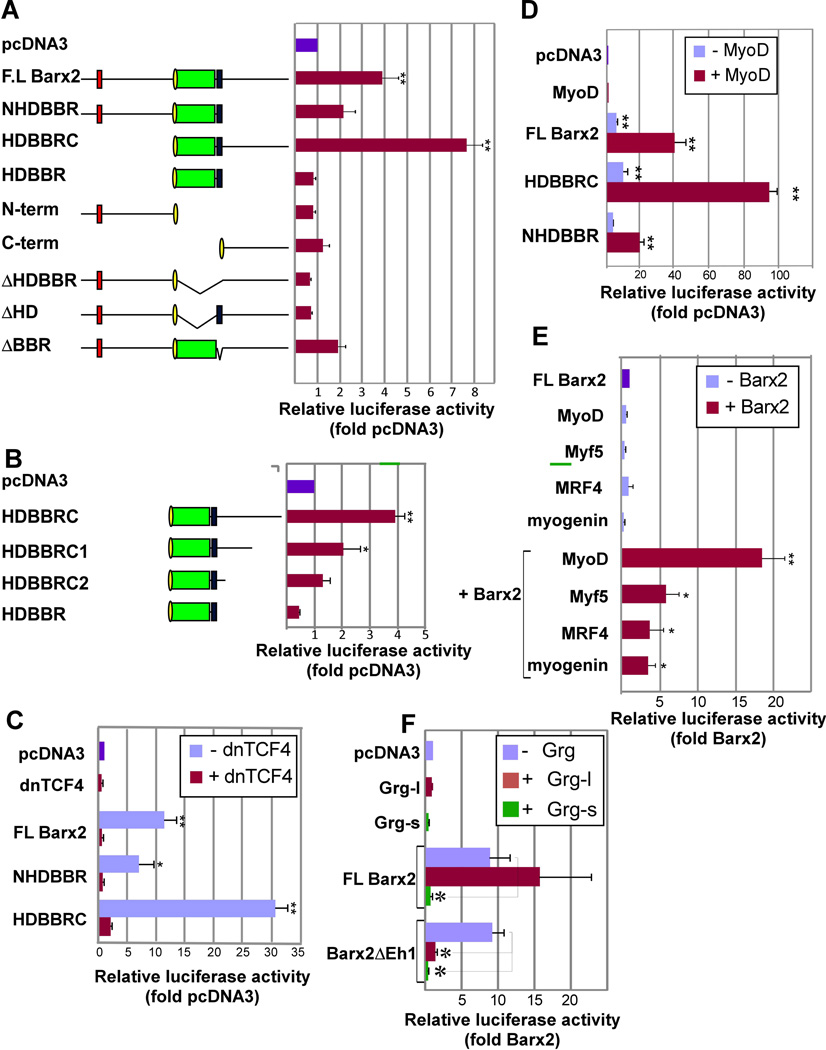
Figure 2.
Barx2 regulates the TOPflash reporter gene alone and synergistically with MRFs in C2C12 cells. A. Analysis of Barx2 deletion constructs shows that the homeodomain and BBR are required for activation of the TOPflash reporter gene. Left: schematic of Barx2 deletion constructs. Green – homeodomain; blue – BBR; yellow – nuclear localization sequence; red- Eh-1 motif. Right: TOPflash luciferase activity after cotransfection of Barx2 constructs in C2C12 cells. B. Analysis of Barx2 deletion constructs shows that the Barx2 activation function is distributed throughout the C-terminal domain. Left: schematic of Barx2 deletion constructs. Right: TOPflash luciferase activity. C. Co-transfection of Barx2 constructs with dnTCF4 shows that activation of the TOPflash reporter by Barx2 is inhibited by dnTCF4. D. Cotransfection of Barx2, constructs with MyoD shows that these factors synergize in regulation of TOPflash activity. E. Cotransfection of FL-Barx2 with MyoD, Myf5, MRF4, or myogenin shows that Barx2 can synergize with all four MRFs. F. Cotransfection with FL-Barx2 or the variant lacking the Eh-1 motif (Barx2ΔEh1), with or without full length (Grg-l) or truncated (Grg-s) Groucho isoforms in C2C12 cells shows that Groucho is involved in regulation of TOPflash; Grg-l blocks activation by Barx2ΔEh1 but not wildtype Barx2, while Grg-s blocks activation by both wildtype Barx2 and Barx2ΔEh1. Luiciferase activity was assayed 48 hours post-transfection; all data were normalized to a Renilla luciferase internal control and then to pcDNA3 transfection. Data were collected in triplicate and at least 2 assays were performed for each condition with similar results. In panels A, B, C, D, and F, ** and * indicate changes relative to the control (pcDNA) that are significant at P<0.01 and P<0.05 respectively. In panel E, ** and * indicate changes relative to the control (Barx2) that are significant at P<0.01 and P<0.05 respectively.
Deletion of the Barx2 homeodomain (ΔHDBBR and ΔHD) prevented activation of TOPflash, while deletion of the BBR alone reduced activation (Figure 2A). Consistent with a critical role for these domains in TOPflash activation, Barx1, which shows 87% homology with Barx2 within the HDBBR, also activated TOPflash (Supplemental Figure S3). We also tested Msx factors, which have a related homeodomain but no BBR domain: Msx2 (but not Msx1) modestly activated TOPflash (Supplemental Figure S3); Msx2 was previously shown to induce β-catenin nuclear localization, which may contribute to its effect on TOPflash [32].
The TOPflash promoter contains no identifiable homeodomain binding motifs suggesting that the response to Barx2 is mediated by TCF/LEF sites. In support of this idea, Barx2 did not activate the FOPflash promoter, which contains mutated TCF/LEF motifs (Supplemental Figure S4A). Moreover, when Barx2 constructs were cotransfected with dominant-negative TCF4 (dnTCF4) that cannot recruit β-catenin, Barx2 failed to activate (Figure 2C), indicating a requirement for β-catenin in Barx2-mediated activation.
Barx2 activates TOPflash synergistically with MyoD
Barx2 and MyoD can physically interact [13], as can MyoD andβ-catenin [33]. To examine whether Barx2 and MyoD might cooperate in regulation of TOPflash, we cotransfected Barx2 constructs witha MyoD expression plasmid. The Barx2/MyoD combination activated TOPflash 8-15 fold more than Barx2 alone (Figure 2D), whereas MyoD alone did not activate TOPFlash (Figure 2D).The NHDBBR and HDBBRC constructs also synergized with MyoD, although HDBBRC was more potent (Figure 2D). Barx2 could also synergize with Myf5, MRF4 and myogenin, but less so than with MyoD (Figure 2E). TOPflash activation is unlikely to be due to a differentiation-promoting effect of Barx2 and MyoD, because cells transfected with TOPFlash alone did not show significant promoter induction even after 48hrs in differentiation media (Supplemental Figure S4B). Moreover, both Barx2 and the Barx2/MyoD combination induced TOPflash activity in COS7 cells, which are unable to undergo myogenic conversion (Supplemental Figure S4C).
Groucho/TLE1 proteins are involved in regulation of TOPflash by Barx2
TLE family co-repressors interact with TCF/LEFs and mediate repression of Wnt targets [34]. Barx2 also interacts with Groucho/TLE1, nominally via an Eh1 motif at the Barx2 N-terminus[31]. We hypothesized that excess Groucho/TLE1 might attenuate activation of TOPflash by Barx2, and that this may be dependent on the Barx2Eh1 motif. However, co-expression of full-length Groucho/TLE1 (Grg-l) had no effect on the activation mediated by full-length Barx2 (Figure 2F). More unexpectedly, Grg-l inhibited activation by a variant of Barx2 in which the Eh1 motif was mutated (Barx2ΔEh1) (Figure 2F).These data suggest that the Barx2 Eh1 motif may in fact block the ability of Grg-l to act as a corepressor for Barx2 in the context of TOPflash regulation.
