Abstract
Proper formation of ureteral smooth muscle cells (SMCs) during embryogenesis is essential for ureter peristalsis that propels urine from the kidney to the bladder in mammals. Currently the molecular factors that regulate differentiation of ureteral mesenchymal cells into SMCs are incompletely understood. A recent study has reported that Smad4 deficiency reduces the number of ureteral SMCs. However, its precise role in the ureteral smooth muscle development remains largely unknown. Here, we used Tbx18:Cre knock-in mouse line to delete Smad4 to examine its requirement in the development of ureteral mesenchyme and SMC differentiation. We found that mice with specific deletion of Smad4 in Tbx18-expressing ureteral mesenchyme exhibited hydroureter and hydronephrosis at embryonic day (E) 16.5, and the mutant mesenchymal cells failed to differentiate into SMCs with increased apoptosis and decreased proliferation. Molecular markers for SMCs including alpha smooth muscle actin (α-SMA) and smooth muscle myosin heavy chain (SM-MHC) were absent in the mutant ureters. Moreover, disruption of Smad4 significantly reduced the expression of genes, including Sox9, Tbx18 and Myocardin associated with SMC differentiation. These findings suggest that Smad4 is essential for initiating the SMC differentiation program during ureter development.
Introduction
Congenital malformations of the urinary tract resulted from embryonic ureter obstruction lead to hydroureter and/or hydronephrosis with dilated ureter and/or renal pelvis, and are one of the main causations of renal failure among children and young adults [1]–[3]. Currently the molecular mechanisms underlying these congenital defects are poorly understood.
The urinary tract is composed of three important cell types: the inner epithelial cells (urothelium), the outer smooth muscle cells (SMCs) that provide contractility to evacuate urine from kidney to bladder [4], and fibroblasts. During ureter development (from E14.5 in mice), the SMCs start to differentiate from mesenchymal cells surrounding ureter pelvis [5], [6]. Failure in differentiation of ureteral mesenchymal cells into SMCs results in obstruction and hydronephrosis with atrophy of kidney parenchyma [2], [7], [8]. Therefore, it is important to understand molecular factors regulating ureteral SMC differentiation during embryonic development.
Molecular regulation of ureteral SMC differentiation is extremely complex. Previous studies revealed several genes, including Tbx18, Six1, Sox9, Tszh3 and β-catenin, are essential for SMC differentiation during ureter development [2], [7]–[11]. TGF-β super family signals play vital roles in ureter development [10], [12], [13]. In mice, Bmp4 promotes ureter growth and elongation [10], [12]. Decreased Bmp4 signaling results in loss of ureteral smooth muscle formation [14]. As a central mediator in TGF-β signaling [15], Smad4 has been found crucial for vascular SMC differentiation and proliferation [16]. Deletion of Smad4 in the ureteral mesenchyme leads to a reduced number of SMCs [17]. However, whether Smad4 is essential for ureteral SMC differentiation remains unclear. In particular, the downstream genes through which Smad4 regulates ureter development are largely unknown. In this study, we generated Tbx18:Cre knock-in mice to ablate Smad4 in the ureteral mesenchyme. Our data revealed that Smad4 acts as upstream of several key genes associated with ureteral SMC differentiation, and plays critical roles for ureter development during mouse embryogenesis.
Materials and Methods
Animals
Smad4:floxed (denoted as Smad4f/f), Rosa26:tdTomato (denoted as Rosa26tdTomato), and Tbx18:nlacZ (denoted as Tbx18nlacZ/+) mice were described previously [18]–[20]. A new Tbx18:Cre (denoted as Tbx18Cre/+) mouse model was generated by inserting a Cre-polyA- FRT-Neo-FRT cassette into the start codon of Tbx18 locus, with disruption of endogenous ATG. Long range PCR was applied to screen targeted ES cells with 5′ primer P1: 5′-GTGTCCCTGAGTTCAGCTGACTGC-3′ and 3′ primer P2: 5′-CCGGTTATTCAACTTGCACCATGC-3′ (Fig. S1 A). Fragment amplified from the positive ES cells was further confirmed by DNA sequencing. Tbx18Cre-FRT-Neo-FRT/+ mice generated from positive ES cells were crossed to Flippase mice [21] to produce Tbx18Cre/+ animals (Neo cassette is removed) (Fig. S1). Smad4f/f mice were bred with Tbx18Cre/+ mice to generate Tbx18Cre/+; Smad4f/+ doubly heterozygous mice. Mutant Tbx18Cre/+; Smad4f/f mice were obtained by mating Tbx18Cre/+; Smad4f/+ with Smad4f/f mice. Genomic DNA was prepared from yolk sacs or tail biopsies for genotyping. Cervical dislocation and carbon dioxide inhalation were applied to euthanize mice. Mouse husbandry was carried out according to an approved IACUC protocol at the Icahn School of Medicine at Mount Sinai (Permit LA09-00494), and is in compliance with institutional and governmental regulation (PHS Animal Welfare Assurance A3111-01).
X-gal staining
Ureters from Tbx18nlaZ/+ mouse were checked for β-galactosidase activity by X-gal staining. Mouse ureters were treated with fixation solution (4% paraformaldehyde in PBS) for 30 min at 4°C. The fixed ureters were washed twice with PBS and then stained with X-gal solution (5 mM Potassium Ferricyanide, 5 mM Potassium Ferrocyanide, 2 mM MgCl2, 1 mg/ml X-gal) for 12 hours at room temperature. Tissues were visualized with a Leica steromicroscope.
Histology
Mouse ureters were washed in PBS and fixed with 4% paraformaldehyde overnight at 4°C, dehydrated in an ascending ethanol series (25%, 50%, 75%, 100%) followed by two changes of 100% xylene. The tissues were then immersed in liquid paraffin for 2 hours and left on a cold plate until wax was solidified. Paraffin blocks were cut into 6 µm in thickness on a microtome. The sections were stained with Hematoxylin and Eosin (H&E) using standard procedures.
Immunofluorescence and RNA in situ hybridization
Mouse ureters were fixed in 4% paraformaldehyde for 30 min and embedded in Optimal Cutting Temperature compound (Tissue-Tek). Frozen samples for immunohistochemistry were cut into 6 µm in thickness. Primary antibodies used in this study were as follows: rabbit anti-Smad4 (1∶100, Millipore), rabbit anti-Sox9 (1∶300, Millipore), mouse anti-αSMA (1∶100, Sigma), rabbit anti-SM-MHC (1∶100, Biomedical Technologies), rabbit anti-Uroplakin (1∶100, a generous gift from Dr. Tung-Tien Sun, NYU) [22]. Alexa Flour 488 or 594 conjugated secondary antibodies (1∶500; Invitrogen) were applied to detect the corresponding primary antibodies. Section RNA in situ hybridization was carried out on 12-µm cryosections with methods described previously [23], [24].
