Abstract
In the past two decades advances in the development of targeted nanoparticles have facilitated their application as molecular imaging agents and targeted drug delivery vehicles. Nanoparticle-enhanced molecular imaging of the angiogenic tumor vasculature has been of particular interest. Not only because angiogenesis plays an important role in various pathologies, but also since endothelial cell surface receptors are directly accessible for relatively large circulating nanoparticles. Typically, nanoparticle targeting towards these receptors is studied by analyzing the contrast distribution on tumor images acquired before and at set time points after administration. Although several exciting proof-of-concept studies demonstrated qualitative assessment of relative target concentration and distribution, these studies did not provide quantitative information on the nanoparticle targeting kinetics. These kinetics will not only depend on nanoparticle characteristics, but also on receptor binding and recycling. In this study, we monitored the in vivo targeting kinetics of αvβ3-integrin specific nanoparticles with intravital microscopy and dynamic contrast enhanced magnetic resonance imaging, and using compartment modeling we were able to quantify nanoparticle targeting rates. As such, this approach can facilitate optimization of targeted nanoparticle design and it holds promise for providing more quantitative information on in vivo receptor levels. Interestingly, we also observed a periodicity in the accumulation kinetics of αvβ3-integrin targeted nanoparticles and hypothesize that this periodicity is caused by receptor binding, internalization and recycling dynamics. Taken together, this demonstrates that our experimental approach provides new insights in in vivo nanoparticle targeting, which may proof useful for vascular targeting in general.
Keywords: DCE-MRI, Intravital microscopy, Targeted nanoparticles, Targeting kinetics, Vascular targeting
Introduction
The application of nanoparticles as targeted contrast agents in molecular imaging and image-guided drug delivery has matured to an established research field [1, 2]. Several papers have reported exciting proof-of-concept studies [3-6], but detailed knowledge on in vivo nanoparticle-target interactions and its effect on nanoparticle accumulation and in vivo detection remains limited. Such knowledge would not only contribute to the field of nanoparticle-enhanced molecular imaging, but will have a much broader impact on the nanoparticle targeting field in general and may contribute to pave the way for the clinical translation of nanomedicines [7-9].
The conventional way of studying target-specific nanoparticle contrast agents in vivo is by acquiring images of the pathological site before and at set time points after administration and subsequently evaluating contrast enhancement distribution patterns. An example of this approach was described by Mulder et al. [4]. While this approach allows for qualitative assessment of relative target concentration and distribution, it does not provide quantitative insight in the nanoparticle targeting kinetics and accumulation rates. These kinetics will not only depend on nanoparticle characteristics, like target affinity and pharmacokinetics, but also on receptor binding and recycling kinetics [10]. Since little is known about these targeting kinetics and how they affect nanoparticle accumulation and detection, their real-time in vivo monitoring could provide valuable information on nanoparticle targeting, but also on the properties of the molecular target itself.
One conventional dynamic imaging technique, which is extensively used in both pre-clinical and clinical studies, is dynamic contrast-enhanced magnetic resonance imaging (DCE-MRI) [11, 12]. In this technique the signal enhancement in a tissue is recorded as a function of time during and after a bolus-injection of an MRI contrast agent. Quantification of contrast agent dynamics through compartment modeling provides quantitative information on a variety of features of the pathological tissue, such as vascular volume and permeability [13-15]. Moreover, DCE-MRI and compartment modeling may also be employed to study the in vivo accumulation kinetics and targeting rates of ligand-functionalized nanoparticles.
One focus in in vivo nanoparticle targeting lies on the targeting of endothelial cell surface receptors. Not only because levels of these receptors are significantly altered in several pathological processes, but also because they are readily accessible for relatively large circulating nanoparticles. For example, vascularization and angiogenesis are crucial for solid tumor growth and therefore important diagnostic and therapeutic targets in oncology [16-18]. One target that has been extensively studied in this context is αvβ3-integrin, which is highly up-regulated on angiogenic endothelial cells [19-21]. αvβ3-integrin is a member of the integrin family which consists of 24 known trans-membrane α-β heterodimers [19, 22]. Their major role on endothelial cells is coupling to the extracellular matrix and neighboring cells, facilitating signaling, cell survival and migration [19, 22]. Recent in vitro studies with human endothelial cells have shown that αvβ3-integrin specific nanoparticles are internalized [4, 23], while at the same time this integrin is known to be recycled back to the plasma membrane after internalization [22, 24].
