Abstract
Signal propagation through enzyme cascades is a critical component of information processing in cellular systems. Although such systems have potential as biomolecular computing tools, rational design of synthetic protein networks remains infeasible. DNA strands with catalytic activity (DNAzymes) are an attractive alternative, enabling rational cascade design through predictable base-pair hybridization principles. Here we report multi-layered DNAzyme signaling and logic cascades. We achieve signaling between DNAzymes using a structured chimeric substrate (SCS) that releases a downstream activator after cleavage by an upstream DNAzyme. The SCS can be activated by various upstream DNAzymes, can be coupled to DNA strand displacement devices, and is highly resistant to interference from background DNA. This work enables rational design of synthetic DNAzyme regulatory networks, with potential applications in biomolecular computing, biodetection, and autonomous theranostics.
Keywords: DNAzymes, signaling cascades, strand displacement, isothermal amplification, regulatory networks
Cells use enzymatic signaling pathways for a number of critical functions, including detection of environmental stimuli, signal amplification, and regulated information propagation through the intracellular environment. Cells typically implement these functions using proteins, but the complexity of protein folding makes the rational design of protein-based signaling cascades infeasible. [1] Though prior work on biocomputing devices using naturally occurring proteins shows promise,[2] this approach is limited by the possible protein-protein interactions. DNA, on the other hand, is an ideal alternative engineering material for de novo design of synthetic enzymatic cascades, thanks to predictable Watson-Crick base pairing and secondary structure formation. Synthetic analogs of some basic cellular processes have been implemented in DNA, including computation,[3] self-assembly,[4] locomotion,[5] small molecule sensing,[6] and catalysis[7]. Here we focus on DNAzymes[8] (also known as deoxyribozymes), which are single-stranded DNA molecules that can catalyze many of the same reactions as protein enzymes[9] and have been used for computation in parallel gate arrays.[3c, 10] We report a DNAzyme cascade system that uses structured, single-stranded substrates to sequester activating sequences and to propagate an activating signal to a downstream DNAzyme when cleaved by an upstream DNAzyme. We develop multi-layer signaling cascades and logic circuits, in which a conformational change in a molecule propagates information downstream, mimicking biological systems that rely on modifications such as phosphorylation of downstream enzymes to propagate information.
We based our designs on the most widely used family of DNAzymes: RNA-cleaving DNAzymes. With appropriate metal cation cofactors, these DNAzymes cleave RNA or chimeric DNA/RNA substrates in a multiple-turnover reaction, providing built-in signal amplification capabilities. For a given catalytic motif, DNAzyme-substrate pairs can be designed by simply choosing appropriate complementary sequences for the substrate and the substrate-binding arms of the DNAzyme. This is considerably simpler than designing enzyme-substrate pairs de novo by protein engineering. Here we use the 8-17 RNA-cleaving DNAzyme due to its compact size and efficient catalytic rate.[11]
We build on previous work on ribozyme circuits[12] and on DNAzyme signaling cascades that either sequestered the downstream effector sequence in a partially complementary complex,[13] or built two-layer cascades where the downstream DNAzyme generates a colorimetric readout.[14] In the first case, the use of a multi-strand complex as the mediator increases the number of strands and the complexity of circuit preparation. In the second case, the downstream DNAzyme cannot propagate the signal further within the molecular circuit. While signal amplification has also been demonstrated using DNA strand displacement[7c, 15] and catalytic hairpin assembly,[16] these circuits must be specifically designed to obtain catalysis, for example, using seesaw gates.[3a, 3b, 17] We use DNAzyme displacement reactions,[18] which combine the advantages of strand displacement to program reaction pathways with the inherent catalytic ability of DNAzymes. This reduces the number of DNA strands needed to achieve signal amplification.