Proliferation and apoptosis analysis
For cell proliferation assay, pregnant mice were intraperitoneally injected with 10 mM EdU (Invitrogen) in PBS (5 mg per 100 g body weight). Embryos were harvested 4 hours later and fixed in 4% paraformaldehyde for 30 min at 4°C, and were embedded in OCT compound. Cell proliferation was assessed on 6 µm frozen sections using Click-iT EdU Cell Proliferation Assay Kit (Invitrogen). To quantify cell proliferation, 8 ureter sections were prepared from each embryo, and sections are from comparable locations in the control and mutant. Two embryos of each genotype were analyzed. Proliferation index was calculated as the percentage of EdU positive cells relative to ureteral mesenchymal cells and ureteric epithelial cells, respectively. Apoptosis assay was performed on 6 µm frozen sections using in situ Cell Death Detection Kit (Roche) according to the manufacturer's instructions. Nine representative ureter sections from each of three embryos with control or mutant genotype at comparable locations were used for apoptosis assay. Apoptosis index was calculated as percentage of TUNEL positive cells relative to ureteral mesenchymal cells.
Quantitative real-time PCR
Total RNA was isolated from ureters with Trizol reagent (Invitrogen) according to the manufacturer's instructions. QuantiTect Reverse Transcription Kit (Qiagen) was used to synthesize First-strand cDNA from total RNA. Quantitative PCR was performed using StepOnePlus PCR system and SYBR green detector (Qiagen). All transcripts were normalized to β-actin. The relative amounts of mRNA were calculated using the comparative Ct (threshold cycle) method. Primers used in this study are listed in Table 1. Data are presented as mean ± SD. Statistical analysis was performed by t-test and a value of p<0.05 was considered significant.
Table 1. Primer sequences for quantitative PCR.
| Gene | Primer sequence (5′-3′) |
| Myocd | CATTCGCCTTTGAGGATGAC |
| CTGAGCCAGGAGTGAGATCC | |
| Acta2 | GAGGCACCACTGAACCCTAA |
| CATCTCCAGAGTCCAGCACA | |
| Myh11 | AACACAGACCAGGCATCCAT |
| CTTTGGTCTGAGCCTTCTGC | |
| Mycn | AGCACCTCCGGAGAGGATAC |
| CGCACAGTGATCGTGAAAGT | |
| Sox9 | TGCAGCACAAGAAAGACCAC |
| CCCTCTCGCTTCAGATCAAC | |
| Tbx18 | TGGGGAGACTTGGATGAGAC |
| GGACAGATCATCTCCGCAAT | |
| Tshz3 | AAGCATCATGCCGAGGAG |
| CTCCATCTGCCGCTTGTT | |
| Ptch1 | TACGTGGAGGTGGTTCATCA |
| AGGCATAGGCAAGCATCAGT | |
| β-actin | TGTTACCAACTGGGACGACA |
| GGGGTGTTGAAGGTCTCAAA |
Results
Tbx18Cre/+ mice mediate specific recombination in the ureteral mesenchyme and SMCs
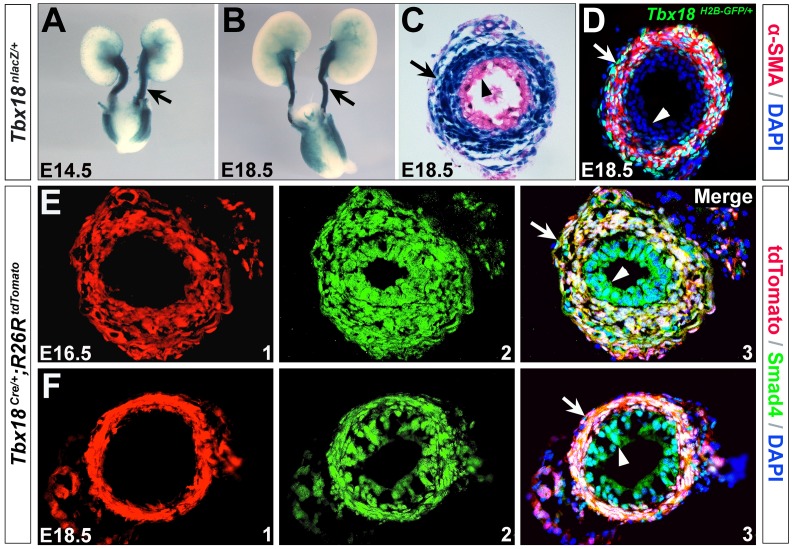
Previous studies showed that nuclear lacZ (nlacZ) expression in the Tbx18nlacZ/+ mice, and H2B-GFP expression in the Tbx18H2B-GFP/+ mice faithfully recapitulated endogenous Tbx18 expression in various organs, including the heart, limb, somite and hair follicle during embryogenesis in mice [19], [25], [26]. We utilized Tbx18nlacZ/+ and Tbx18H2B-GFP/+ mice to examine Tbx18 expression in the developing ureter during mouse embryogenesis. X-gal staining revealed that Tbx18 is highly expressed in the ureter tube at E14.5-18.5 (Fig. 1A-C). Immunostaining indicated Tbx18 is confined to the developing ureteral mesenchyme and SMCs (Fig. 1D). Given the specific expression of Tbx18 during embryogenesis, we generated Tbx18Cre-FRTNeoFRT/+ knock-in mice by inserting a Cre-polyA-FRT-Neo-FRT cassette into the start codon of Tbx18 locus (Fig. S1). Tbx18Cre/+ mice were obtained by removing the Neo cassette through crossing Tbx18Cre-FRTNeoFRT/+ mice to Flippase mice [21] (Fig. S1). Subsequently, we determined Tbx18 cell fates by crossing Tbx18Cre/+ mice to Rosa26tdTomato reporter line [20]. Lineage analysis of Tbx18Cre/+; Rosa26tdTomato/+ doubly heterozygous animals revealed that Tbx18 progeny encompass ureteral mesenchyme and its corresponding smooth muscles, but not epithelial cells (urothelium) (Fig. 1E,F).
Figure 1. Expression and lineage analysis of Tbx18 in the developing ureters.
(A-C) Tbx18 expression in the developing ureters. A and B are whole-mount X-gal staining of Tbx18nlacZ/+ urinary system at E14.5 and E18.5, respectively. C is a transverse section of the ureter tube at E18.5. (D) Immunostaining of α-SMA (red) on Tbx18H2BGFP/+ ureter (transverse section) at E18.5. (E,F) Immunofluorescence analysis of Smad4 with Tbx18 lineage on Tbx18Cre/+;Rosa26tdTomato ureter at E16.5 and E18.5. Arrowheads indicate urothelium and arrows indicate ureter tube (A and B) or ureteral SMCs (C-F).