In the present work we used intravital microscopy (IVM) and DCE-MRI to study the tumor vasculature targeting kinetics of c(Asp-(d)Phe-Lys(S-Ata)-Arg-Gly) (RGD) conjugated nanoemulsion (RGD nanoemulsion). Conjugation of this cyclic pentapeptide to nanoparticles results in high affinity and selectivity towards αvβ3-integrin [25]. As control a non-targeted nanoemulsion (CTRL nanoemulsion) was used, which is known to accumulate in tumor tissue through a non-specific mechanism termed the enhanced permeability and retention (EPR) effect [26, 27]. In a recent study we have optimized the RGD nanoemulsion to most effectively target the tumor vasculature [26]. Our experimental set-up with a window chamber model and bimodal (MR and optical) nanoparticles offers a unique platform to assess different aspects of the in vivo nanoemulsion as well as αvβ3-integrin behavior [26]. A special feature in this study was the use of IVM in the window chamber model to obtain high quality vascular input functions (VIFs) for the DCE-MRI contrast agent kinetic processing.
Using the above mentioned experimental set-up we were able to monitor nanoemulsion accumulation dynamics in the tumor in real-time and quantify nanoparticle targeting rates. Interestingly, in case of the RGD nanoemulsion a periodicity in the accumulation kinetics of the agent was observed, which was absent when administering CTRL nanoemlusion. Herein we hypothesize that this periodicity is caused by integrin binding, and internalization and recycling dynamics. Taken together, our approach may both serve as a potent tool to increase our understanding of nanoparticle-receptor interactions as well as provide more quantitative information about receptor expression levels in vivo.
Methods
Nanoparticle synthesis
Nanoemulsions (see Fig. 1a for a nanoemulsion schematic) were prepared using a lipid film hydration approach as previously described [26]. The nanoemulsion was composed of 3.3 % (v/v) soybean oil and 0.83 % (w/v) of an amphiphilic lipid mixture in HBS buffer (2.38 g/L Hepes and 8 g/L NaCl, pH 7.4). The amphiphilic lipid mixture consisted of 37 mol% 1,2-distearoyl-sn-glycero-3-phosphocholine (DSPC), 33 mol% cholesterol, 2.5 mol% 1,2-distearoyl-sn-glycero-3-phosphoethanolamine-N-[methoxy(polyethylene glycol)-2000 (PEG2000-DSPE), 2.5 mol% maleimide-PEG2000-DSPE, and 25 mol% gadolinium- diethylene-triaminepentacetate-bis(stearylamide) (Gd-DTPA-DSA) (all purchased from Avanti Polar Lipids). The amount of soybean oil added was 3 mg/μmol amphiphilic lipid.
Fig. 1.
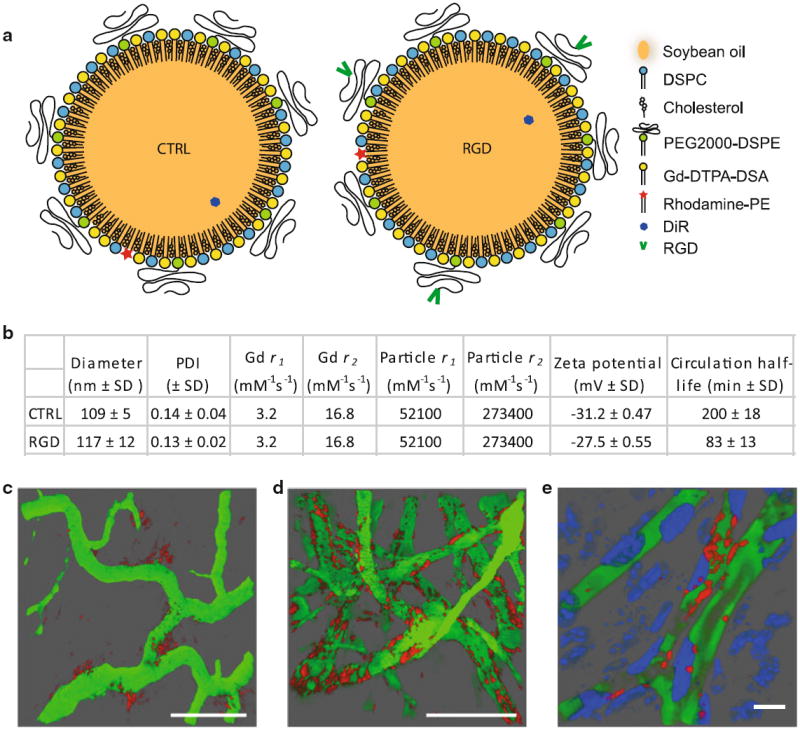
Nanoemulsion characteristics. a Nanoemulsion cartoon with legend. b Nanoemulsion characteristics. Relaxivities were measured at 7T and room temperature. SD standard deviation. c–e IVM of the tumor vasculature (green) 8 h post injection of CTRL nanoemulsion (red) (c), 2 h post injection of RGD nanoemulsion (red) (d), and 3 h post injection of RGD nanoemulsion (red) (e). In e cell nuclei were labeled with an intravenous injection of Hoechst (blue). The scale bars in c and d represent 100 μm and in e 10 μm. c, d Were reprinted (adapted) with permission from [27]. Copyright (2012) American Chemical Society