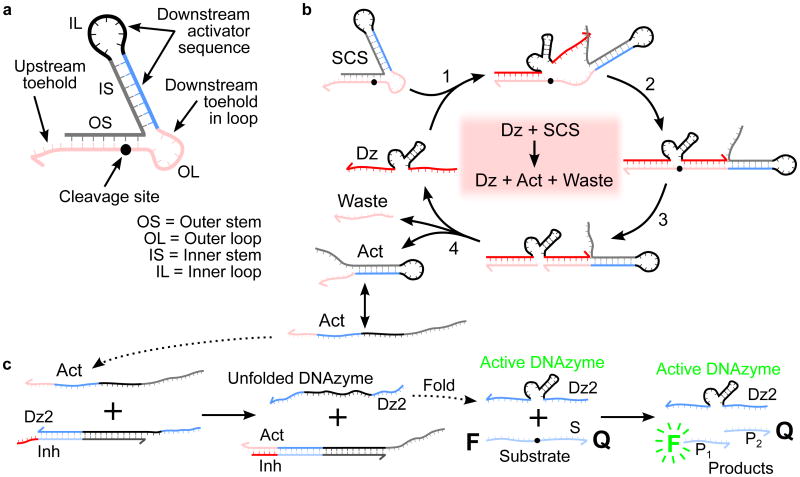
In cellular enzymatic signaling cascades, an activation signal is typically passed from one enzyme to another via chemical modifications. Here we achieve information propagation between enzymatic units through the covalent modification of a structured chimeric substrate (SCS). The SCS uses a metastable dual stem-loop design[19] (Figure 1a) and comprises several domains that make up interchangeable input and output modules. The use of a modular intermediary simplifies the design process by removing the need for direct enzyme-enzyme interactions, as are often found with protein-based cascades, e.g., phosphorylation in the MAPK pathway.[20] The inner 7bp stem and 8bp loop constitute the output module, whose secondary structure weakly sequesters a downstream activator. The outer 7bp stem and 6bp loop stabilize the structure and protect the activator toehold in the outer loop to prevent unwanted interactions with the downstream DNAzyme before cleavage. The outer stem and loop also constitute the input module, with a substrate binding and cleavage domain for an upstream DNAzyme. We minimized the size of the outer loop to better protect the toehold, which led to a 5bp overlap between the upstream DNAzyme binding arm and the downstream inhibitor toehold sequences. As shown in Figure 1b, an upstream DNAzyme interacts with the SCS when one of the 8bp substrate binding arms hybridizes with the 4bp outermost toehold and opens the outer stem via a toehold-mediated strand displacement reaction.[15b] The second arm binds the outer loop, linearizing the substrate domain and correctly positioning the SCS cleavage site opposite the catalytic core of the DNAzyme. The subsequent cleavage reaction causes the outer stem to dissociate as waste, freeing the protected toehold in the outer loop of the SCS, which can now hybridize to its complement more effectively. The relatively weak secondary structure in the activator released by SCS cleavage allows it to interconvert between hairpin and linear structural forms. Thus, downstream interactions are not impeded by secondary structure in the activator.
Figure 1.
SCS design and mechanisms for SCS cleavage and DNAzyme displacement. a) Design of a structured chimeric substrate (SCS) to enable signaling between DNAzymes. The SCS consists of an outer stem and loop, which make up the upstream DNAzyme binding domain (red), and an inner stem and loop, which sequester a downstream activator sequence (blue and black). The cleavage site is located towards the inner end of the outer stem. The grey cage sequence is chosen to fold into the desired structure, producing a topological constraint on the downstream reaction kinetics that is undone when the SCS is cleaved by the upstream DNAzyme. b) Mechanism of cleavage of the SCS by an upstream DNAzyme (Dz). The upstream DNAzyme binds to the outer stem and loop by toehold-mediated strand displacement. The cleavage reaction produces a waste strand and an activator strand (Act). In the activator structure, the outer loop has been released from the topological constraint previously imposed by the outer stem, making the downstream toehold in the outer loop available to bind with a downstream circuit element. c) DNAzyme displacement reaction mechanism. The catalytic activity of the downstream DNAzyme strand (Dz2) is inhibited by hybridization to a partially complementary inhibitor strand (Inh) with a short overhanging toehold. Activation is by a toehold-mediated strand displacement reaction: an input strand (Act) binds to the complex (Dz2-Inh) via the toehold. The input initiates a branch-migration reaction that eventually displaces a catalytically active downstream DNAzyme strand (Dz2), leaving an inert waste complex (Act-Inh). The DNAzyme strand then folds into a catalytically active conformation and proceeds to bind to substrate molecules (S) and cleave them, producing shorter cleavage products (P1 and P2). The cleavage reaction causes separation of the fluorophore-quencher pair attached to the two ends of the substrate, observed as an increase in bulk fluorescence due to loss of FRET.
This mechanism is particularly suited for use with our previously reported DNAzyme displacement logic gates,[18] in which DNAzyme catalysis is controlled using toehold-mediated DNA strand displacement reactions.[15b] The activator released by SCS cleavage binds to the toehold of the downstream DNAzyme-inhibitor complex and undergoes branch migration to displace a catalytically active DNAzyme strand, producing an inert waste complex (Figure 1c). The displaced DNAzyme refolds into a catalytically active conformation and can then cleave its own substrate. Thus, activation of one DNAzyme species causes the activation of a second DNAzyme species, implementing signal propagation.