Disruption of Smad4 in the ureteral mesenchyme causes hydroureter and hydronephrosis
To investigate the potential role of Smad4 in ureter development, we first performed immunostaining and found that Smad4 is universally expressed in both ureteral mesenchyme and epithelial cells, and is co-expressed with Tbx18 lineages in the mesenchyme and SMCs (Fig. 1E,F, and data not shown for E13.5-E15.5). We examined Tbx18Cre/+ ureters found they developed normally without defects at birth (P0) when compared to their wild type littermates (Fig. S2A,B). Further examination of Tbx18 mRNA expression in the ureters by quantitative RT-PCR (qRT-PCR) showed no significant difference between the wild type and Tbx18Cre/+ mice at this stage (Fig. S2C).
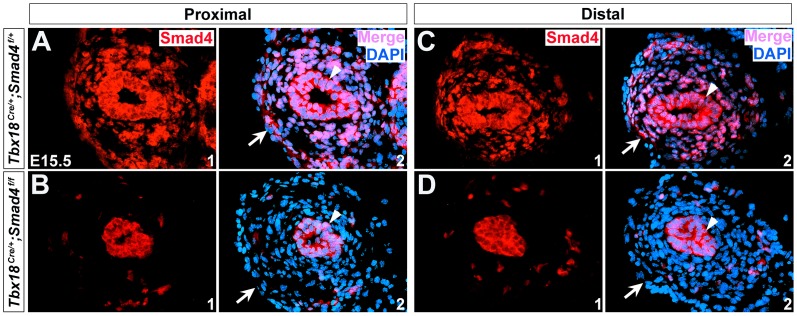
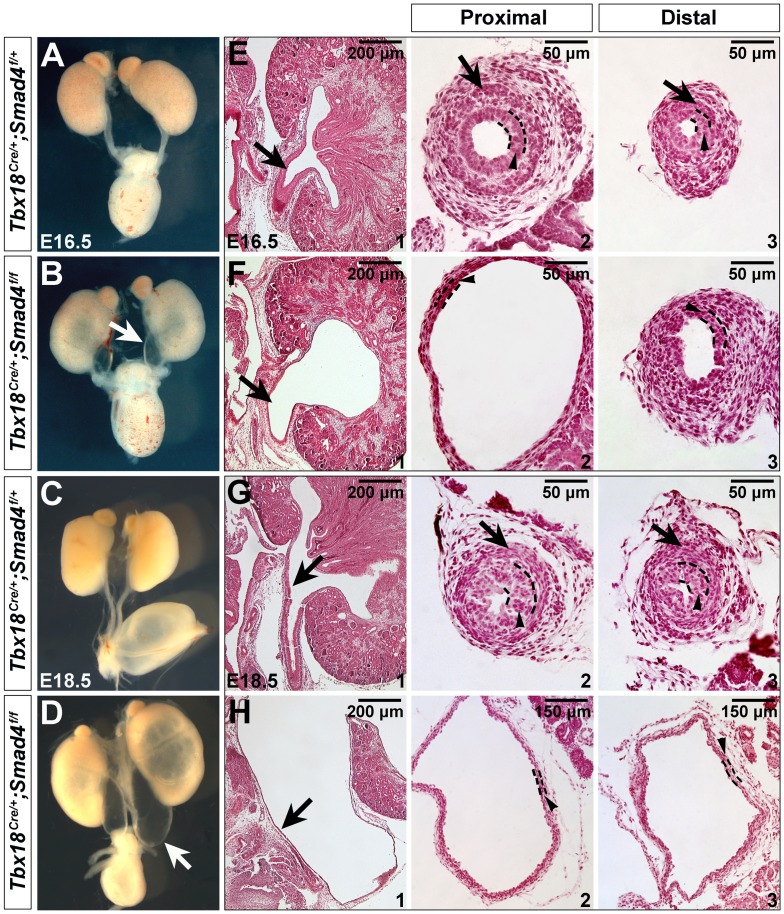
We crossed Tbx18Cre/+;Smad4f/+ double heterozygous mice to Smad4f/f mice and Tbx18Cre/+; Smad4f/f mutant animals (denoted as Smad4CKO) were collected for analysis. Littermates with Tbx18Cre/+;Smad4f/+ genotype were utilized as controls. To determine if Tbx18Cre/+ mediates efficient recombination in the ureteral mesenchyme and SMCs, we performed immunostaining in the mutant tissues. It showed that Smad4 expression was absent in the ureteral mesenchyme and smooth muscles (notched arrows in Fig. 2, data not shown for E14.5), but was unaffected in ureteric epithelium (arrowheads in Fig. 2A-D), suggesting that Smad4 was specifically disrupted in ureteral mesenchyme and SMCs on Smad4CKO embryos. At E16.5, Smad4CKO embryos displayed a prominent bilateral hydroureter and hydronephrosis (Fig. 3A, B), and the defects became more severe at E18.5 (Fig. 3C,D). Histological analysis of these embryos showed dilated and thin-walled ureters with renal parenchymal atrophy and dilation of the renal pelvis (Fig. 3E-H). The mutant mice cannot survive beyond 24 hours after birth, whereas control mice with Tbx18Cre/+; Smad4f/+ genotype develop normally and can survive to adulthood without any apparent morphogenetic defects.
Figure 2. Inactivation of Smad4 in Smad4CKO ureter.
(A-D) Immunostaining of Smad4 in the control (A and C, Tbx18Cre/+;Smad4f/+) and mutant ureters (B and D, Smad4CKO) in the proximal and distal positions at E15.5. Arrowheads indicate urothelium and arrows indicate ureteral mesenchyme and SMCs.
Figure 3. Disruption of Smad4 leads to hydroureter and hydronephrosis.
(A-D) Morphology of urinary system in the control and mutant mice at E16.5 (A and B) and E18.5 (C and D). (E-H) Hematoxylin and Eosin staining of kidney sagittal sections (E1/F1/G1/H1), and transverse sections of ureter tube in the proximal (E2/F2/G2/H2) and distal positions (E3/F3/G3/H3) at E16.5 and E18.5. Arrows in B/D and E1/F1/G1/H1 indicate ureter tube, and in E2/F2/G2/H2 and E3/F3/G3/H3 indicate ureteral SMCs. Arrowheads indicate urothelium.