For fluorescent detection, 1,2-dipalmitoyl-sn-glycero-3-phosphoethanolamine-N-lissamine rhodamine B sulfonyl ammonium salt (rhodamine-PE, Avanti Polar Lipids) or 1,1′-Dioctadecyl-3,3,3′,3′-Tetramethylindotricarbocyanine Iodide (DiR, Invitrogen) was incorporated at 0.1 mol% of the amphiphilic lipid mixture. After preparation, half of each nanoemulsion suspension was conjugated with c(RGDf(-S-acetylthioacetyl)K) (RGD, 13.5 μg RGD per μmol lipid), as described previously (Hak 2012 #37).
Nanoparticle characteristics
The hydrodynamic diameter and polydispersity indexes (PDI, defined as standard deviation divided by the mean) of the nanoemulsions were measured using dynamic light scattering (DLS, Malvern, Zetasizer Nano PS). Reported values were the average of 3 batches of nanoemulsions, where the result of each batch was an average of approximately 50 measurements. In previous work, we have measured the T1 and T2 relaxivity (r1 and r2, respectively) of the Gd in the nanoemulsion to be 3.2 mM−1s−1 and the zeta potential to be highly negative [26].
Circulation half-life determination
The circulation half-lives of the nanoemulsions were determined by measuring fluorescent labeled nanoemulsion in blood samples [28]. In short, male athymic Balb/c Nu/nu mice were injected with CTRL nanoemulsion (n = 3) or RGD nanoemulsion (n = 3) according to the same injection regime and dose used for DCE-MRI. Blood samples (10–30 μl) were collected from the saphenous vein before and at 5, 15, 30, 60 min and 2, 4, 6, and 24 h post injection. DiR fluorescence intensity in the blood samples was measured using an optical imager (Pearl Impulse, Licor) and normalized to the amount of blood taken. The obtained fluorescence intensity versus time curves were most accurately fitted by mono-exponentials, from which the circulation half-lives were obtained.
Animals and tumors
For MRI, xenografts of the ovarian cancer cell line TOV112D (obtained after inoculation of 5 × 106 tumor cells in the flank) in 12 weeks old female athymic Balb/c Nu/nu mice were used. Three to 5 weeks after inoculation the xenografts were 0.5–1 cm in diameter and used for experiments.
For measuring VIFs, mice with dorsal window chambers were used. In-house built window chambers, made of polyoxymethylene, were implanted as previously described [29] in male athymic Balb/c Nu/nu mice (22–24 g of weight). 24 h after chamber implantation the animals were used for experiments. All animals were kept under pathogen-free conditions at a temperature of 19–22 °C, 50–60 % humidity, and 65 air changes per h, and were allowed food and water ad libitum. All experiments were approved by the institutional ethics committee and were in accordance with the national and institutional guidelines.