We also co-expressed Barx2variants with a naturally occurring Groucho/TLE1 splice variant (Grg-s) that is known to antagonize β-catenin activity [35]. Grg-s attenuated the ability of both full-length Barx2 and Barx2ΔEh1 to activate TOPflash, suggesting different roles for the two Grg splice variants in regulation by Barx2.
Barx2 interacts physically with β-catenin and TCF family members
To determine whether Barx2 might form a complex with core Wnt effectors, we performed co-immunoprecipitation using myc-epitope tagged Barx2 in COS7 cells. Expressed Barx2 was efficiently co-immunoprecipitated with endogenous β-catenin (Figure 3A). Co-immunoprecipitation also occurred when lysates were sonicated and treated with ethidium bromide to disrupt protein-DNA interactions, suggesting that the interaction does not require DNA (Supplemental Figure S5A). We also demonstrated interaction of endogenous Barx2 and β-catenin in primary myoblasts (Figure 3B) and embryonic limb mesenchymal cells (Supplemental Figure S5B).
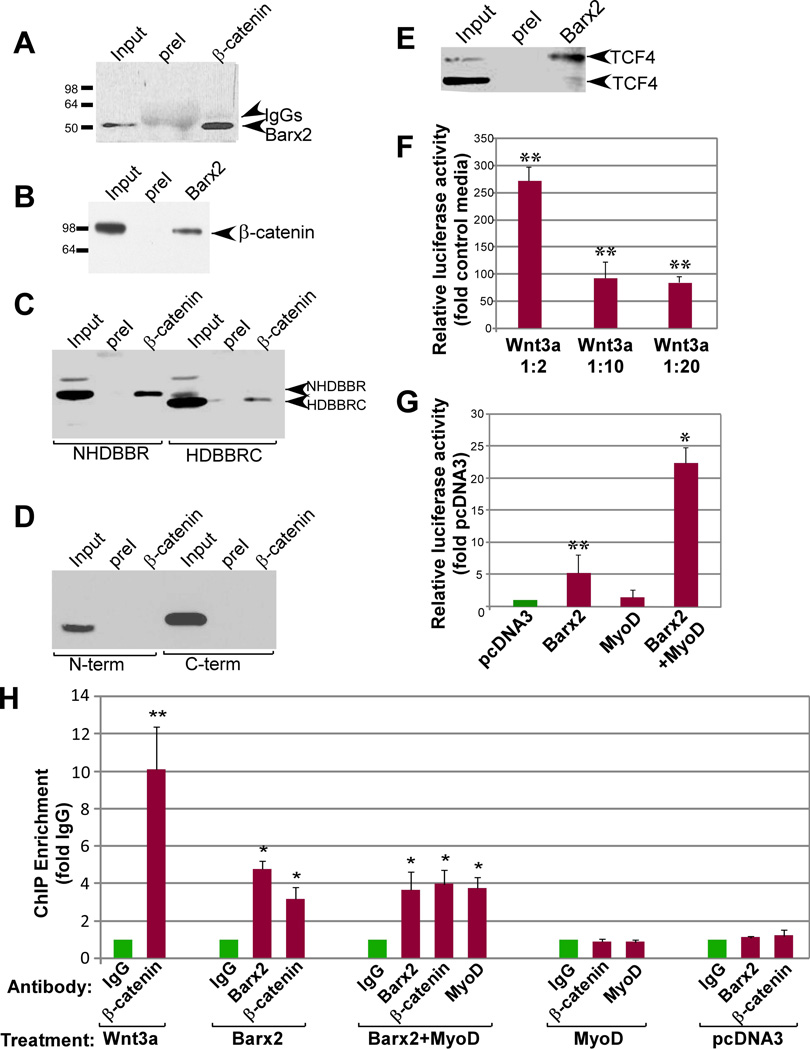
Figure 3.
Barx2 interacts with β-catenin and TCF proteins in vitro and is recruited to the TOPflash promoter in cells. A. Myc-epitope tagged FL-Barx2 was expressed in COS7 cells and immunoprecipitated using polyclonal antibodies to endogenous β-catenin. After blotting the SDS-PAGE resolved proteins, the membrane was probed with monoclonal antibodies to the myc-tag. Barx2 was robustly co-immunoprecipitated with β-catenin (arrow at right). B. Endogenous Barx2 was immunoprecipitated from primary mouse myoblasts using polyclonal antibodies to Barx2. SDS-PAGE resolved proteins were blotted and probed with an antibody to β-catenin. β-catenin was co-immunoprecipitated by the Barx2 antibody (arrow at right). C. The NHDBBR and HDBBR domains of Barx2 were expressed in COS7 cells, immunoprecipitated using polyclonal antibodies to β-catenin and blotted with antibodies to the myc-tag. Both domains were co-immunoprecipitated with β-catenin (arrows at right). D. The isolated N-term and C-term domains of Barx2 were expressed in COS7 cells, immunoprecipitated using polyclonal antibodies to β-catenin and blotted with antibodies to the myc-tag. Neither domain was co-immunoprecipitated with β-catenin. E. Myc-epitope tagged TCF4 and Barx2 were expressed in COS7 cells and immunoprecipitated using polyclonal antibodies to Barx2. SDS-PAGE resolved proteins were blotted and probed with monoclonal antibodies to the myc-tag. Two TCF4 species were co-immunoprecipitated with Barx2 (arrows at right). All immunoprecipitation assays were repeated at least twice. F, G. A stable C2C12 cell line carrying an integrated TOPflash plasmid shows induction of luciferase after treatment with Wnt3a (F) or transfection of Barx2 and MyoD (G). H. ChIP was performed on chromatin from the stable TOPflash C2C12 cell line after the indicated treatments. ChIP using antibodies to β-catenin showed that the TOPflash promoter recruited β-catenin after treatment with Wnt3a, transfection with Barx2 alone, or combined transfection of Barx2 and MyoD, but not after transfection with pcDNA3 or MyoD alone. ChIP using antibodies to Barx2 showed that the TOPflash promoter recruited Barx2 after transfection with Barx2 alone or the combination of Barx2 and MyoD but not after transfection with pcDNA3 or MyoD alone. ChIP using antibodies to MyoD shows enrichment of TOPflash promoter DNA after transfection with Barx2 and MyoD, but not after transfection with pcDNA3 or with MyoD alone. Data are PCR amplification values for the TOPflash-promoter normalized to amplification values for a control non-target locus, with enrichment values for each antibody subsequently normalized to the mock ChIP with preimmune IgG. The data are the mean of between 3 and 8 independent transfection experiments per condition. In panels F-H, asterisks indicate values significantly different from the control condition with either P<0.05 (*) or P<0.01 (**).
To delineate interaction domains, we expressed the Barx2 NHDBBR, HDBBRC, N-term, and C-term constructs in COS7 cells. NHDBBR and HDBBRC proteins co-immunoprecipitated with endogenous β-catenin (Figure 3C) while the N-term and C-term proteins lacking homeodomain and BBR (HDBBR) were not precipitated (Figure 3D), showing that the HDBBR is necessary for interaction. We could not test the sufficiency of the HDBBR because it showed poor stability in cells.