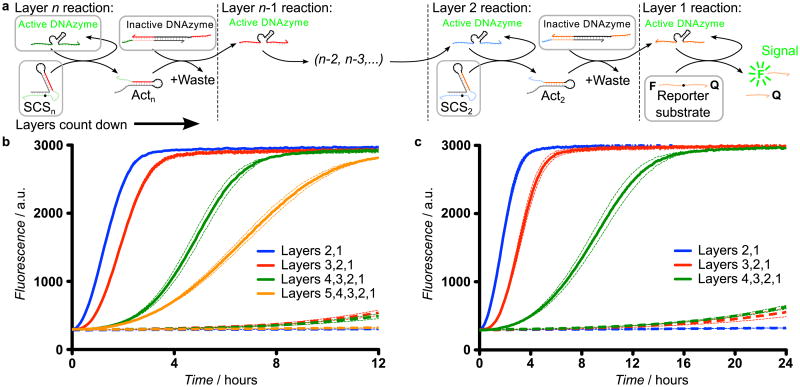
For correct behavior in synthetic multi-enzyme systems, each enzyme must interact with its intended substrate with high specificity. In protein-based enzymatic cascades, specificity is derived from complex interactions between the secondary and tertiary conformations of both enzyme and substrate, rendering rational design of such interactions infeasible. DNAzyme-based cascades achieve specificity through sequence-specific hybridization to substrates. We can modify the SCS input and output modules to enable signaling between DNAzymes with different substrate binding arms while keeping the SCS structure intact. Since the SCS does not need to be redesigned for each subsequent layer, this enables rapid construction of two-, three-, four-, and five-layer linear DNAzyme signaling cascades, each initiated by the addition of active top-layer DNAzymes (Figure 2). Each cascade uses the same reporter layer (layer 1), with layer n+1 added upstream of layer n to extend the cascade. This naming system reflects the sequence commonality between each layer n, irrespective of cascade length. Our five-layer cascade is the longest DNAzyme signaling cascade implemented to date. Development of extended catalytic signaling cascades with a high signal-to-noise ratio is challenging because unwanted signal generated in the absence of input (leakage) is also amplified by downstream circuit elements. Kinetic traces of multi-layer cascades (Figure 2b) show that the time taken for cascade execution increase with the number of layers (Supplementary Discussion 1). Lower DNAzyme concentrations reduce leakage at the expense of activation speed by relying on multiple-turnover cleavage for signal amplification (Figure S1). In particular, using lower concentrations in the upstream layers of the cascade with increasing concentrations in each downstream layer can reduce leakage without affecting the maximum output level or a significant sacrifice in speed (Figure 2c). Additional controls using uncleavable SCS molecules demonstrate that cleavage is necessary for signal propagation (Figure S2). Thus we have demonstrated that chemical modification of a structured substrate by a DNAzyme can be used to propagate information in a signaling cascade.
Figure 2.
Demonstration of DNAzyme signaling cascades. a) Schematic of multi-layer DNAzyme signaling cascades using DNAzyme displacement reactions. Initial species for each layer of the cascade are highlighted in grey boxes. In each layer, an active DNAzyme cleaves the corresponding SCS, producing an activator that releases the downstream DNAzyme from its catalytically inactive enzyme-inhibitor complex via a DNAzyme displacement reaction, thereby propagating the activating signal to the next layer of the cascade. b) The mean fluorescence signal (solid lines) from two-layer (blue), three-layer (red), four-layer (green) and five-layer (orange) linear DNAzyme signaling cascades with equimolar (100 nM) DNAzyme concentrations in each layer. The dashed line represents the same reaction without the top-layer active DNAzyme, which measures the non-specific activation (leakage) of the downstream circuit. The dotted lines represent the 95% confidence interval from three replicate experiments. c) Kinetic traces for multi-layer linear DNAzyme signaling cascades with increasing DNAzyme concentrations in each layer (25 nM in layer 4, 50 nM in layer 3, 75 nM in layer 2, and 100 nM in layer 1) to demonstrate signal amplification. In both plots, dotted lines represent the 95% confidence interval from three replicate experiments.
Because DNA interactions are sequence-specific, the SCS can interact with any upstream or downstream circuit components with the correct sequence. We implemented signaling cascades between a variety of DNA logic components in both the upstream and downstream positions, including various DNAzyme logic gates and a strand displacement reporter gate (Figure S3). This demonstrates the flexibility of DNAzyme-based interactions via the SCS, which enables development of hybrid DNA circuits comprising components from multiple architectures, which is currently a significant challenge.