Measurement of the input functions with intravital microscopy
Confocal laser scanning microscopy (CLSM, Zeiss LSM 510 META) was used to measure VIFs in window chamber mice. The mice were anesthetized by subcutaneous injections of 12 mg/kg midazolam/fentanyl/Haldol/water (3/3/2/4), cannulated in the tail veins, and placed on a custom built temperature controlled imaging stage. The vasculature was imaged using a Plan-Neofluor 20×/0.5 objective (in plane spatial resolution of 2.5 × 2.5 μm) at a temporal resolution of 3.94 s; using 325 frames this sequence lasted 21 min and 14 s. Rhodamine-PE (excited at 543 nm and detected at 569–643 nm) labeled CTRL nanoemulsions (n = 5) or RGD nanoemulsions (n = 3) were injected at the start of the 10th repetition according to the same injection regime and dose used for DCE-MRI. Linearity of fluorescence intensity with lipid concentration in the lipid concentration range studied was confirmed, see supporting info S1. Directly after the dynamic imaging session blood samples (~50 μl) were obtained from the saphenous vein, in which the Gd concentration was determined with inductively coupled plasma mass spectrometry. Fluorescence intensity versus time curves were generated from regions of interest (ROIs) drawn inside the vasculature using ImageJ (NIH). The fluorescence intensities of the last 4 intravital fluorescence measurements were averaged and this average intensity was used to determine the Gd concentration to fluorescence intensity ratio at the end of imaging. The entire fluorescence intensity vs time curve was subsequently multiplied by this ratio which resulted in a Gd versus time curve, the input function.
MRI
Magnetic resonance imaging was performed on a 7.05 T horizontal bore magnet (Biospec, Bruker) with a 72 mm volume resonator for RF transmission and a quadrature mouse brain surface coil for reception. Mice were anesthetized with isoflurane (2 % in 67 % N2/33 % O2) and the tail vein was cannulated. Respiration rate and body temperature were monitored using pressure-sensitive and rectal temperature probes (SA Instruments, New York, NY, USA). Gas anesthesia was adjusted accordingly and warm water flow in the animal bed maintained the body temperature at 37 °C.
Pre-injection T1 values were measured using a fast T1 mapping method [rapid acquisition with relaxation enhancement (RARE)] pulse sequence: echo time (TE) of 7 ms, repetition time (TR) of 150, 750, 1,500, 2,500, 4,500, 12,500 ms, RARE factor of 2, zero fill acceleration of 1.34, FOV of 25.2 × 14.4 mm, matrix size (MTX) of 56 × 32, slice thickness of 1 mm, 1 slice, 1 average, lasting 4 min and 28 s.
A dynamic series of 60 T1-weighted images was obtained at a temporal resolution of 21.6 s with identical geometry as the T1 map. (RARE: TE of 7 ms, TR of 300 ms, RARE factor 2, zero fill acceleration of 1.34, 6 averages).
Either RGD nanoemulsions (n = 4) or CTRL nanoemulsions (n = 4) were administered 1 min and 48 s after the start of imaging, at the beginning of the recording of the 6th frame, as a bolus lasting approximately 20 s at an amphiphilic lipid dose of 80 μmol/kg, equaling a Gd dose of 20 μmol/kg. At a 10 mM amphiphilic lipid concentration of the nanoemulsion this resulted in an injection volume of 160 μl per mouse of 20 g. 25 min after DCE-MRI an anatomical high resolution image was obtained with the same slice planning as the T1 map and DCE-MRI series (fast low angle shot (FLASH) TE of 5.4 ms, TR of 350 ms, MTX of 224 × 128, 8 averages).
In vitro targeting kinetic measurements
To investigate the integrin targeting dynamics in vitro, αvβ3-integrin expressing human umbilical vein endothelial cells (HUVEC, Lonza) were incubated with rhodamine-PE labeled RGD or CTRL nanoemulsion at 1 mM lipid in growth medium under various conditions. HUVEC were grown to approximately 80 % confluency in 12-well plates in EGM 2 growth medium (CC-3124 EGM BulletKit (CC-3121 & CC4133)—Lonza) at 37 °C and 5 % CO2. Cells were incubated with nanoemulsion containing medium two times for 4 min (referred to as first and second nanoemulsion incubation in the remainder). The time in between the first and second nanoemulsion incubation was varied and is referred to as ‘pause’. During this pause the cells were exposed to nanoemulsion free medium. Directly after the first nanoemulsion incubation the cells were washed 3 times with growth medium. Directly after the second nanoemulsion incubation the cells were washed 3 times with PBS, trypsinized, resuspended in ice-cold 10 % FBS in PBS and kept on ice until analysis. As a control we also included a sample in which the HUVEC were incubated with nanoemulsion only once for 4 min, after which the cells were prepared for anaylsis as described above. Table 1 shows the 7 incubation conditions which were defined (n = 3 for each RGD and CTRL sample).
Table 1.