Smad4 is essential for ureteral mesenchymal cell differentiation
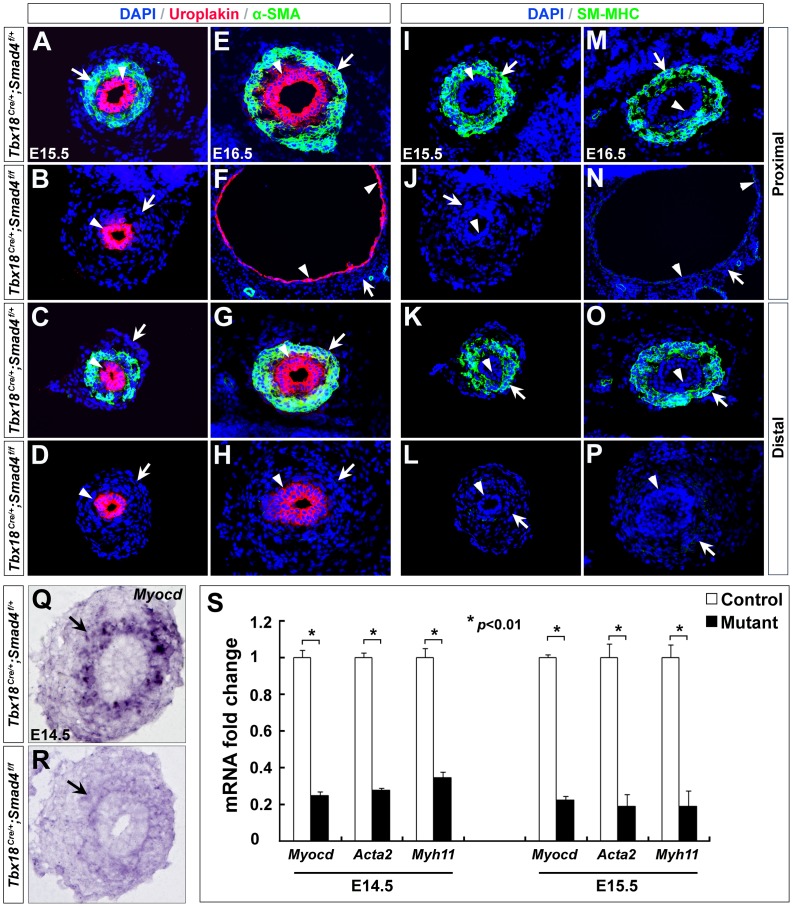
SMCs are essential for ureter peristalsis that propels urine towards bladder. Previous studies suggested that hydronephrosis and hydroureter could primarily result from malformed SMCs [2], [7], [8]. To determine whether the hydronephrosis and hydroureter defects in Smad4CKO embryos were due to insufficient differentiation of the ureteral SMCs, we examined mutant ureters with SMC markers. During embryonic development, ureteral SMC differentiation is characterized by increased expression of contractile SMC markers, such as α-SMA and SM-MHC [27], from E14.5 to E16.5. While we found α-SMA and SM-MHC were highly expressed in the ureteral SMCs at E15.5 and E16.5 in the controls, they could be barely detected in the mutant embryos (notched arrows in Fig. 4A-P). Similar observations were made for Myocardin (Myocd), a key regulator and marker of SMC differentiation, on E14.5 mutant ureters (notched arrows in Fig. 4Q,R). Further qRT-PCR showed that Myocd, Acta2 and Myh11 were down-regulated in the Smad4CKO ureters by 75%, 72% and 65% at E14.5, and by 78%, 81% and 81% at E15.5, respectively (Fig. 4S, p<0.01). Moreover, we investigated whether Smad4 deficiency in the ureteral mesenchyme also affects urothelium formation. Uroplakin, a marker for urothelium, was still expressed in the mutant embryos, suggesting that urothelial development was unaffected in Smad4CKO mice (arrowheads in Fig. 4A-H).
Figure 4. SMC differentiation is perturbed in Smad4CKO ureters.
(A-H) Immunostaining of α-SMA and Uroplakin in the control (A/C/E/G) and mutant ureters (B/D/F/H) at E15.5 and E16.5. Arrowheads indicate Uroplakin (urothelium) and arrows indicate α-SMA staining (SMCs). (I-P) Immunostaining of SM-MHC in the control (I/K/M/O) and mutant embryos (J/L/N/P) at E15.5 and E16.5. Arrowheads indicate urothelium and arrows indicate SMCs. (Q,R) RNA in situ hybridization revealed Myocd expression is downregulated in the mutants (arrows) at E14.5. (S) Relative expression of Myocd, Acta2 and Myh11 was determined by qRT-PCR in the mutant and control littermate ureters at E14.5 and E15.5.
Increased apoptosis and decreased proliferation in Smad4CKO ureteral mesenchyme
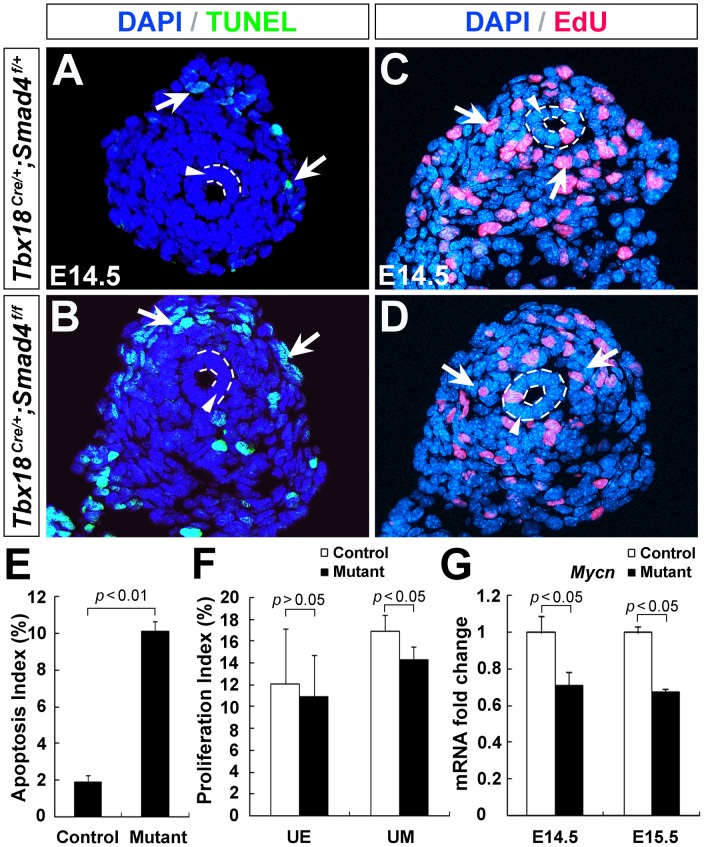
Cell apoptosis and proliferation are highly associated with hydroureter during ureter development [2], [5], [28], [29]. We examined the effect of Smad4 deletion on cell apoptosis and proliferation in the ureteral mesenchyme. TUNEL analysis revealed no change of apoptosis in ureteric epithelium between control and mutant at E14.5. In contrast, we found a increased number of apoptotic cells in the mutant ureteral mesenchyme (Fig. 5A,B,E, control: 1.88±0.38%; mutant: 10.1±0.58%; p<0.01). Moreover, we assessed mesenchymal cell proliferation by EdU incorporation. The ureteral mesenchymal cell proliferation rate was ∼15% less in the Smad4CKO mutants at E14.5 (control: 16.93±1.44%; mutant: 14.32±1.22%; p<0.05), yet cell proliferation rate in the epithelium was unaffected (Fig. 5C,D,F, p>0.05). Consistent with these observations, expression of Mycn, a pro-proliferative factor, was significantly reduced in Smad4CKO ureters at E14.5 and E15.5 (Fig. 5G, p<0.05).