Multi-layer synthetic DNAzyme logic cascades offer a route to increasing the sophistication of biomolecular logic circuits, with the long-term aim of enabling robust, isothermal detection of disease states via sequence-specific nucleic acid detection[14, 21] or aptamer-based detection of small molecules.[6a-c] Incorporating logic into enzymatic cascades enables the integration of multiple input signals, which can reduce false positives in bioassays and enable detection of disease states where a single target is insufficient for an accurate diagnosis. To illustrate the potential of DNAzyme logic cascades for detecting multiple pathogenic targets in extracted DNA, we implemented multi-layer circuits for typing representative pathogen signatures from all four dengue virus serotypes (DEN1-4). Dengue is a major global health concern,[22] and accurate serotyping is important because sequential infection with different serotypes is a risk factor for dengue hemorrhagic fever and dengue shock syndrome, both of which can be fatal.[23]
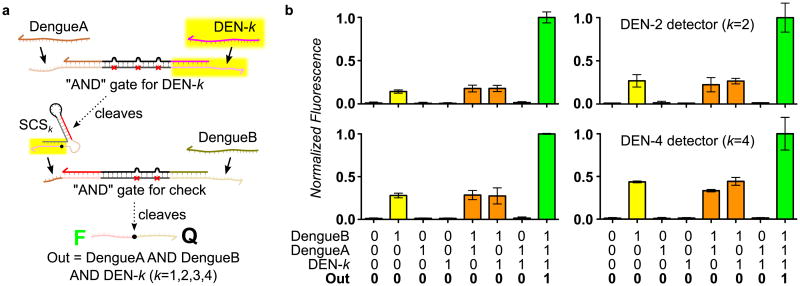
We exploited the modularity of the SCS to design a two-layer, three-input “AND” circuit template, in which two DNA oligomers derived from conserved sequences within the ssRNA dengue genomes and a serotype-specific DNA oligomer must be present to produce a fluorescent output. As shown in Figure 3a, each layer of the circuit is an “AND” gate activated by two inputs in a cooperative displacement reaction.[24] The use of mismatches in the inhibitor is required for rapid release of the DNAzyme, because the catalytic core is not displaced by either input.[18] One of the inputs of the downstream gate is released upon cleavage of the SCS. We replicated this template, modifying the highlighted parts of the upstream “AND” gate and the SCS, to produce four circuits, each sensitive to a different serotype-specific target sequence (Figure S4). We observed strong positive responses from all four circuits in the presence of all three signatures, at least 2.5 times the maximum response seen in the absence of one or more signatures (Figure 3b). Higher leakage is seen in the presence of the downstream DengueB input, suggesting that there is some interaction between the SCS and the downstream AND gate prior to SCS cleavage. Misfolding of SCS or enzyme strands reduced system performance (Figure S5), showing that optimization of the predicted secondary structure is important for efficient circuit operation.
Figure 3.
Example application of the SCS in a multi-layer diagnostic logic circuit. a) Design of multi-layer diagnostic logic circuits for detection of sequences from the genomes of all four dengue serotypes. The circuit template for serotype DEN-k (k=1,2,3,4) requires the presence of two conserved sequences from the dengue viral genome (DengueA and DengueB) and one sequence specific to the serotype of interest (DEN-k). This is implemented by DNAzyme displacement “AND” gates with mismatched inhibitors,[18] which are connected by a SCS molecule. When both upstream inputs are present, the active upstream DNAzyme cleaves the SCS, producing an activator that serves as one input to the downstream gate. If the second input to the downstream gate is also present, the downstream DNAzyme is activated and we observe this via loss of FRET following substrate cleavage. The upstream “AND” gate uses the 3 mismatch design characterized in Supplementary Figure 1, whereas the downstream gate uses an asymmetric pattern of mismatches because the activator produced by the SCS can displace into the catalytic core. We derived detection circuits for all four dengue serotypes (DEN1-4) by modifying only the domains highlighted in yellow. b) Demonstrations of serotyping circuits for DEN1-4, which show correct operation of all four instantiations of the three-input “AND” circuit template. Each serotyping circuit was characterized using all eight combinations of the two conserved sequences and the correct serotype-specific sequence. Variations in the normalized fluorescence levels (i.e., different levels of activation and leakage) may be attributed to variations in the stability of the corresponding SCSk structure in each case. Error bars represent the 95% confidence interval from three replicate experiments.