Barx2 antibodies also co-immunoprecipitated heterologously-expressed TCF4 in COS7 cells. Two TCF4 species of ~60 and 80kDa were observed on immunoblots; the larger variant is consistent with a post-translationally modified form [36] that is reported to confer the majority of transcriptional activity in other cells [37]. Barx2 preferentially co-immunoprecipitated the larger molecular weight TCF4 form (Figure 3E).
To determine whether these interactions were direct, we also attempted co-immunoprecipitation using proteins generated by in vitro transcription/translation. While TCF4 co-immunoprecipitated with β-catenin, we were unable to co-immunoprecipitate either TCF4 or β-catenin with Barx2 (not shown). It is possible that additional proteins or post-translational modifications are required to mediate or stabilize the interaction. In support of the latter idea, a variant form of Barx2 with disrupted phosphorylation sites within the homeodomain was unable to activate TOPflash (not shown).
Barx2 binds to the TOPflash promoter and promotes recruitment of β-catenin
To test the hypothesis that Barx2 is recruited to the TOPflash promoter via interactions with TCF/LEF and/orβ-catenin, we performed ChIP using a stable C2C12 cell line that carries an integrated TOPflash promoter/luciferase reporter. This line showed induction of luciferase activity by Wnt3a (Figure 3F),and by Barx2 and MyoD (Figure 3G). ChIP with β-catenin antibodies in Wnt3a-treated cells produced ~10-fold enrichment of TOPflash promoter DNA, indicating recruitment of β-catenin (Figure 3H). In cells transfected with Barx2 or the combination of Barx2 and MyoD (Barx2/MyoD), ChIP with Barx2 antibodies produced 3-4 fold enrichment of TOPflash promoter DNA (Figure 3H), suggesting that Barx2 binds the promoter. Moreover, in cells transfected with Barx2 or Barx2/MyoD, ChIP with β-catenin antibodies produced 3-4 fold enrichment of TOPflash promoter DNA (Figure 3H) indicating recruitment of β-catenin. No β-catenin recruitment was seen with either Barx2 or β-catenin antibodies when cells were transfected with empty vector.
We also examined recruitment of MyoD to TOPflash after transfection of C2C12 cells with pcDNA3, MyoD, or Barx2/MyoD. ChIP with antibodies to MyoD showed that MyoD was only recruited to the TOPflash promoter after Barx2/MyoD co-transfection, and not when MyoD was expressed alone. Transfection of MyoD alone was also unable to induce recruitment of β-catenin to TOPflash (Figure 3H). Efficacy of the MyoD antibody in ChIP was demonstrated by the recruitment of MyoD to an E-box-containing promoter (desmin) (Supplemental Figure S6). Overall, these data suggest that Barx2 recruits both MyoD and β-catenin to the TOPflash promoter.
Pax7 antagonises Barx2 and β-catenin in regulation of TOPflash activity
Pax7 inhibits myoblast differentiation while Barx2 promotes differentiation [7, 9, 13]. To examine whether these opposing effects of Barx2 and Pax7 may be related to differential effects on Wnt target regulation, we tested the ability of Pax7 to modulate TOPflash activity. We used the Pax7D cDNA, which is the most abundant isoform in skeletal muscle. In contrast to the effect of Barx2, Pax7 repressed basal TOPflash activity by approximately 2-fold (Figure 4A). Moreover, co-transfection of Pax7 blocked activation of TOPflash by Barx2 (Figure 4A) and by the combination of Barx2 and MyoD (not shown).Conversely, co-transfection of siRNA directed against Pax7 (leading to ~50% reduction in Pax7 mRNA) increased activation of the TOPflash promoter by Barx2 and MyoD (Figure 4B).
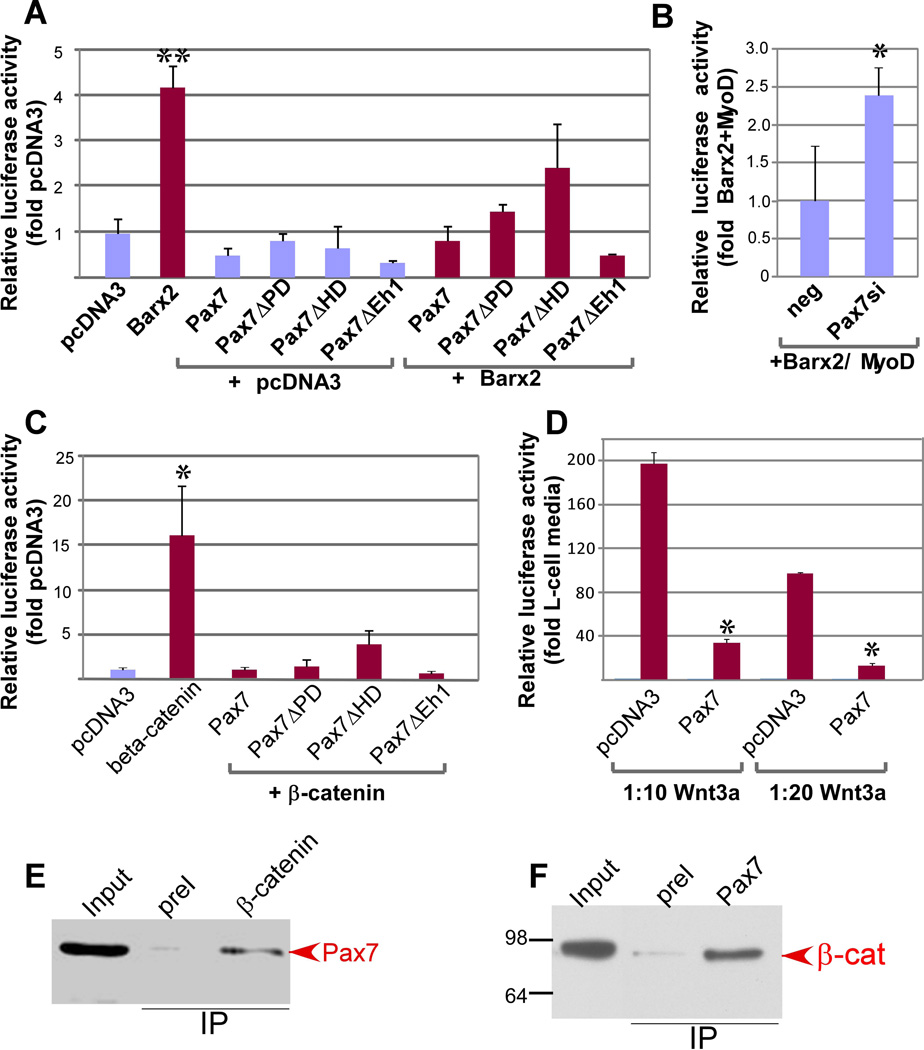
Figure 4.