The 7 incubation conditions defined in the in vitro incubation experiment
| First nanoemulsion incubation (min) | 4 | 4 | 4 | 4 | 4 | 4 | 4 |
| Pause (emulsion free growth medium, min) | – | 0 | 1 | 3 | 6 | 12 | 30 |
| Second nanoemulsion incubation (min) | – | 4 | 4 | 4 | 4 | 4 | 4 |
Cellular uptake was quantified with flow cytometry (Beckman Coulter, Gallios), using excitation at 560 nm and detection at 578–592 nm. 10,000 cells in each sample were counted and analyzed and the reported values are the median fluorescence intensities per cell normalized to the median autofluorescence intensity of non-incubated HUVEC.
MR image analysis
Magnetic resonance image data analysis and compartment analysis was performed with in-house developed software in Matlab (The Mathworks inc.). ROIs containing the tumors were manually defined in the high resolution anatomical images and downsampled to obtain low resolution ROIs corresponding to the tumor region in the DCE-MRI and T1 -mapping data. DCE-MRI analysis was performed on data from ROIs containing only tumor rim (0.5–1 mm), which were generated using ROI erosion and subtraction. From the DCE-MRI signal intensities, Gd concentrations were calculated using the pre-injection T1 values, T1 values from the signal equation of the sequence and assuming constant r1 of the Gd in the nanoemulsions. Where applicable, the signal intensity was normalized to the average signal intensity from the 5 pre-injection images. Fourier transformation was performed using standard Matlab functions.
To study nanoemulsion distribution in the tumor, the distance from each pixel in the tumor to the tumor core was calculated. These distances were subsequently binned in 10 bins between 0 and maximum distance and the average signal enhancement in each ‘distance bin’ was calculated. Pixels with negative enhancement were excluded from this analysis. As the tumors differ in size, the distances were normalized to the maximum distance (the radius) in each tumor, which made it possible to compute averages for both groups.
Histology
Two hours after injection of the nanoemulsion the mice were anesthetized with isofluorane and a lethal dose of Pentobarbital (0.3 ml/mouse) was injected intraperitone-ally. 2 min later, the mice were perfused with 20 ml saline via a needle in the left ventricle while the heart was still beating. Then the tumors were excised, snap frozen in liquid nitrogen and stored at −80 °C.
The tumors were cut into 4 μm thick sections, mounted on glass slides and air dried for 30 min before fixation in 4 % paraformaldehyde for 10 min at room temperature. Next, the slides were transferred into a DAKO Autostainer. Serum-free Protein Block (Dako) was applied for 10 min. Endothelial cells were stained with rabbit anti-CD31 (ab28364, Abcam) at 1:20 dilution with 1 % bovine serum albumin in Tris Buffered Saline (TBS) for 1 h followed by incubation with goat anti-rabbit FITC (F0382, Sigma) at 1:80 dilution in TBS for 30 min. Nuclei were counter-stained with VECTASCHIELD® Mounting Medium with DAPI (Vector laboratories) and coverslipped. Between each step the slides were rinsed twice with TBS.
Images of sections with either CTRL or RGD nanoemulsion were recorded with identical settings on an inverted fluorescence microscope Olympus IX71 using a 20× objective. DAPI, FITC and rhodamine were excited with a Xe–Hg lamp using the following excitation filters: 325–375, 493–509 and 541–569 nm respectively. The respective emission filters were 432–482, 507–545 and 581–654 nm.
Kinetic modeling
Dynamic contrast-enhanced magnetic resonance imaging in combination with small molecular contrast agents is a powerful technique to study contrast agent dynamics and as such assess and characterize tumor vasculature and function [11, 12]. At the basis of this success lies a large body of research developing and validating various compartment models quantifying contrast agent kinetics in terms of physiological relevant parameters. All the available models make basic assumptions about various concepts in tracer kinetics and NMR theory [30]:
The tracer is well mixed in uniform concentration throughout each compartment
Time invariance of the determined parameters during data collection
The intercomparment fluxes of the tracer are assumed to be linear in concentration, i.e. the fluxes are proportional to concentration differences.
The tracer has a constant r1 throughout the tissue and fast exchange of all mobile protons is occurring, such that a single T1 exists in the tissue of interest.