To test our approach in a minimal biological background, we implemented a two-layer DNAzyme cascade using the SCS with increasing amounts of random background DNA (Figure S6). This models a common detection scenario in which all the nucleic acids have been extracted from a sample for analysis. We showed that the SCS design is sufficient for this minimal assay detection environment. Furthermore, all experiments described herein were performed using minimal oligonucleotide purification techniques, which is essential for the development and use of low-cost bioassays. Thus we have demonstrated two key properties for a practical bioassay: robust operation in background and straightforward preparation.
In summary, we have developed a method to design extended DNAzyme signaling cascades that exhibit many of the functionalities of cellular cascades: integration of multiple input signals, signal amplification, transduction, and propagation. The combination of DNAzymes, strand displacement and rationally designed, structured chimeric substrates enabled us to implement synthetic signaling cascades compatible with a variety of DNA logic gates, including the longest DNAzyme signaling cascade demonstrated to date. These DNAzyme cascades hold promise for practical applications such as pathogen detection. We illustrated this by demonstrating that our circuits resist background interference and can implement multi-input, multi-layer detection of multiple pathogen signatures.
Future work will explore the operation of DNAzyme cascades in physiologically relevant conditions[25] such as cell lysate[26] or serum, which may be challenging due to the presence of nucleases that may degrade circuit components, or due to insufficient concentrations of the metal ion cofactors required for efficient DNAzyme catalysis.[27] Furthermore, the modular design of the SCS should allow the implementation of increasingly complex synthetic DNAzyme signaling networks, incorporating network motifs such as feedforward and feedback cycles.[28] These circuits could exhibit non-trivial dynamic behaviors to enable more sophisticated decision-making for diagnostic and therapeutic applications, possibly connected to alternative readout technologies such as gold nanoparticles[29] or paperfluidic devices.[21a, 30]
Experimental Section
Materials
All oligonucleotides were purchased from Integrated DNA Technologies (Coralville, IA). Oligonucleotide sequences are listed in Supplementary Tables S1-8. DNAzymes and inhibitors were purchased with standard desalting whenever possible, with the exception of oligonucleotides that exceeded 60 base pairs in length (which were PAGE purified by the manufacturer, in accordance with the manufacturer's recommended procedures). All DNA/RNA chimeric substrates (SCS molecules and fluorescent reporter substrates) were purified by RNase-free HPLC by the manufacturer. The fluorescent reporter substrates were labeled with a 5′ FAM quenched by a 3′ TAMRA fluorophore. Oligonucleotides were resuspended in RNase-free H2O (Sigma-Aldrich) in accordance with the manufacturer-provided specifications at a stock concentration of 50 μM. Working stocks were made by adding 50 μL of the resuspended oligonucleotide solution into 950 μL buffer.
Preparation of DNAzyme-inhibitor complexes and SCS molecules
DNAzyme strands and inhibitor strands were pre-complexed by heating the DNAzyme and inhibitor strands together at 95 °C for 3 minutes on a heat block, and subsequently annealing by cooling to room temperature over a minimum of 90 minutes. In many cases, an excess of inhibitor relative to DNAzyme was used, to ensure complete inhibition of the DNAzymes – in these cases, the resulting solution of DNAzyme-inhibitor complexes and excess free inhibitor strands was used without further purification. Single-stranded SCS molecules (and loop-inhibited DNAzymes) were prepared using the same heating and annealing protocol.
Assay conditions and instrumentation
All assays were performed at room temperature (23 °C) in a buffer of 1M NaCl, 50 mM HEPES, 1 mM ZnCl2, pH 7.0. Fluorescence was read either on a Quantamaster 40 fluorimeter (PTI, Binghamton, NJ) in a 300 μL reaction volume or Spectramax M2e fluorescent plate reader (Molecular Devices, Sunnyvale, CA) in a 200 μL reaction volume. In all cases, fluorescein emission was monitored at 492 nm excitation and 518 nm emission wavelengths. Error bars indicate two standard deviations from the mean of three replicates, representing the 95% confidence interval. Full details of assay conditions for individual experiments are listed in the Supporting Information.
Supplementary Material
Footnotes
This material is based upon work supported by the National Science Foundation under grants 1027877 and 1028238. C.W.B. gratefully acknowledges support from INCBN IGERT DGE-0549500. M.R.L. gratefully acknowledges support from the New Mexico Cancer Nanoscience and Microsystems Training Center (NIH/NCI grant 5R25CA153825).
Supporting information for this article is available on the WWW under http://www.angewandte.org.
Contributor Information
Carl W. Brown, III, Center for Biomedical Engineering, Department of Chemical and Nuclear Engineering, University of New Mexico, Albuquerque, NM 87131 (USA).