Pax7 antagonizes the activating effect of Barx2 and β-catenin on TOPflash and interacts with β-catenin. A. Analysis of Pax7 deletion constructs shows that the Pax7 HD is important for repression of TOPflash activity. Luciferase activity was assayed after transfection with FL-Barx2, FL-Pax7, or Pax7 deletion constructs (Pax7ΔPD, Pax7ΔHD, and Pax7ΔEh). Cells were also cotransfected with FL-Barx2 in combination with each of the Pax7 constructs showing that Pax7 represses Barx2-mediated activation. B. Knock-down of Pax7 increases the ability of Barx2 and MyoD to activate TOPflash activity. Luciferase activity was assayed after co-transfection with FL-Barx2 and MyoD and either negative control siRNA or the Pax7siRNA. Data is normalized to the activation generated by Barx2 and MyoD with negative control siRNA. C. TOPflash activity was assayed after transfection of a constitutively active β-catenin construct alone or in combination with various Pax7 deletion constructs showing that Pax7 represses β-catenin-mediated activation. D. TOPflash activity was assayed after transfection of Pax7 and treatment with diluted Wnt3a-CM showing that Pax7 represses Wnt-mediated activation. All luciferase data were collected 48 hours post-transfection, normalized to a Renilla luciferase internal control and then to pcDNA3 transfection. Data were collected in triplicate and at least 3 assays were performed for each condition with similar results. In panels A, C and D, asterisks indicate changes relative to the control condition (pcDNA in A, C, and D and Barx2+MyoD in B) that are significant at P<0.01 (**) or P<0.05 (*). E. Myc-epitope tagged Pax7 protein was expressed in COS7 cells and complexes were immunoprecipitated with antibodies to endogenous β-catenin. SDS-PAGE resolved proteins were blotted and probed with antibodies to the myc-tag showing that Pax7 was immunoprecipitated with β-catenin. F. Endogenous Pax7 was immunoprecipitated from primary mouse myoblasts using polyclonal antibodies to Pax7. SDS-PAGE resolved proteins were blotted and probed with an antibody to β-catenin. β-catenin was co-immunoprecipitated by the Pax7 antibody (arrow at right).
To delineate the functional domains of Pax7, we generated mutant expression constructs: Pax7ΔPD lacked the N-terminal paired domain, Pax7ΔHD lacked the homeodomain, and Pax7ΔEh1 had mutations within the Eh1 motif. These constructs were cotransfected with Barx2 and TOPflash in C2C12 cells. Mutation of the Pax7 Eh1 domain had no effect on the ability of Pax7 to block activation by Barx2 (Figure 4A); deletion of the paired domain also had no effect. However, deletion of the homeodomain (Pax7ΔHD) moderately impaired the ability of Pax7 to inhibit Barx2-mediated activation of TOPflash.
To assess whether Pax7 is a specific antagonist of Barx2-mediated TOPflash activation, we examined its ability to antagonize activation by either constitutively active β-catenin or Wnt3a ligand. We titrated both the β-catenin plasmid and Wnt3a to induce modest activations of TOPflash. Pax7 abolished activation by β-catenin (Figure 4B), and greatly reduced activation by Wnt3a (Figure 4C); the Pax7ΔHD variant was less repressive than wildtype Pax7 in both contexts. Pax7 could also attenuate but not abolish activation of the Axin2 and CyclinD1 promoters by β-catenin (not shown). Overall, these data indicate a previously unreported role for Pax7 as a repressor of canonical Wnt signaling.
To determine whether the effects of Pax7 may be mediated by interaction with β-catenin, myc-epitope tagged Pax7 was expressed in COS7 cells and complexes immunoprecipitated with antibodies to endogenous β-catenin. Immunoblotting showed that Pax7 co-immunoprecipitated with β-catenin (Figure 4D). We were also able to demonstrate interaction of endogenous Pax7 and β-catenin in primary myoblasts (Figure 4E). These data support the idea that Pax7 acts as an antagonist of the core Wnt effector complex.
Barx2 regulates genes within both the Wnt and Notch signaling pathways
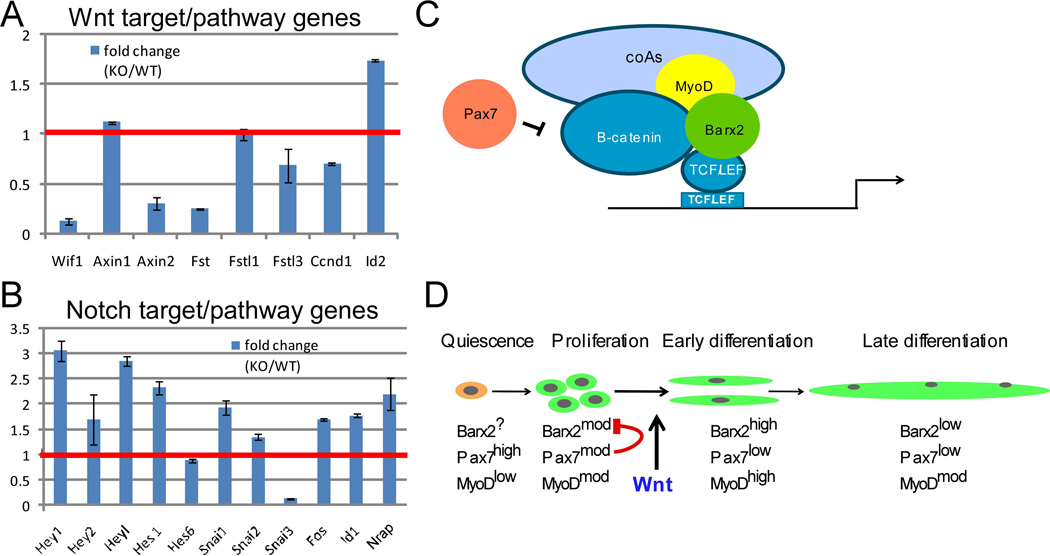
To obtain further insight into regulation of signalling pathways by Barx2, we generated RNA-seq data from Barx2−/− and Barx2+/+ primary myoblasts. We mined this data for genes involved in the Wnt and Notch signalling pathways. As discussed above, multiple Wnt pathway genes were downregulated in Barx2−/− myoblasts (Supplemental Table S1 and Figure 5A). In contrast, several Notch target and pathway genes were upregulated in Barx2−/− myoblasts including Notch3, Dll1, Hes1, Hey1, Heyl and Snai1 (Figure 5B). The mechanism underlying the misregulation of Notch target and pathway genes is as yet unknown; Barx2 had no effect on the activity of Notch (RBPj) promoter-reporter constructs (not shown). Overall, our data suggest a model in which Barx2 inhibits Notch signalling whilst promoting Wnt signalling in myoblasts.
Figure 5.
A, B. Relative expression of a representative set of Wnt and Notch target and pathway genes in Barx2+/+ (WT) and Barx2−/− (KO) primary myoblasts obtained from RNA-seq analysis. The red line denotes the level in Barx2+/+ (WT) myoblasts which is set to 1. All gene expression changes greater than 1.5 fold are significant at P<0.05 (see also Supplemental Table S1). C. Model of the transcriptional complexes identified in this study. Barx2 interacts with β-catenin and TCF/LEF factors as well as MyoD. This complex recruits co-activators that promote transcription via TCF/LEF sites. Pax7 inhibits the function of this complex, likely via interaction with β-catenin which could lead to sequestering of β-catenin, and/or failure to dismiss co-repressors and recruit coactivators. D. Barx2 and Pax7 may control the switch from proliferation to differentiation via antagonistic regulation of Wnt signaling. In proliferating myoblasts both Barx2 and Pax7 are expressed at moderate levels, thus Pax7 may inhibit the ability of Barx2 to activate via β-catenin-TCF/LEF complexes. After induction of differentiation by canonical Wnt signals, Barx2 is upregulated and Pax7 is downregulated. This may allow Barx2 to activate TCF/LEF target genes associated with early differentiation events. Later in mature myofibers, expression of both Barx2 and Pax7 is low or absent.
Discussion
Muscle growth and repair is regulated by a variety of cell-intrinsic regulatory molecules such as homeobox and bHLH transcription factors, as well as extrinsic signals from soluble factors including Wnts. The functional integration of these intrinsic and extrinsic regulators is not well understood. Here we have demonstrated a novel pathway for translation of Wnt signals into gene expression changes in myoblasts that involves homeobox proteins and MRFs acting in concert with core Wnt effector proteins (Figure 5C).