Figure 5. Apoptosis and cell proliferation in Smad4CKO ureters.
(A, B) Apoptosis was assessed by TUNEL. Arrows indicate TUNEL positive cells. (C, D) Cell proliferation was analyzed by EdU labeling. Arrows indicate proliferating cells. (E) Statistical analysis of TUNEL positive cells. (F) EdU positive cells were quantified. (G) Relative Mycn mRNA expression was measured by qRT-PCR.
Sox9 and Tbx18 expression is down-regulated in Smad4CKO ureter
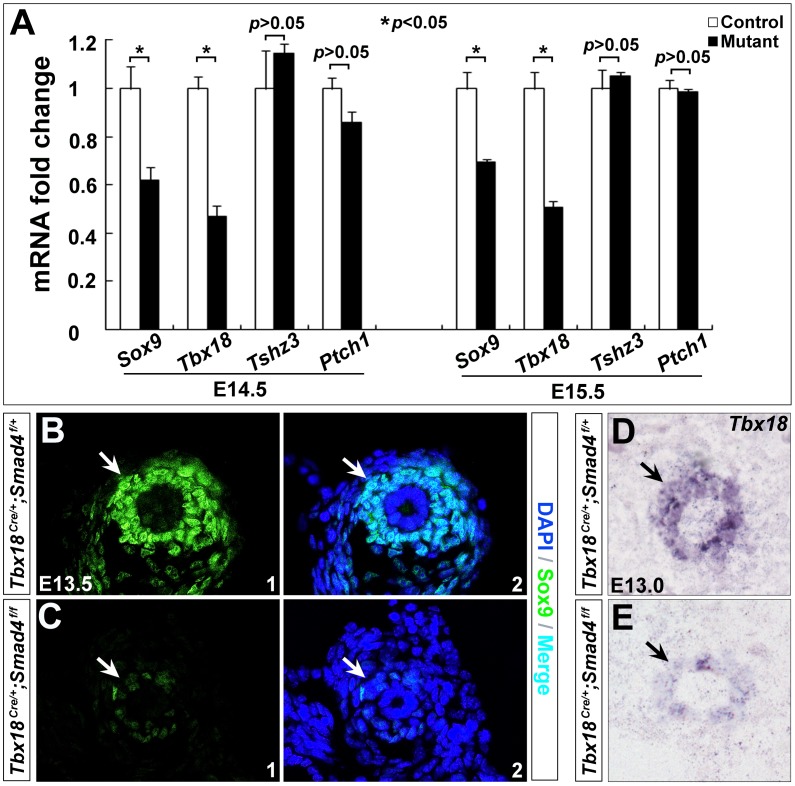
We next performed qRT-PCR to identify genes as potential downstream of Smad4 signaling for ureteral SMC differentiation. Given that Sox9, Tbx18, Tszh3 and Shh have been shown important in regulating ureteral SMC differentiation [2], [5], [7], [8], we assessed the effect of Smad4 deficiency on these genes during mouse embryonic development. qRT-PCR revealed ∼38% reduction in Sox9 and ∼53% reduction in Tbx18 on the mutant ureters at E14.5, and ∼31% reduction in Sox9 and ∼50% reduction in Tbx18 at E15.5 (p<0.05), yet Tszh3 and Ptch1 (receptor of Shh) expression was not significantly changed (Fig. 6A, p>0.05). Further immunostaining and RNA in situ hybridization showed that Sox9 was down-regulated in the mutant ureteral mesenchyme as early as E13.5 (Fig. 6B,C), and Tbx18 mRNA was significantly decreased in the mutant mesenchyme at E13.0 (Fig. 6D,E). These data suggest that Sox9 and Tbx18 may act as downstream of Smad4 to govern ureteral smooth muscle differentiation during mouse embryogenesis.
Figure 6. Reduced Sox9 and Tbx18 expression in Smad4CKO ureters.
(A) qRT-PCR was performed to determine expression of Sox9, Tbx18, Tshz3 and Ptch1. (B,C) Immunostaining of Sox9 in control (B) and mutant embryos (C) at E13.5. (D,E) RNA in situ hybridization of Tbx18 in the control (D) and mutant embryos (E) at E13.0, respectively. Arrows indicate positive staining of cells in the controls with corresponding regions in the mutants.
Discussion
Although a few genes (e.g., Tbx18, Sox9, Tszh3 and β-catenin) have been identified crucial for ureter development [2], [7]–[9], the interaction network of these genes with other factors in regulating ureteral SMC development remains to be determined. In this study, we found loss of Smad4 in the ureteral mesenchyme led to failure of SMC differentiation, resulting in severe hydroureter and hydronephrosis during mouse embryogenesis. Smad4 acts as upstream of Sox9, Tbx18 and Myocardin, and is required for their normal expressions in ureter development [2], [8].
Tbx18Cre/+ mouse model as a robust genetic tool to study ureteral mesenchyme and SMC development
The expression analysis of Tbx18 with Tbx18nlacZ/+ and Tbx18H2B-GFP/+ mice revealed that Tbx18 was specifically expressed in the ureteral mesenchyme and SMCs (Fig.1A-D), consistent with the RNA in situ hybridization[2]. Lineage tracing experiments with Tbx18Cre/+; Rosa26tdTomato animals revealed Tbx18 progeny include undifferentiated ureteral mesenchyme and its SMC derivatives, but not ureteric epithelium. Therefore, the Tbx18Cre/+ animal could be utilized as an excellent genetic tool to dissect functions of genes in the developing ureteral mesenchyme and/or SMCs during mouse embryogenesis with Cre-loxP technology. Comparing to Tbx18Cre/+ knock-in mice, Pax3-Cre transgenic line introduces recombination in ureteral mesenchyme (including SM layers and the adventitia) and metanephric mesenchyme (including the glomeruli and the proximal and distal tubules) [28]. The BAC transgenic Tbx18-Cre line was also generated [30]. However, it may not fully recapitulate endogenous Tbx18 expression thus it is not clear whether it introduces effective recombination in all the ureteral mesenchymal cells. Evidence in supporting this argument is thatTbx18-Cre mouse seems not able to mediate effective recombination in the somites of mouse embryos [30].