Dr. Matthew R. Lakin, Department of Computer Science, Center for Biomedical Engineering, University of New Mexico, Albuquerque, NM 87131 (USA)
Eli K. Horwitz, Center for Biomedical Engineering, Department of Chemical and Nuclear Engineering, University of New Mexico, Albuquerque, NM 87131 (USA)
M. Leigh Fanning, Department of Computer Science, Center for Biomedical Engineering, University of New Mexico, Albuquerque, NM 87131 (USA).
Hannah E. West, Center for Biomedical Engineering, Department of Chemical and Nuclear Engineering, University of New Mexico, Albuquerque, NM 87131 (USA)
Prof. Darko Stefanovic, Email: darko@cs.unm.edu, Department of Computer Science, Center for Biomedical Engineering, University of New Mexico, Albuquerque, NM 87131 (USA).
Prof. Steven W. Graves, Email: graves@unm.edu, Center for Biomedical Engineering, Department of Chemical and Nuclear Engineering, University of New Mexico, Albuquerque, NM 87131 (USA).
References
- 1.Martin CH, Nielsen DR, Solomon KV, Prather KL. Chem Biol. 2009;16:277. doi: 10.1016/j.chembiol.2009.01.010. [DOI] [PubMed] [Google Scholar]
- 2.a) Katz E, editor. Biomolecular Information Processing - From Logic Systems to Smart Sensors and Actuators. Wiley-VCH; Weinheim, Germany: 2012. [Google Scholar]; b) Katz E, Privman V. Chem Soc Rev. 2010;39:1835. doi: 10.1039/b806038j. [DOI] [PubMed] [Google Scholar]; c) Katz E, Wang J, Privman M, Halamek J. Anal Chem. 2012;84:5463. doi: 10.1021/ac3007076. [DOI] [PubMed] [Google Scholar]; d) Wang J, Katz E. Anal Bioanal Chem. 2010;398:1591. doi: 10.1007/s00216-010-3746-0. [DOI] [PubMed] [Google Scholar]; e) Zhou J, Arugula MA, Halamek J, Pita M, Katz E. J Phys Chem B. 2009;113:16065. doi: 10.1021/jp9079052. [DOI] [PubMed] [Google Scholar]
- 3.a) Qian L, Winfree E. Science. 2011;332:1196. doi: 10.1126/science.1200520. [DOI] [PubMed] [Google Scholar]; b) Qian L, Winfree E, Bruck J. Nature. 2011;475:368. doi: 10.1038/nature10262. [DOI] [PubMed] [Google Scholar]; c) Stojanovic MN, Stefanovic D. Nat Biotechnol. 2003;21:1069. doi: 10.1038/nbt862. [DOI] [PubMed] [Google Scholar]
- 4.a) Woo S, Rothemund PW. Nat Chem. 2011;3:620. doi: 10.1038/nchem.1070. [DOI] [PubMed] [Google Scholar]; b) Yin P, Choi HMT, Calvert CR, Pierce NA. Nature. 2008;451:318. doi: 10.1038/nature06451. [DOI] [PubMed] [Google Scholar]
- 5.a) Bath J, Green SJ, Allen KE, Turberfield AJ. Small. 2009;5:1513. doi: 10.1002/smll.200900078. [DOI] [PubMed] [Google Scholar]; b) Yin P, Yan H, Daniell XG, Turberfield AJ, Reif JH. Angew Chem Int Ed Engl. 2004;43:4906. doi: 10.1002/anie.200460522. [DOI] [PubMed] [Google Scholar]; c) Yurke B, Turberfield AJ, Mills AP, Simmel FC, Neumann JL. Nature. 2000;406:605. doi: 10.1038/35020524. [DOI] [PubMed] [Google Scholar]; d) Lund K, Manzo AJ, Dabby N, Michelotti N, Johnson-Buck A, Nangreave J, Taylor S, Pei R, Stojanovic MN, Walter NG, Winfree E, Yan H. Nature. 2010;465:206. doi: 10.1038/nature09012. [DOI] [PMC free article] [PubMed] [Google Scholar]; e) Pei R, Taylor SK, Stefanovic D, Rudchenko S, Mitchell TE, Stojanovic MN. J Am Chem Soc. 2006;128:12693. doi: 10.1021/ja058394n. [DOI] [PubMed] [Google Scholar]; f) Gu H, Chao J, Xiao SJ, Seeman NC. Nature. 2010;465:202. doi: 10.1038/nature09026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.a) Huizenga DE, Szostak JW. Biochemistry. 1995;34:656. doi: 10.1021/bi00002a033. [DOI] [PubMed] [Google Scholar]; b) Stojanovic MN, Kolpashchikov DM. J Am Chem Soc. 2004;126:9266. doi: 10.1021/ja032013t. [DOI] [PubMed] [Google Scholar]; c) Teller C, Shimron S, Willner I. Anal Chem. 2009;81:9114. doi: 10.1021/ac901773b. [DOI] [PubMed] [Google Scholar]; d) Achenbach JC, Nutiu R, Li Y. Analytica Chimica Acta. 2005;534:41. [Google Scholar]; e) Li Y, Lu Y. Functional Nucleic Acids for Analytical Applications. Springer; 2009. [Google Scholar]