Antagonism between Barx2 and Pax7 in regulation of Wnt signaling
Pax7 is expressed in quiescent and proliferating satellite cells/myoblasts, and is downregulated when cells begin to differentiate [6]. Barx2 is also expressed in satellite cells nd myoblasts [10], but in contrast to Pax7, Barx2 is induced during differentiation and promotes pro-differentiation events such as cell spreading and migration [13]. We propose a model in which high levels of Pax7 counteract the pro-differentiation effects of Barx2 in proliferating cells; subsequent downregulation of Pax7 in cells committed to differentiation allows Barx2 to activate target genes in cooperation with β-catenin and MRFs (Figure 5D). The latter would be favoured by the Wnt-driven increase in Barx2, MyoD and stable β-catenin. Thus antagonism between Pax7 and Barx2 with respect to Wnt signalling may be a key component of the Wnt-induced switch from proliferation to differentiation [21]. Interestingly, we recently observed interaction between Barx2 and Pax6 in regulation of a Wnt reporter in epithelial cells, suggesting that Barx-Pax interaction is a broadly applicable paradigm.
This is the first report that MRFs can promote transcription from TCF/LEF sites. Although interaction of β-catenin and MyoD was previously reported, this was only shown to enhance transcription via E-box motifs [33]. Interestingly, in our study MRFs only activated TOPflash in concert with Barx2 and not alone, and MyoD was not recruited to the TOPflash promoter in the absence of Barx2. Intriguingly, Barx2-MRF synergy did not occur on the Axin2 or cyclinD1 promoters (not shown); the reason for this difference is currently unknown but may relate to the density of TCF/LEF motifs. Similarly, Pax7 only partially inhibited Barx2 and β-catenin mediated activation of the Axin2 and cyclinD1 promoters (not shown), suggesting that the function of Pax7 is also influenced by the density of TCF/LEF sites. Pax7 is known to destabilize MyoD protein [5], and this may in part explain the ability of Pax7 to inhibit Barx2/MyoD-mediated activation of TOPflash. However, the ability of Pax7 to block activation by β-catenin in both myogenic and non-myogenic cells (Supplemental data Figure S4)indicates a role within the Wnt effector complex that is independent of MRFs.
Homeodomain-β-catenin interactions
The Barx2 homeodomain was necessary for interaction with β-catenin, and activation of TOPflash required the homeodomain in conjunction with either an N-terminal or C-terminal activation domain. Repression of TOPflash by Pax7 was also partly dependent on the homeodomain. A handful of other homeodomain proteins have been reported to engage in functional interactions with β-catenin or TCF/LEF proteins [38–40]. In particular, Pitx2 interacts with β-catenin via its homeodomain [41], raising the interesting possibility that Barx2 and Pitx2 could compete for this interaction. The role of post-translational modification in the interaction of Barx2 with β-catenin also requires further investigation, particularly given that mutation of phosphorylation sites within the Barx2 homeodomain blocked activation of TOPflash (not shown).
Mechanisms of activation and repression byBarx2
Activation of TOPflash by the Barx2 C-terminal domain (plus HDBBR) is consistent with our previous work [30]. However, the Barx2 N-terminal domain also activated TOPflash, whereas previously a repression function had been shown for this domain in association with TLE1 and NCOR[30, 31]. Groucho/TLE1 (Grg-l) was predicted to inhibit Barx2-mediated activation of TOPflash; however, unexpectedly it only did so in the absence of the Barx2 Eh1 motif. One model to explain this result is that Grg-l interacts with Barx2 downstream of the Eh1 motif, but is inhibited by other as yet unknown factors that bind to the Eh1. This is consistent with reports that Barx2 recruits other corepressors downstream of the Eh1 motif [31], but also suggests novel functions for the Barx2 Eh1. Groucho/TLE1 short (Grg-s, also termed AES or GRG5) is a natural splice variant that lacks the C-terminal WD40 protein-interaction domain[42]. Grg-s inhibited Barx2-mediated activation regardless of the presence or absence of the Barx2 Eh1, suggesting that Grg-s also interacts with Barx2 downstream of the Eh1, but that it is unaffected by factors that bind to the Barx2 Eh1. Our data complements previous reports of divergent Groucho activities in association with the TLX-1 homeodomain protein [43] and estrogen-related receptor gamma [44]. The transducin-beta like factors Tbl1 and TblR1 also interact with Barx2 [31], and are known to promote recruitment of β-catenin to TOPflash [45]; the role of TLE and other transducin-like factors in regulation of Wnt-target genes by Barx2 requires further investigation.
Altered Wnt and Notch signalling in the muscle phenotype of Barx2 null mice
Barx2 null mice show defective muscle growth and repair [10, 46]. Many of the Wnt and Notch target genes misregulated in Barx2 null myoblasts have known roles in myogenesis that could underlie aspects of this phenotype. For example, Dlk1, which is both a Wnt target gene and a Notch ligand, controls postnatal muscle growth [47] and its ablation in the myogenic lineage results in postnatal growth retardation, reduced body and muscle weight, and impaired regeneration [48]; a phenotype that resembles that of Barx2 null mice. Fst is another Wnt target critical for muscle growth and regeneration, [49–53]. Notch3, which was elevated in Barx2−/− myoblasts is suggested to be an inhibitor of satellite cell proliferation and self-renewal via inhibition of Notch1 [54]. Notch target Snail1, which was also elevated in Barx2−/− myoblasts, has been reported to inhibit binding of MyoD to target genes and thus myogenesis. Overall, the spectrum of misregulated genes in Barx2−/− myoblasts is consistent with their reduced proliferation and differentiation in culture and the reduced capacity of Barx2−/− mice to grow, maintain, and repair muscle [55]; although further work is required to assess the contribution of each of these genes to the null phenotype.
Although the expression of many Wnt and Notch target genes was significantly altered in cultured myoblasts, we were only able to observe changes in a subset of genes in whole muscle from postnatal (P21) mice. This may relate to the fact that Barx2 is only expressed in muscle progenitors and not the myofibres that make up the bulk of the muscle. Consistent with this idea, cyclinD1, like Barx2, is primarily expressed in proliferating myoblasts and cyclinD1 was consistently downregulated in Barx2 null muscle. Variability of gene expression in P21 muscle may also reflect the heterogeneity of developmental states of muscle progenitor cells in vivo, relative to cultures where they are deprived of developmental signals. For example, Axin2 mRNA has a short half-life [56] and shows ultraradian oscillation in somitogenesis [57]. Similar oscillations may occur during muscle growth as progenitors are repeatedly activated and progress through proliferation and differentiation phases and it is unlikely that all parts of the muscle are synchronous in these waves. Thus although Axin2 showed very high variance in whole muscle, misregulation of Axin2 in cultured cells using two different perturbation models (Barx2 knockout and over-expression) as well as regulation at the level of promoter activity, strongly suggests that Axin2 is a Barx2 target gene. The complex mechanisms of Axin2 regulation are a subject of ongoing study.
Barx2, Pax7, Wnt and Notch in the proliferation-differentiation ‘switch’
Overall, our studies show that Barx2 is a target of canonical Wnt signaling, interacts with β-catenin/TCF, and regulates components and targets of both the Wnt and Notch signalling cascades. Moreover, Pax7 is a functional antagonist of Barx2 with respect to Wnt signaling. Pax7 is also known to be a direct target of Notch signaling and may promote myoblast proliferation and inhibit differentiation downstream of Notch signals [5, 7–9]. It was previously postulated that a temporal switch from Notch to Wnt signalling may control the switch from myoblast proliferation to differentiation [21]. We now propose that functional antagonism between Barx2 and Pax7 may be an intermediate control point in this Notch-Wnt signalling switch, the precise mechanisms of which will be explicated further in future studies.