Smad4 is required for ureteral SMC differentiation from mesenchyme
Previous studies demonstrated that Bmp4 plays an important role in ureter development [10], [12], [31]. Smad4 is a central mediator of TGF-β/Bmp signaling pathway [32], [33] and it regulates morphogenesis of various organs during embryonic development [34]–[36]. Our study showed that Smad4 is expressed in both ureteral mesenchyme and epithelium during ureter development. Mice with genetic deletion of Smad4 in ureteral mesenchyme displayed hydroureter and hydronephrosis as early as E16.5, and the mutant mesenchymal cells failed to differentiate into SMCs at E15.5. Myocd, a critical transcription factor required for expression of SMC contractile proteins (αSMA, SM22α and SM-MHC) [37], is dramatically down-regulated in the mutant ureters. Ureter SMC developmental defects are highly associated with defective cell proliferation and survival [9], [10], [28]. In Smad4CKO embryos, we also detected less proliferative ureteral mesenchymal cells with apoptosis at E14.5. The increased mesenchymal cell death in Smad4CKO mice may be attributed to the reduced Myocd expression, given that Myocd is required for cell survival [38]. Furthermore, loss of Smad4 resulted in down-regulation of Mycn in the ureter. Mycn (Nmyc) is a Myc transcription factor and it plays critical roles in cell proliferation [16], [39], [40]. In the developing hearts, Mycn is a direct downstream target of Smad4 and inactivation of Smad4 down-regulates Mycn expression [39]. Mycn may play a key role in Smad4-mediated signaling cascades for the ureteral mesenchymal cell proliferation. Collectively, these observations suggested that Smad4 signals are crucial for SMC differentiation, proliferation and survival during ureter development.
A recent study has attempted to disrupt Smad4 with Tbx18-Cre transgenic line, and the mutant embryos displayed hydronephrosis at E17.5 [17]. The defects in these animals were not as severe as our Smad4CKO mice: although the ureteral SMC number was reduced, SMC differentiation still occurred and thickness of smooth muscle layer was not affected. Moreover, no significant difference was found in cell survival and proliferation in the mesenchyme or SMCs. The discrepancies may be due to Cre excision efficiency: Tbx18-Cre transgenic line may not able to mediate efficient excision in the ureteral mesenchymal cells [17]. Some of the mesenchymal cells in the mutant ureters may still maintain Smad4 expression and thus develop normally into SMCs. Tbx18Cre/+ knock-in mouse line used in our study can mediate efficient ablation in almost all the ureteral mesenchymal cells (Fig. 2A-D), and therefore SMC differentiation does not occur in Smad4CKO ureters.
Sox9 and Tbx18 act as downstream of Smad4 during ureteral mesenchymal cell development
Genetic regulation of ureteral SMC differentiation is a complex process. Pathways underlying this process are still largely to be determined. To date, a few genes, including Sox9 and Tbx18, were identified as key regulators for ureteral SMC differentiation [2], [8]. Sox9 is a transcription factor highly expressed in ureteral mesenchyme at E12.5 and E13.5 [41]–[43]. Deletion of Sox9 in ureteral mesenchyme results in hydroureter and hydronephrosis [8], mimicking defects in Smad4CKO embryos (Fig. 3A-D). Mesenchymal cells in Sox9−/− ureters do not differentiate into SMCs [8]. Sox9 and Tshz3 regulate smooth muscle differentiation program by controlling the activity of Myocd [44], a key regulator for SMC differentiation [45]–[47]. Previous studies also showed that Smad4 regulates Sox9 expression in limb and heart development [48], [49]. In Smad4CKO embryos, we also found Sox9 expression was down-regulated (Fig. 6A-C). Tbx18 is a transcription factor highly expressed in ureteral mesenchyme and is essential for SMC differentiation [2], [50]. Loss of Tbx18 in mice results in defective mesenchymal cell proliferation and SMC differentiation [2], similar to Smad4CKO embryos. Disruption of Smad4 down-regulates Tbx18 expression (Fig. 6A,D,E). Based on these observations, we speculate that Smad4 may act as upstream of both Sox9 and Tbx18, and that Smad4-Sox9/Tbx18 signaling cascades may regulate ureteral mesenchymal cell development through Myocd.
In addition, microRNA-21 (miR-21) has been shown to promote vascular SMC differentiation by inducing expression of SMC contractile proteins including αSMA, calponin and SM22a [51]. MiR-21 also regulates vascular SMC growth and survival by silencing expression of PTEN and increasing expression of BCL2 [52]. TGF-β and Bmps (Bmp4) signaling elevates the expression of mature miR-21 [51], [53]. It is not certain whether miR-21 mediates ureteral SMC differentiation as downstream of Smad4, although miR-21 biogenesis in the vascular SMCs is only regulated by ligand-specific Smad proteins [51].
In the future, it will be of interest to determine whether Smad4 directly activates the expression of Sox9/Tbx18/Myocd by binding their regulatory elements, and whether miR-21 is affected by Smad4 during ureteral development. Furthermore, given the critical roles of Smad4 in ureter development, it will be also of interest to perform genetic studies to determine whether Smad4 and its associated downstream genes Sox9, Tbx18, and Myocd are mutated in human patients with congenital urinary tract malformations.
Supporting Information
Generation of Tbx18Cre/+ knock-in mouse. (A) Schematic representation of targeting strategy. A Cre-polyA-FRT-Neo-FRT cassette was introduced into the Tbx18 genomic locus (6 bp upstream of the ATG). The Neo cassette is flanked by two FRT sites. Tbx18Cre-FRT-Neo-FRT/+ mice were generated from the positive ES cells. Flippase deleter mice were crossed to Tbx18Cre-Neo mice to remove Neo cassette. (B) Long range PCR analysis of genomic DNA from targeted ES cells. A 4.2-kb fragment was amplified with 5′ primer external to the 5′ arm (P1) and 3′ primer within Neo cassette (P2).
(TIF)
Normal ureter development in Tbx18Cre/+ mice. (A,B) Comparisons of the urinary system from Tbx18Cre/+ knock-in mice and their wild type littermates at birth (P0). (C) Tbx18 mRNA expression in ureters measured by qRT-PCR does not show significant difference between Tbx18Cre/+ and wild type mice. β-actin was used as an internal reference gene.
(JPG)
ARRIVE Checklist.
(DOC)
Acknowledgments
The authors thank Dr. Chuxia Deng (NIDDK, USA) for his generosity in providing Smad4f/f mice, and Dr. Tung-Tien Sun (NYU) for providing Uroplakin antibody in this study.