- 7.a) Seelig G, Yurke B, Winfree E. J Am Chem Soc. 2006;128:12211. doi: 10.1021/ja0635635. [DOI] [PubMed] [Google Scholar]; b) Stojanovic MN, Mitchell TE, Stefanovic D. J Am Chem Soc. 2002;124:3555. doi: 10.1021/ja016756v. [DOI] [PubMed] [Google Scholar]; c) Zhang DY, Turberfield AJ, Yurke B, Winfree E. Science. 2007;318:1121. doi: 10.1126/science.1148532. [DOI] [PubMed] [Google Scholar]
- 8.Li YF, Breaker RR. Curr Opin Struct Biol. 1999;9:315. doi: 10.1016/S0959-440X(99)80042-6. [DOI] [PubMed] [Google Scholar]
- 9.a) Breaker RR, Joyce GF. Chem Biol. 1995;2:655. doi: 10.1016/1074-5521(95)90028-4. [DOI] [PubMed] [Google Scholar]; b) Chandra M, Sachdeva A, Silverman SK. Nat Chem Biol. 2009;5:718. doi: 10.1038/nchembio.201. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Cuenoud B, Szostak JW. Nature. 1995;375:611. doi: 10.1038/375611a0. [DOI] [PubMed] [Google Scholar]; d) Li Y, Breaker RR. Proc Natl Acad Sci U S A. 1999;96:2746. doi: 10.1073/pnas.96.6.2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.a) Macdonald J, Li Y, Sutovic M, Lederman H, Pendri K, Lu W, Andrews BL, Stefanovic D, Stojanovic MN. Nano Lett. 2006;6:2598. doi: 10.1021/nl0620684. [DOI] [PubMed] [Google Scholar]; b) Pei R, Matamoros E, Liu M, Stefanovic D, Stojanovic MN. Nat Nanotechnol. 2010;5:773. doi: 10.1038/nnano.2010.194. [DOI] [PubMed] [Google Scholar]
- 11.a) Li J, Zheng W, Kwon AH, Lu Y. Nucleic Acids Res. 2000;28:481. doi: 10.1093/nar/28.2.481. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Santoro SW, Joyce GF. Proc Natl Acad Sci U S A. 1997;94:4262. doi: 10.1073/pnas.94.9.4262. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Schlosser K, Li Y. Chembiochem. 2010;11:866. doi: 10.1002/cbic.200900786. [DOI] [PubMed] [Google Scholar]
- 12.a) Penchovsky R. ACS Synth Biol. 2012;1:471. doi: 10.1021/sb300053s. [DOI] [PubMed] [Google Scholar]; b) Penchovsky R, Breaker RR. Nat Biotechnol. 2005;23:1424. doi: 10.1038/nbt1155. [DOI] [PubMed] [Google Scholar]
- 13.a) Elbaz J, Lioubashevski O, Wang F, Remacle F, Levine RD, Willner I. Nat Nanotechnol. 2010;5:417. doi: 10.1038/nnano.2010.88. [DOI] [PubMed] [Google Scholar]; b) Elbaz J, Moshe M, Shlyahovsky B, Willner I. Chemistry. 2009;15:3411. doi: 10.1002/chem.200802004. [DOI] [PubMed] [Google Scholar]
- 14.Gerasimova YV, Cornett EM, Edwards E, Su X, Rohde KH, Kolpashchikov DM. ChemBioChem. 2013;14:2087. doi: 10.1002/cbic.201300471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.a) Seelig G, Soloveichik D, Zhang DY, Winfree E. Science. 2006;314:1585. doi: 10.1126/science.1132493. [DOI] [PubMed] [Google Scholar]; b) Zhang DY, Seelig G. Nat Chem. 2011;3:103. doi: 10.1038/nchem.957. [DOI] [PubMed] [Google Scholar]; c) Zhang DY, Winfree E. J Am Chem Soc. 2008;130:13921. doi: 10.1021/ja803318t. [DOI] [PubMed] [Google Scholar]