Materials and Methods
Animals
Barx2-LacZ knockin mice obtained from Dr. Geoff Rosenfeld were maintained and genotyped according to [31].
Plasmids and Cell lines
The Axin2 promoter/intron reporter construct and the TOPflash and FOPflash reporter plasmids were obtained from Addgene (plasmids 21275, 12456 and 12457). The cyclinD1 promoter was a kind gift of Dr. Johan Auwerx. Full-length Barx2 cDNA isolated from E14 mouse tongue was cloned into the pcDNA3 vector in frame with an N-terminal myc-tag. Barx2 deletion constructs were made by fusion of PCR fragments using ligation or the Infusion kit (Clontech). The Barx2 Eh1 motif (FMI, amino acids 27-29) was replaced with alanines (AAA) using the Site Directed Mutagenesis Kit (Clontech).
Pax7D, Myf5 and MRF4 cDNAs were isolated from postnatal mouse muscle and cloned into the pcDNA3 vector in frame with the myc-tag. Pax7D deletions were made with the Infusion kit (Clontech). MyoD/pcDNA3 and myogenin/pcDNA were described previously [58]. cDNAs encoding constitutively active β-catenin and dominant-negative TCF4 (dnTCF4) were obtained from Victor Korinek and subcloned into the pcDNA3 vector. All constructs were shown to express by immunoblot analysis. Sequences of PCR primers used for cloning are in Supplementary table S3.
All cell lines were obtained from ATCC. COS7 cells were grown in DMEM supplemented with 10% FBS; C2C12 cells were grown in DMEM with 20% FBS. L-cells stably expressing Wnt5a and Wnt3a (developed by R. Nusse) were cultured for 4 days in DMEM with 10% FBS and appropriate antibiotics and conditioned media prepared as recommended by ATCC. Conditioned media was filtered and diluted with appropriate media before adding to cultures.
Primary myoblast cultures
Satellite cells were isolated as previously described [59] from postnatal day 4-5 TOPEGFP or Barx2-LacZ knockin mice, and cultured as myoblasts on collagen-coated plates in DMEM/F10 with 20% FBS and 5ng/ml bFGF. Differentiation was induced by switching to DMEM with 2% horse serum. Primary myoblast transfections were performed using a Nucleofector (Amaxa).
Barx2-TET-ON C2C12 stable lines
C2C12 lines were generated that carry the TET-activator plasmid (Clontech) and screened for maintenance of differentiation capacity. The Barx2 cDNA was cloned into the TET-ON vector and introduced into C2C12 lines; clones were again screened for differentiation capacity. Two lines were selected that differentiated efficiently and showed induction of Barx2 protein by Doxycycline (Dox). These lines were used for RNA analysis after Dox and Wnt3a treatment; data from one line is shown but results were substantially similar in the two lines.
Co-immunoprecipitation
Co-immunoprecipitations (coIP) were performed essentially as described in[60, 61] using either recombinant proteins expressed in COS7 cells or endogenous proteins in mouse embryonic limb cells or primary myoblasts as detailed in Supplemental Methods.
Chromatin Immunoprecipitation (ChIP)
We generated a stable C2C12 cell line carrying the TOPflash promoter/luciferase construct that was robustly activated by Wnt3a and by Barx2/MyoD transfection. ChIP was performed using a modified MicroChIP protocol [62] and analysed by genomic qPCR with primers that amplify the TOPflash promoter, the desmin proximal promoter, or a control non-target locus (β2-microgloubulin). Details are provided in Supplemental Methods and primer sequences are in Supplementary Table 1. Between 3 and 8 independent transfection/ChIP experiments were performed per condition and results averaged; significance was assessed using Student’s T-test.
Transfections and promoter assays in C2C12 and COS7 cells
C2C12 or COS7 cells were seeded in 24-well plates at 2 × 104 cells/well and co-transfected with TOPflash and combinations of expression plasmids (0.5µg for each plasmid; 50 ng for β-catenin). The Renilla Luciferase reporter pRLCMV (Promega) was included as an internal reference. Luciferase assays were performed 48 hours after transfection using the Promega Dual luciferase assay kit. All transfections were performed in duplicate and repeated 2–8 times; significance was assessed using Student’s T-test. Pax7 siRNA was designed based on [5], adapted for double stranded siRNAs. Pax7 or universal negative control siRNAs (IDT) were cotransfected at 30nM with luciferase reporter and effector plasmids in C2C12 cells as described above and results assayed after 48 hours. Efficacy of siRNA-mediated Pax7 knockdown was assessed in separate transfection experiments as about 50% by RT-PCR and from ~90% by immunoblot using goat anti-Pax7 antibody (Santa Cruz Biotechnologies).
Barx2 loss- and gain-of-function in primary myoblasts
Satellite cells were isolated from Barx2−/− and Barx2+/+ mouse pups (4-5 days postnatal) and cultured as previously described [59]. Cells from at least 5 pups per genotype (approximately equal cell contributions from each) were pooled. For gain-of-function studies, wildtype primary myoblasts were transfected with Barx2-pcDNA3 or empty pcDNA3 plasmid using the AmaxaNucleofector. Cells were harvested after 48 hours for RNA preparation.
Quantitative RT-PCRs (qRT PCRs) and RT Profiler PCR Arrays
RNA was prepared from cells or from whole muscle tissue (after grinding with plastic pestles), using Trizol (Invitrogen); after DNAse treatment, cDNA was synthesized using MMuLV reverse transcriptase and random primers (NEB). Real time RT-PCR was performed using a Corbett Rotogene and GoTaq SYBR green (Promega). Primers are listed in Supplementary Table S3. For PCR arrays, RNA was prepared from pooled primary myoblast cultures using the RNeasyPlus Kit (Qiagen). cDNA was synthesized using RT2 First Strand Kit (SABiosciences) and applied to RT2 Profiler™ Mouse WNT Signaling Targets (PAMM-243A) and Wnt Signaling Pathway (PAMM-043A) arrays (these arrays share some genes). Analysis used an Applied Biosystems 7300 Real-Time PCR System and SABiosciences online software to calculate fold-change and p-value (each sample was run three times for each array).
Supplementary Material
Acknowledgements
This work was supported by National Institutes of Health Grant 5R01AR053163 (to H.P.M. and R.M.) and by a grant from the Association Francaise contre les Myopathies (to H.P.M.). The study was also supported by an Australian Research Council Fellowship FT110100573 (to R.M.), and a Flinders Foundation grant (to R.M.).
Footnotes
Author contribution summary: LZ, JH and AG jointly performed all cloning and gene/reporter expression analyses, TN generated and analysed stable cell lines, MD, RY and RE generated and analysed RNA-seq data, CL performed bioinformatic analysis, RM and HM jointly conceived and directed the project and also performed in vivo analyses.