One extensively used model was introduced by Larsson [13] and Tofts and Kermode [15] and has later been developed further into the so called extended Tofts model by them and others [30-32]. The model describes a tissue containing an intravascular compartment and an extravascular leakage space to which the agent has access. Bi-directional contrast agent exchange between these two compartments is taken into account. The model is described by [30, 33]:
| (1) |
where
| (2) |
with Ct being the total tissue concentration (as determined from the DCE-MRI data), Cp the plasma concentration (as determined from the VIFs), Vp the plasma volume fraction (in ml per 100 g tissue), Ki the unidirectional influx constant (in ml min−1 100 g−1), which is a measure for the transfer of contrast agent from the plasma to the extravascular compartment, Hct the blood hematocrit, set to 0.38, kep the rate constant between the extravascular space and the plasma (in min−1), which is a measure for contrast agent backflow from the extravascular compartment to the plasma, and Ve the volume fraction of the extravascular space of distribution (in ml 100 g−1). In case of conventional small molecular contrast agents, Ve represents the total extravascular extracellular space. However in this study it represents the extravascular space over which the nanoemulsion distributes. No discrimination between arterial and venous plasma concentration of the contrast agent is made in this model. In case of low vascular permeability for the agent, as is likely the case for the large nanoemulsions, this is valid. When backflow of the contrast agent from the extravascular compartment to the plasma is ignored, which may also be the case for the relatively large nanoemulsions, Eq. 1 reduces to:
| (3) |
When dividing Eq. 3 by Cp(t) this results in:
| (4) |
Equation 4 represents a graphical method for analysis of dynamic contrast imaging data introduced by Patlak et al. [14, 34]. It comprises one region, which includes the intravascular compartment, over which the agent can distribute freely and an extravascular compartment which is irreversibly connected to the other compartments. Contrast agent which has entered the irreversible compartment is assumed to stay there during the measurement. A plot of versus will result in a straight line after the initial vascular phase, in which no steady state between the blood plasma and the reversible region is obtained yet. Ki is the slope of this line and Vp is the intercept with the y-axis. Vp is a measure for the plasma volume fraction [33]. Importantly, when back diffusion does occur, the Patlak plot will not be linear, but will rather level off at later time points [33]. In this case, determination of Ki and Vp from the Patlak plot will result in an underestimation of Ki and an overestimation of Vp.
Furthermore, it is important to note that the basic assumptions of the nanoemulsion having a constant r1 throughout the tissue and that fast exchange of all mobile protons is occurring, may be violated. Moreover, although not considered in existing literature, constant r2 is also assumed in the presented compartment models. If the dynamics of the contrast agent include displacement from the intravascular and/or interstitial compartments into an intracellular compartment, r1 and r2 may be affected. The r1 may decrease due to limited water diffusion through the cell membrane and the intracellular membrane compartment (e.g. endosomes). The r2 may increase due to increased susceptibility effects upon compartmentalization.
Results
Nanoparticle characteristics
See Fig. 1 for an overview of particle characteristics. The diameter of the CTRL nanoemulsions was 109 ± 5 nm (mean ± SD) and of the RGD nanoemulsion it was 117 ± 12 nm, both with a PDI below 0.2. The measured circulation half-lives were 200 ± 18 min (mean ± SD) for CTRL nanoemulsion and 83 ± 13 min for the RGD nanoemulsion. We have previously demonstrated that the RGD nanoemlsion efficiently targets angiogenic tumor vasculature, using amongst others IVM (Fig. 1c, d). Using IVM, we also observed a substantial portion of the RGD nanoemulsion to localize in close vicinity of cell nuclei of cells lining the blood vessels (most likely endothelial cells), being indicative for in vivo cellular uptake of the RGD nanoemulsion (Fig. 1e).
Vascular input function
For measuring the VIFs, a novel approach was successfully employed. Fluorescently labeled nanoemulsions were intravenously injected in window chamber mice and the vasculature was imaged using CLSM before, during and after injection. Snapshots from a resulting dynamic imaging series (movie in supportive information) are shown in Fig. 2a, visualizing the nanoemulsion wash in into the vasculature. The obtained average VIFs for the RGD and the CTRL nanoemulsion are plotted in Fig. 2b. After the wash in, for both nanoemulsions the concentration hardly decreased over the 20 min time course of the measurement, which can be explained by their long circulation half-lives (Fig. 1b). These averaged VIFs were used for analysis of the DCE-MRI data presented below.
Fig. 2.