- 16.a) Chen X, Briggs N, McLain JR, Ellington AD. Proc Natl Acad Sci U S A. 2013;110:5386. doi: 10.1073/pnas.1222807110. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Li B, Ellington AD, Chen X. Nucleic Acids Res. 2011;39:e110. doi: 10.1093/nar/gkr504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Qian L, Winfree E. J R Soc Interface. 2011;8:1281. doi: 10.1098/rsif.2010.0729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brown CW, III, Lakin MR, Stefanovic D, Graves SW. ChemBioChem. 2014;15:950. doi: 10.1002/cbic.201400047. [DOI] [PubMed] [Google Scholar]
- 19.a) Bois JS, Venkataraman S, Choi HM, Spakowitz AJ, Wang ZG, Pierce NA. Nucleic Acids Res. 2005;33:4090. doi: 10.1093/nar/gki721. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Turberfield AJ, Mitchell JC, Yurke B, Mills AP, Blakey MI, Simmel FC. Phys Rev Lett. 2003;90 doi: 10.1103/PhysRevLett.90.118102. [DOI] [PubMed] [Google Scholar]
- 20.Seger R, Krebs EG. FASEB J. 1995;9:726. [PubMed] [Google Scholar]
- 21.a) Allen PB, Arshad SA, Li B, Chen X, Ellington AD. Lab Chip. 2012;12:2951. doi: 10.1039/c2lc40373k. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Lu CH, Wang F, Willner I. J Am Chem Soc. 2012;134:10651. doi: 10.1021/ja3037838. [DOI] [PubMed] [Google Scholar]; c) Wang F, Elbaz J, Teller C, Willner I. Angew Chem Int Ed Engl. 2011;50:295. doi: 10.1002/anie.201005246. [DOI] [PubMed] [Google Scholar]; d) Wang F, Elbaz J, Willner I. J Am Chem Soc. 2012;134:5504. doi: 10.1021/ja300616w. [DOI] [PubMed] [Google Scholar]; e) Weizmann Y, Patolsky F, Willner I. Analyst. 2001;126:1502. doi: 10.1039/b106613g. [DOI] [PubMed] [Google Scholar]
- 22.Simmons CP, Farrar JJ, Nguyen VVC, Wills B. New Engl J Med. 2012;366:1423. doi: 10.1056/NEJMra1110265. [DOI] [PubMed] [Google Scholar]
- 23.Sangkawibha N, Rojanasuphot S, Ahandrik S, Viriyapongse S, Jatanasen S, Salitul V, Phanthumachinda B, Halstead SB. Am J Epidemiol. 1984;120:653. doi: 10.1093/oxfordjournals.aje.a113932. [DOI] [PubMed] [Google Scholar]
- 24.Zhang DY. J Am Chem Soc. 2011;133:1077. doi: 10.1021/ja109089q. [DOI] [PubMed] [Google Scholar]
- 25.Achenbach JC, Chiuman W, Cruz RP, Li Y. Curr Pharm Biotechnol. 2004;5:321. doi: 10.2174/1389201043376751. [DOI] [PubMed] [Google Scholar]
- 26.Kahan-Hanum M, Douek Y, Adar R, Shapiro E. Sci Rep. 2013;3 doi: 10.1038/srep01535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.a) Fokina AA, Meschaninova MI, Durfort T, Venyaminova AG, Francois JC. Biochemistry. 2012;51:2181. doi: 10.1021/bi201532q. [DOI] [PubMed] [Google Scholar]; b) Young DD, Lively MO, Deiters A. J Am Chem Soc. 2010;132:6183. doi: 10.1021/ja100710j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Alon U. An Introduction to Systems Biology: Design Principles of Biological Circuits. Chapman & Hall / CRC; 2007. [Google Scholar]
- 29.a) Zhao W, Lam JC, Chiuman W, Brook MA, Li Y. Small. 2008;4:810. doi: 10.1002/smll.200700757. [DOI] [PubMed] [Google Scholar]; b) Liu J, Lu Y, Fluoresc J. 2004;14:343. doi: 10.1023/b:jofl.0000031816.06134.d3. [DOI] [PubMed] [Google Scholar]; c) Liu J, Lu Y. J Am Chem Soc. 2004;126:12298. doi: 10.1021/ja046628h. [DOI] [PubMed] [Google Scholar]; d) Liu J, Lu Y. Anal Chem. 2004;76:1627. doi: 10.1021/ac0351769. [DOI] [PubMed] [Google Scholar]
- 30.Zhao W, Ali MM, Aguirre SD, Brook MA, Li Y. Anal Chem. 2008;80:8431. doi: 10.1021/ac801008q. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.