Funding Statement
This study is supported by grants to C.L.C. from the NHLBI (1R01HL095810 and 1K02HL094688), the American Heart Association (0855808D) and the March of Dimes Foundation (5-FY07-642), and grant to P-X.X. from the NIDDK (R01 DK064640). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Pope JC 4th, Brock JW 3rd, Adams MC, Stephens FD, Ichikawa I (1999) How they begin and how they end: classic and new theories for the development and deterioration of congenital anomalies of the kidney and urinary tract, CAKUT. J Am Soc Nephrol 10: 2018–2028. [DOI] [PubMed] [Google Scholar]
- 2. Airik R, Bussen M, Singh MK, Petry M, Kispert A (2006) Tbx18 regulates the development of the ureteral mesenchyme. J Clin Invest 116: 663–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Chen F (2009) Genetic and developmental basis for urinary tract obstruction. Pediatr Nephrol 24: 1621–1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Woolf AS, Davies JA (2013) Cell biology of ureter development. J Am Soc Nephrol 24: 19–25. [DOI] [PubMed] [Google Scholar]
- 5. Yu J, Carroll TJ, McMahon AP (2002) Sonic hedgehog regulates proliferation and differentiation of mesenchymal cells in the mouse metanephric kidney. Development 129: 5301–5312. [DOI] [PubMed] [Google Scholar]
- 6. Airik R, Kispert A (2007) Down the tube of obstructive nephropathies: the importance of tissue interactions during ureter development. Kidney Int 72: 1459–1467. [DOI] [PubMed] [Google Scholar]
- 7. Caubit X, Lye CM, Martin E, Core N, Long DA, et al. (2008) Teashirt 3 is necessary for ureteral smooth muscle differentiation downstream of SHH and BMP4. Development 135: 3301–3310. [DOI] [PubMed] [Google Scholar]
- 8. Airik R, Trowe MO, Foik A, Farin HF, Petry M, et al. (2010) Hydroureternephrosis due to loss of Sox9-regulated smooth muscle cell differentiation of the ureteric mesenchyme. Hum Mol Genet 19: 4918–4929. [DOI] [PubMed] [Google Scholar]
- 9. Trowe MO, Airik R, Weiss AC, Farin HF, Foik AB, et al. (2012) Canonical Wnt signaling regulates smooth muscle precursor development in the mouse ureter. Development 139: 3099–3108. [DOI] [PubMed] [Google Scholar]
- 10. Miyazaki Y, Oshima K, Fogo A, Hogan BL, Ichikawa I (2000) Bone morphogenetic protein 4 regulates the budding site and elongation of the mouse ureter. J Clin Invest 105: 863–873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Nie X, Sun J, Gordon RE, Cai CL, Xu PX (2010) SIX1 acts synergistically with TBX18 in mediating ureteral smooth muscle formation. Development 137: 755–765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Miyazaki Y, Oshima K, Fogo A, Ichikawa I (2003) Evidence that bone morphogenetic protein 4 has multiple biological functions during kidney and urinary tract development. Kidney Int 63: 835–844. [DOI] [PubMed] [Google Scholar]
- 13. Bush KT, Sakurai H, Steer DL, Leonard MO, Sampogna RV, et al. (2004) TGF-beta superfamily members modulate growth, branching, shaping, and patterning of the ureteric bud. Dev Biol 266: 285–298. [DOI] [PubMed] [Google Scholar]
- 14. Wang GJ, Brenner-Anantharam A, Vaughan ED, Herzlinger D (2009) Antagonism of BMP4 signaling disrupts smooth muscle investment of the ureter and ureteropelvic junction. J Urol 181: 401–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ten Dijke P, Arthur HM (2007) Extracellular control of TGFbeta signalling in vascular development and disease. Nat Rev Mol Cell Biol 8: 857–869. [DOI] [PubMed] [Google Scholar]
- 16. Mao X, Debenedittis P, Sun Y, Chen J, Yuan K, et al. (2012) Vascular smooth muscle cell Smad4 gene is important for mouse vascular development. Arterioscler Thromb Vasc Biol 32: 2171–2177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Tripathi P, Wang Y, Casey AM, Chen F (2012) Absence of canonical Smad signaling in ureteral and bladder mesenchyme causes ureteropelvic junction obstruction. J Am Soc Nephrol 23: 618–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Yang X, Li C, Herrera PL, Deng CX (2002) Generation of Smad4/Dpc4 conditional knockout mice. Genesis 32: 80–81. [DOI] [PubMed] [Google Scholar]
- 19. Cai CL, Martin JC, Sun Y, Cui L, Wang L, et al. (2008) A myocardial lineage derives from Tbx18 epicardial cells. Nature 454: 104–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Madisen L, Zwingman TA, Sunkin SM, Oh SW, Zariwala HA, et al. (2010) A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat Neurosci 13: 133–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Farley FW, Soriano P, Steffen LS, Dymecki SM (2000) Widespread recombinase expression using FLPeR (flipper) mice. Genesis 28: 106–110. [PubMed] [Google Scholar]
- 22. Deng FM, Ding M, Lavker RM, Sun TT (2001) Urothelial function reconsidered: a role in urinary protein secretion. Proc Natl Acad Sci U S A 98: 154–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wilkinson DG (1998) In Situ Hybridization: A Practical Approach. 2nd edition. New York: Oxford University Press.