References
- 1.Lepper C, Partridge TA, Fan CM. An absolute requirement for Pax7-positive satellite cells in acute injury-induced skeletal muscle regeneration. Development. 2011;138(17):3639–3646. doi: 10.1242/dev.067595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McCarthy JJ, et al. Effective fiber hypertrophy in satellite cell-depleted skeletal muscle. Development. 2011;138(17):3657–3666. doi: 10.1242/dev.068858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Murphy MM, et al. Satellite cells, connective tissue fibroblasts and their interactions are crucial for muscle regeneration. Development. 2011;138(17):3625–3637. doi: 10.1242/dev.064162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sambasivan R, et al. Pax7-expressing satellite cells are indispensable for adult skeletal muscle regeneration. Development. 2011;138(17):3647–3656. doi: 10.1242/dev.067587. [DOI] [PubMed] [Google Scholar]
- 5.Olguin HC, Olwin BB. Pax-7 up-regulation inhibits myogenesis and cell cycle progression in satellite cells: a potential mechanism for self-renewal. Dev Biol. 2004;275(2):375–388. doi: 10.1016/j.ydbio.2004.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zammit PS. Muscle satellite cells adopt divergent fates: a mechanism for self-renewal? J. Cell Biol. 2004;166:347–357. doi: 10.1083/jcb.200312007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Olguin HC, et al. Reciprocal inhibition between Pax7 and muscle regulatory factors modulates myogenic cell fate determination. J. Cell Biol. 2007;177(5):769–779. doi: 10.1083/jcb.200608122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Otto A, Collins-Hooper H, Patel K. The origin, molecular regulation and therapeutic potential of myogenic stem cell populations. Journal of Anatomy. 2009;9999:9999. doi: 10.1111/j.1469-7580.2009.01138.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zammit PS, et al. Pax7 and myogenic progression in skeletal muscle satellite cells. J Cell Sci. 2006;119(9):1824–1832. doi: 10.1242/jcs.02908. [DOI] [PubMed] [Google Scholar]
- 10.Meech R, et al. Barx2 is expressed in satellite cells and is required for normal muscle growth and regeneration. Stem cells. 2012;30(2):253–265. doi: 10.1002/stem.777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Makarenkova HP, et al. Barx2 Controls Myoblast Fusion and Promotes MyoD-mediated Activation of the Smooth Muscleα-Actin Gene. Journal of Biological Chemistry. 2009;284(22):14866–14874. doi: 10.1074/jbc.M807208200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Meech R, et al. The Homeobox Transcription Factor <italic>Barx2</italic>Regulates Plasticity of Young Primary Myofibers. PLoS ONE. 2010;5(7):e11612. doi: 10.1371/journal.pone.0011612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Makarenkova HP, et al. Barx2 controls myoblast fusion and promotes MyoD-mediated activation of the smooth muscle alpha actin gene. J Biol Chem. 2009 doi: 10.1074/jbc.M807208200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Meech R, et al. The homeodomain protein Barx2 promotes myogenic differentiation and is regulated by myogenic regulatory factors. J Biol Chem. 2003;278(10):8269–8278. doi: 10.1074/jbc.M207617200. [DOI] [PubMed] [Google Scholar]
- 15.Tajbakhsh S, et al. Differential activation of Myf5 and MyoD by different Wnts in explants of mouse paraxial mesoderm and the later activation of myogenesis in the absence of Myf5. Development. 1998;125(21):4155–4162. doi: 10.1242/dev.125.21.4155. [DOI] [PubMed] [Google Scholar]
- 16.Takata H, et al. Involvement of Wnt4 signaling during myogenic proliferation and differentiation of skeletal muscle. Developmental Dynamics. 2007;236(10):2800–2807. doi: 10.1002/dvdy.21327. [DOI] [PubMed] [Google Scholar]
- 17.Tsivitse S. Notch and Wnt signaling, physiological stimuli and postnatal myogenesis. Int J Biol Sci. 2010;6(3):268–281. doi: 10.7150/ijbs.6.268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pansters N, et al. Segregation of myoblast fusion and muscle-specific gene expression by distinct ligand-dependent inactivation of GSK-3β. Cellular and Molecular Life Sciences. 2011;68(3):523–535. doi: 10.1007/s00018-010-0467-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bernardi H, et al. Wnt4 activates the canonical β-catenin pathway and regulates negatively myostatin: functional implication in myogenesis. American Journal of Physiology - Cell Physiology. 2011;300(5):C1122–C1138. doi: 10.1152/ajpcell.00214.2010. [DOI] [PubMed] [Google Scholar]
- 20.Tanaka S, Terada K, Nohno T. Canonical Wnt signaling is involved in switching from cell proliferation to myogenic differentiation of mouse myoblast cells. Journal of Molecular Signaling. 2011;6(1):12. doi: 10.1186/1750-2187-6-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brack AS, et al. A Temporal Switch from Notch to Wnt Signaling in Muscle Stem Cells Is Necessary for Normal Adult Myogenesis. Cell Stem Cell. 2008;2(1):50–59. doi: 10.1016/j.stem.2007.10.006. [DOI] [PubMed] [Google Scholar]
- 22.Braun T, Gautel M. Transcriptional mechanisms regulating skeletal muscle differentiation, growth and homeostasis. Nat Rev Mol Cell Biol. 2011;12(6):349–361. doi: 10.1038/nrm3118. [DOI] [PubMed] [Google Scholar]
- 23.Abu-Elmagd M, et al. Wnt/Lef1 signaling acts via Pitx2 to regulate somite myogenesis. Dev Biol. 2010;337(2):211–219. doi: 10.1016/j.ydbio.2009.10.023. [DOI] [PubMed] [Google Scholar]
- 24.Wodarz A, Nusse R. Mechanisms of Wnt signaling in development. Annual review of cell and developmental biology. 1998;14:59–88. doi: 10.1146/annurev.cellbio.14.1.59. [DOI] [PubMed] [Google Scholar]
- 25.Hecht A, et al. The p300/CBP acetyltransferases function as transcriptional coactivators of [beta]-catenin in vertebrates. EMBO J. 2000;19(8):1839–1850. doi: 10.1093/emboj/19.8.1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Otto A, et al. Canonical Wnt signalling induces satellite-cell proliferation during adult skeletal muscle regeneration. J Cell Sci. 2008;121(17):2939–2950. doi: 10.1242/jcs.026534. [DOI] [PubMed] [Google Scholar]
- 27.Botrugno OA, et al. Synergy between LRH-1 and β-Catenin Induces G1 Cyclin-Mediated Cell Proliferation. Molecular Cell. 2004;15(4):499–509. doi: 10.1016/j.molcel.2004.07.009. [DOI] [PubMed] [Google Scholar]
- 28.Jho E-h, et al. Wnt/{beta}-Catenin/Tcf Signaling Induces the Transcription of Axin2, a Negative Regulator of the Signaling Pathway. Mol. Cell. Biol. 2002;22(4):1172–1183. doi: 10.1128/MCB.22.4.1172-1183.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Veeman MT, et al. Zebrafish Prickle, a Modulator of Noncanonical Wnt/Fz Signaling, Regulates Gastrulation Movements. Current Biology. 2003;13(8):680–685. doi: 10.1016/s0960-9822(03)00240-9. [DOI] [PubMed] [Google Scholar]
- 30.Edelman DB, Meech R, Jones FS. The homeodomain protein Barx2 contains activator and repressor domains and interacts with members of the CREB family. J Biol Chem. 2000;275(28):21737–21745. doi: 10.1074/jbc.M909998199. [DOI] [PubMed] [Google Scholar]