Measuring the vascular input function using intravital microscopy. a Snapshots from the dynamic imaging series at the start and different time points after starting the injection of CTRL nanoemulsion. b The obtained vascular input functions for the CTRL (n = 5) and RGD nanoemulsions (n = 3). Error bars represent the standard error of the mean
Nanoemulsion distribution in the tumor
To study nanoemulsion distribution in the tumor, we determined the relative signal enhancement in the tumor as a function of distance to the tumor core at 3 different time points in the DCE-MRI sequence. Although both agents showed relatively low accumulation in the core, the RGD nanoemulsion more predominantly accumulated in the tumor rim (Fig. 3a, lower panel), whereas the CTRL nanoemulsion distributed more evenly throughout the tumor (Fig. 3a, upper panel). As angiogenesis is most prevalent in the tumor rim [35], this confirmed the ability of the RGD nanoemulsion to target angiogenic vasculature. Interestingly, this trend was already observed at 3 min post injection, demonstrating the receptor binding to occur shortly after injection. To illustrate the bar diagrams, relative enhancement maps overlaid on anatomical images are shown in Fig. 3b, c.
Fig. 3.
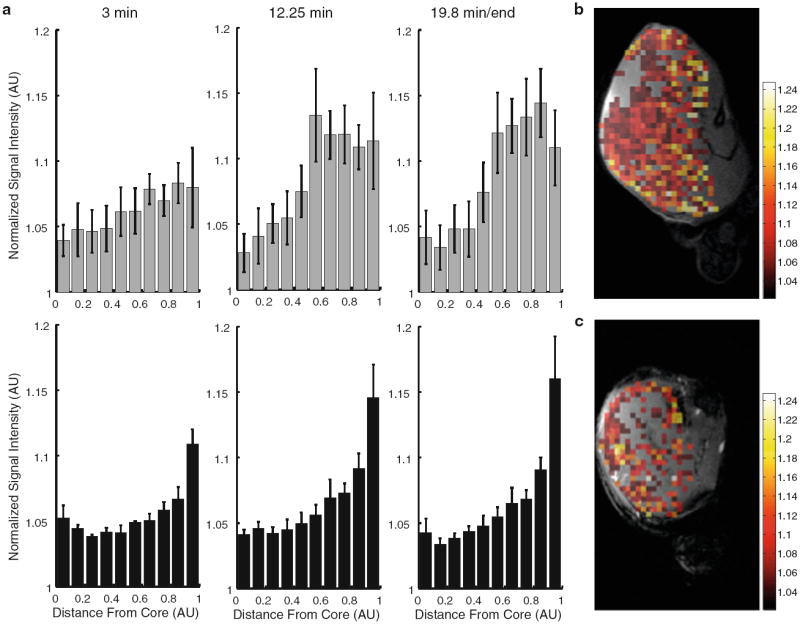
Distribution of nanoemulsion in the tumor. a Normalized signal enhancement (in arbitrary units, AU) in the tumor plotted as a function of the normalized distance from the tumor core for CTRL (top row, n = 4) and RGD (bottom row, n = 4) nanoemulsion. x = 0 is at the tumor core and x = 1 is at the tumor rim. Error bars represent standard error of the mean. b, c Relative enhancement maps overlaid over anatomical images for CTRL (b) and RGD (c) nanoemulsion
Histological analysis
Histologic examination of the tumors confirmed that the RGD (Fig. 4b) and CTRL nanoemulsions accumulated differently in the tumor tissue (Fig. 4a). In line with previous work [26], the RGD nanoemulsion mainly colocalized with vascular endothelial cells and formed circular aggregates, whereas the CTRL nanoemulsion mostly extravasated from the vasculature and accumulated in a more diffuse pattern in the extravascular space.
Fig. 4.