- 24. Cai X, Zhang W, Hu J, Zhang L, Sultana N, et al. (2013) Tbx20 acts upstream of Wnt signaling to regulate endocardial cushion formation and valve remodeling during mouse cardiogenesis. Development 140: 3176–3187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kraus F, Haenig B, Kispert A (2001) Cloning and expression analysis of the mouse T-box gene Tbx18. Mech Dev 100: 83–86. [DOI] [PubMed] [Google Scholar]
- 26. Grisanti L, Clavel C, Cai X, Rezza A, Tsai SY, et al. (2013) Tbx18 targets dermal condensates for labeling, isolation, and gene ablation during embryonic hair follicle formation. J Invest Dermatol 133: 344–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Owens GK (1995) Regulation of differentiation of vascular smooth muscle cells. Physiol Rev 75: 487–517. [DOI] [PubMed] [Google Scholar]
- 28. Chang CP, McDill BW, Neilson JR, Joist HE, Epstein JA, et al. (2004) Calcineurin is required in urinary tract mesenchyme for the development of the pyeloureteral peristaltic machinery. J Clin Invest 113: 1051–1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Xu J, Qi X, Gong J, Yu M, Zhang F, et al. (2012) Fstl1 antagonizes BMP signaling and regulates ureter development. PLoS One 7: e32554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wang Y, Tripathi P, Guo Q, Coussens M, Ma L, et al. (2009) Cre/lox recombination in the lower urinary tract. Genesis 47: 409–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Brenner-Anantharam A, Cebrian C, Guillaume R, Hurtado R, Sun TT, et al. (2007) Tailbud-derived mesenchyme promotes urinary tract segmentation via BMP4 signaling. Development 134: 1967–1975. [DOI] [PubMed] [Google Scholar]
- 32. Massague J (1998) TGF-beta signal transduction. Annu Rev Biochem 67: 753–791. [DOI] [PubMed] [Google Scholar]
- 33. Kawabata M, Miyazono K (1999) Signal transduction of the TGF-beta superfamily by Smad proteins. J Biochem 125: 9–16. [DOI] [PubMed] [Google Scholar]
- 34. Zhou YX, Zhao M, Li D, Shimazu K, Sakata K, et al. (2003) Cerebellar deficits and hyperactivity in mice lacking Smad4. J Biol Chem 278: 42313–42320. [DOI] [PubMed] [Google Scholar]
- 35. Qi X, Yang G, Yang L, Lan Y, Weng T, et al. (2007) Essential role of Smad4 in maintaining cardiomyocyte proliferation during murine embryonic heart development. Dev Biol 311: 136–146. [DOI] [PubMed] [Google Scholar]
- 36. Ko SO, Chung IH, Xu X, Oka S, Zhao H, et al. (2007) Smad4 is required to regulate the fate of cranial neural crest cells. Dev Biol 312: 435–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Huang J, Cheng L, Li J, Chen M, Zhou D, et al. (2008) Myocardin regulates expression of contractile genes in smooth muscle cells and is required for closure of the ductus arteriosus in mice. J Clin Invest 118: 515–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Huang J, Min Lu M, Cheng L, Yuan LJ, Zhu X, et al. (2009) Myocardin is required for cardiomyocyte survival and maintenance of heart function. Proc Natl Acad Sci U S A 106: 18734–18739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Song L, Yan W, Chen X, Deng CX, Wang Q, et al. (2007) Myocardial smad4 is essential for cardiogenesis in mouse embryos. Circ Res 101: 277–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Eilers M, Eisenman RN (2008) Myc's broad reach. Genes Dev 22: 2755–2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Akiyama H, Chaboissier MC, Martin JF, Schedl A, de Crombrugghe B (2002) The transcription factor Sox9 has essential roles in successive steps of the chondrocyte differentiation pathway and is required for expression of Sox5 and Sox6. Genes Dev 16: 2813–2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Akiyama H, Chaboissier MC, Behringer RR, Rowitch DH, Schedl A, et al. (2004) Essential role of Sox9 in the pathway that controls formation of cardiac valves and septa. Proc Natl Acad Sci U S A 101: 6502–6507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Pritchett J, Athwal V, Roberts N, Hanley NA, Hanley KP (2011) Understanding the role of SOX9 in acquired diseases: lessons from development. Trends Mol Med 17: 166–174. [DOI] [PubMed] [Google Scholar]
- 44. Martin E, Caubit X, Airik R, Vola C, Fatmi A, et al. (2013) TSHZ3 and SOX9 Regulate the Timing of Smooth Muscle Cell Differentiation in the Ureter by Reducing Myocardin Activity. PLoS One 8: e63721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Wang Z, Wang DZ, Pipes GC, Olson EN (2003) Myocardin is a master regulator of smooth muscle gene expression. Proc Natl Acad Sci U S A 100: 7129–7134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Raphel L, Talasila A, Cheung C, Sinha S (2012) Myocardin overexpression is sufficient for promoting the development of a mature smooth muscle cell-like phenotype from human embryonic stem cells. PLoS One 7: e44052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Liu ZP, Wang Z, Yanagisawa H, Olson EN (2005) Phenotypic modulation of smooth muscle cells through interaction of Foxo4 and myocardin. Dev Cell 9: 261–270. [DOI] [PubMed] [Google Scholar]
- 48. Benazet JD, Pignatti E, Nugent A, Unal E, Laurent F, et al. (2012) Smad4 is required to induce digit ray primordia and to initiate the aggregation and differentiation of chondrogenic progenitors in mouse limb buds. Development 139: 4250–4260. [DOI] [PubMed] [Google Scholar]
- 49. Nie X, Deng CX, Wang Q, Jiao K (2008) Disruption of Smad4 in neural crest cells leads to mid-gestation death with pharyngeal arch, craniofacial and cardiac defects. Dev Biol 316: 417–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Bohnenpoll T, Bettenhausen E, Weiss AC, Foik AB, Trowe MO, et al. (2013) Tbx18 expression demarcates multipotent precursor populations in the developing urogenital system but is exclusively required within the ureteric mesenchymal lineage to suppress a renal stromal fate. Dev Biol 380: 25–36. [DOI] [PubMed] [Google Scholar]
- 51. Davis BN, Hilyard AC, Lagna G, Hata A (2008) SMAD proteins control DROSHA-mediated microRNA maturation. Nature 454: 56–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Ji R, Cheng Y, Yue J, Yang J, Liu X, et al. (2007) MicroRNA expression signature and antisense-mediated depletion reveal an essential role of MicroRNA in vascular neointimal lesion formation. Circ Res 100: 1579–1588. [DOI] [PubMed] [Google Scholar]
- 53. Kang H, Davis-Dusenbery BN, Nguyen PH, Lal A, Lieberman J, et al. (2012) Bone morphogenetic protein 4 promotes vascular smooth muscle contractility by activating microRNA-21 (miR-21), which down-regulates expression of family of dedicator of cytokinesis (DOCK) proteins. J Biol Chem 287: 3976–3986. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Generation of Tbx18Cre/+ knock-in mouse. (A) Schematic representation of targeting strategy. A Cre-polyA-FRT-Neo-FRT cassette was introduced into the Tbx18 genomic locus (6 bp upstream of the ATG). The Neo cassette is flanked by two FRT sites. Tbx18Cre-FRT-Neo-FRT/+ mice were generated from the positive ES cells. Flippase deleter mice were crossed to Tbx18Cre-Neo mice to remove Neo cassette. (B) Long range PCR analysis of genomic DNA from targeted ES cells. A 4.2-kb fragment was amplified with 5′ primer external to the 5′ arm (P1) and 3′ primer within Neo cassette (P2).
(TIF)
Normal ureter development in Tbx18Cre/+ mice. (A,B) Comparisons of the urinary system from Tbx18Cre/+ knock-in mice and their wild type littermates at birth (P0). (C) Tbx18 mRNA expression in ureters measured by qRT-PCR does not show significant difference between Tbx18Cre/+ and wild type mice. β-actin was used as an internal reference gene.
(JPG)
ARRIVE Checklist.
(DOC)