- 31.Olson LE, et al. Barx2 functions through distinct corepressor classes to regulate hair follicle remodeling. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(10):3708–3713. doi: 10.1073/pnas.0500519102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shao J-S, et al. Msx2 promotes cardiovascular calcification by activating paracrine Wnt signals. The Journal of Clinical Investigation. 2005;115(5):1210–1220. doi: 10.1172/JCI24140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim C-H, et al. {beta}-Catenin Interacts with MyoD and Regulates Its Transcription Activity. Mol. Cell. Biol. 2008;28(9):2941–2951. doi: 10.1128/MCB.01682-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Daniels DL, Weis WI. Beta-catenin directly displaces Groucho/TLE repressors from Tcf/Lef in Wnt-mediated transcription activation. Nat Struct Mol Biol. 2005;12(4):364–371. doi: 10.1038/nsmb912. [DOI] [PubMed] [Google Scholar]
- 35.Lepourcelet M, Shivdasani RA. Characterization of a Novel Mammalian Groucho Isoform and Its Role in Transcriptional Regulation. Journal of Biological Chemistry. 2002;277(49):47732–47740. doi: 10.1074/jbc.M208154200. [DOI] [PubMed] [Google Scholar]
- 36.Yamamoto H, et al. Sumoylation is involved in [beta]-catenin-dependent activation of Tcf-4. EMBO J. 2003;22(9):2047–2059. doi: 10.1093/emboj/cdg204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chesire DR, et al. In vitro evidence for complex modes of nuclear beta-catenin signaling during prostate growth and tumorigenesis. Oncogene. 2002;21(17):2679–2694. doi: 10.1038/sj.onc.1205352. [DOI] [PubMed] [Google Scholar]
- 38.Olson LE, et al. Homeodomain-Mediated2-Catenin-Dependent Switching Events Dictate Cell-Lineage Determination. 2006;125(3):593–605. doi: 10.1016/j.cell.2006.02.046. [DOI] [PubMed] [Google Scholar]
- 39.Vadlamudi U, et al. PITX2, β-catenin and LEF-1 interact to synergistically regulate the LEF-1 promoter. Journal of Cell Science. 2005;118(6):1129–1137. doi: 10.1242/jcs.01706. [DOI] [PubMed] [Google Scholar]
- 40.Park J-S, et al. Six2 and Wnt Regulate Self-Renewal and Commitment of Nephron Progenitors through Shared Gene Regulatory Networks. Developmental Cell. 2012;23(3):637–651. doi: 10.1016/j.devcel.2012.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Amen M, et al. PITX2 and {beta}-Catenin Interactions Regulate Lef-1 Isoform Expression. Mol. Cell. Biol. 2007;27(21):7560–7573. doi: 10.1128/MCB.00315-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Beagle B, Johnson GVW. AES/GRG5: More than just a dominant-negative TLE/GRG family member. Developmental Dynamics. 2010;239(11):2795–2805. doi: 10.1002/dvdy.22439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Riz I, et al. Transcriptional activation by TLX1/HOX11 involves Gro/TLE corepressors. Biochemical and Biophysical Research Communications. 2009;380(2):361–365. doi: 10.1016/j.bbrc.2009.01.099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hentschke M, Borgmeyer U. Identification of PNRC2 and TLE1 as activation function-1 cofactors of the orphan nuclear receptor ERRγ. Biochemical and Biophysical Research Communications. 2003;312(4):975–982. doi: 10.1016/j.bbrc.2003.11.025. [DOI] [PubMed] [Google Scholar]
- 45.Li J, Wang CY. TBL1-TBLR1 and beta-catenin recruit each other to Wnt target-gene promoter for transcription activation and oncogenesis. Nat Cell Biol. 2008;10(2):160–169. doi: 10.1038/ncb1684. [DOI] [PubMed] [Google Scholar]
- 46.Makarenkova HP, Meech R. In: Chapter four - Barx Homeobox Family in Muscle Development and Regeneration, in International Review of Cell and Molecular Biology. Kwang WJ, editor. Academic Press; 2012. pp. 117–173. [DOI] [PubMed] [Google Scholar]
- 47.Fleming-Waddell JN, et al. Effect of DLK1 and RTL1 but not MEG3 or MEG8 on muscle gene expression in Callipyge lambs. PloS one. 2009;4(10):e7399. doi: 10.1371/journal.pone.0007399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Waddell JN, et al. Dlk1 Is Necessary for Proper Skeletal Muscle Development and Regeneration. PLoS ONE. 2010;5(11):e15055. doi: 10.1371/journal.pone.0015055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Amthor H, et al. Follistatin complexes Myostatin and antagonises Myostatin-mediated inhibition of myogenesis. Developmental Biology. 2004;270(1):19–30. doi: 10.1016/j.ydbio.2004.01.046. [DOI] [PubMed] [Google Scholar]
- 50.Benabdallah BF, et al. Inhibiting myostatin with follistatin improves the success of myoblast transplantation in dystrophic mice. Cell transplantation. 2008;17(3):337–350. doi: 10.3727/096368908784153913. [DOI] [PubMed] [Google Scholar]
- 51.Gilson H, et al. Follistatin induces muscle hypertrophy through satellite cell proliferation and inhibition of both myostatin and activin. American journal of physiology. Endocrinology and metabolism. 2009;297(1):E157–E164. doi: 10.1152/ajpendo.00193.2009. [DOI] [PubMed] [Google Scholar]
- 52.Hiroki E, et al. A comparative study of myostatin, follistatin and decorin expression in muscle of different origin. Anatomical science international. 2011;86(3):151–159. doi: 10.1007/s12565-011-0103-0. [DOI] [PubMed] [Google Scholar]
- 53.Zhu J, et al. Follistatin improves skeletal muscle healing after injury and disease through an interaction with muscle regeneration, angiogenesis, and fibrosis. The American journal of pathology. 2011;179(2):915–930. doi: 10.1016/j.ajpath.2011.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kitamoto T, Hanaoka K. Notch3 Null Mutation in Mice Causes Muscle Hyperplasia by Repetitive Muscle Regeneration. Stem Cells. 2010;28(12):2205–2216. doi: 10.1002/stem.547. [DOI] [PubMed] [Google Scholar]
- 55.Meech R, et al. Barx2 Is Expressed in Satellite Cells and Is Required for Normal Muscle Growth and Regeneration. Stem Cells. 2012;30(2):253–265. doi: 10.1002/stem.777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hughes TA, Brady HJM. Expression of axin2 Is Regulated by the Alternative 5′-Untranslated Regions of Its mRNA. Journal of Biological Chemistry. 2005;280(9):8581–8588. doi: 10.1074/jbc.M410806200. [DOI] [PubMed] [Google Scholar]
- 57.Jensen PB, et al. A Wnt Oscillator Model for Somitogenesis. Biophysical journal. 2010;98(6):943–950. doi: 10.1016/j.bpj.2009.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Meech R, et al. The homeobox transcription factor Barx2 regulates chondrogenesis during limb development. Development. 2005;132(9):2135–2146. doi: 10.1242/dev.01811. [DOI] [PubMed] [Google Scholar]
- 59.Rando TA, Blau HM. Methods for myoblast transplantation. Methods Cell Biol. 1997;52:261–272. doi: 10.1016/s0091-679x(08)60382-9. [DOI] [PubMed] [Google Scholar]
- 60.Klenova E, et al. Immunoprecipitation techniques for the analysis of transcription factor complexes. Methods. 2002;26(3):254–259. doi: 10.1016/S1046-2023(02)00029-4. [DOI] [PubMed] [Google Scholar]
- 61.Zorn AM, et al. Regulation of Wnt Signaling by Sox Proteins: XSox17[alpha]/[beta] and XSox3 Physically Interact with [beta]-catenin. Molecular Cell. 1999;4(4):487–498. doi: 10.1016/s1097-2765(00)80200-2. [DOI] [PubMed] [Google Scholar]
- 62.Attema JL, et al. Hematopoietic stem cell ageing is uncoupled from p16 INK4A-mediated senescence. Oncogene. 2009;28:2238–2243. doi: 10.1038/onc.2009.94. [DOI] [PubMed] [Google Scholar]
- 63.Brack AS, et al. Increased Wnt Signaling During Aging Alters Muscle Stem Cell Fate and Increases Fibrosis. Science. 2007;317(5839):807–810. doi: 10.1126/science.1144090. [DOI] [PubMed] [Google Scholar]
- 64.Trapnell C, et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc. 2012;7(3):562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.