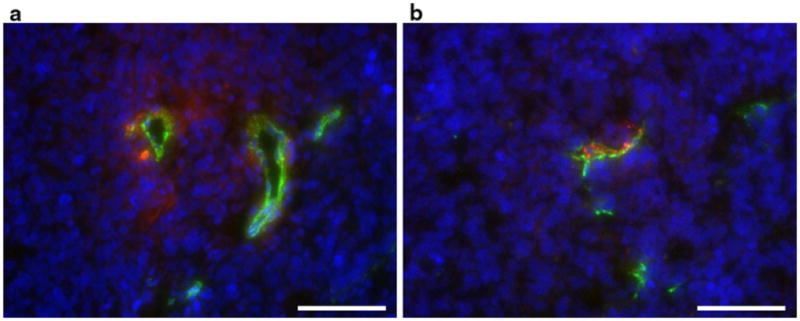
Histology. Tumor tissue containing CTRL (a) and RGD (b) nanoemulsion. Particles are shown in red, endothelial cells in green (CD31) and cell nuclei in blue (DAPI stain). The bars represent 100 μm
DCE-MRI
Figure 5a shows an overlay of the VIFs (from Fig. 2b) and the average Gd concentration in the tumor rim versus time as obtained from the DCE-MRI signal intensities. The graphs illustrate that after the vascular wash-in phase, the Gd concentration in the tissue is increasing while the Gd concentration in the blood is only slightly decreasing. This did not only confirm the accumulation of the nanoemulsion in the tumor, but more importantly demonstrates that these dynamics can be monitored with DCE-MRI. In Fig. 5b the normalized MRI signal intensity in the tumor rim is plotted as a function of time. In case of the CTRL nanoemulsion the signal intensity continuously increased upon injection, reaching a maximum signal enhancement of approximately 7 % at the end of the measurement. Strikingly, after injection of the RGD nanoemulsion, a periodicity in the accumulation was observed. A fast initial increase to 5–6 % signal enhancement at approximately 3 min and 30 s post injection was followed by a slight decrease lasting about 2 min (until approximately 5 min and 30 s post injection). After this it increased again to approximately 9 % signal enhancement at 13 min post injection, after which it again slightly decreased towards the end of the measurement.
Fig. 5.
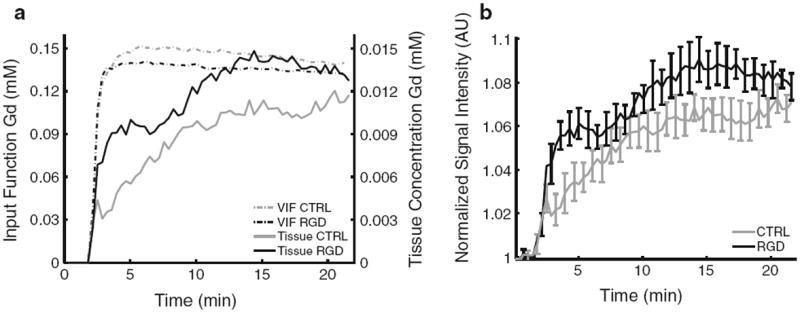
DCE-MRI data. a Overlay of the average vascular input functions (dotted lines, scaling to the left y axis) and the average Gd concentration in the tumor rim (continuous lines, scaling to the y axis on the right). b Averaged normalized signal intensity in the tumor rim plotted as a function of time.Error bars represent the standard error of the mean and for both CTRL and RGD n = 4
Kinetic modeling
Patlak
To study the observed dynamics and periodicity in more detail, Patlak plots were generated. From these plots, no single linear regime after the vascular phase was observed, which indicated that the influx constant was either changing or not unidirectional during the time course of our measurement (Fig. 6a, b) [14]. However, when a linear portion in the Patlak plot is found, the ‘net transfer process is effectively unidirectional for that specific period of time’ [34] and the slope of that period could represent the transfer constant for that specific time period [34]. In the Patlak plot of the RGD nanoemulsion, parts of the plot are approaching linearity (Fig. 6b). Therefore, Ki was determined from these linear regions (Table 2). The first 300 s post injection were excluded from this analysis as this time frame was assumed to constitute the vascular phase. In case of CTRL nanoemulsion, Ki decreased during the course of the experiment, which may suggest backflow is occurring. In contrast, in case of the RGD nanoemulsion the Ki decreased drastically and even became negative during the course of the experiment. Theoretically this implies diffusion from high to low concentration to occur. As this is very unlikely, it may be an indication for a decrease in the r1 or an increase in r2 of the RGD nanoemulsion once the nanoemulsion is bound and internalized, a phenomena which have been observed in human endothelial cells in vitro [23]. Hence, the two nanoemulsions accumulate in a distinctly different manner in the tumor tissue.
Fig. 6.

Kinetic modeling. a, b Patlak plots for the CTRL (a) and the RGD (b) nanoemulsion including the fits to 2 intervals (300–700 s in dark grey, and 700–1,296 s in black) which were considered linear in the RGD nanoemulsion Patlak plot upon visual inspection. The first 300 s (in light grey) were omitted from analysis as this is assumed to be the vascular phase. c, d Tofts model fits (solid lines) to the DCE-MRI data (filled circles). In c the total curves were analyzed and in d the first 4.5 min post injection were fitted
Table 2.
Ki (from both Tofts and Patlak analysis) and Ve (Tofts only) obtained from the fits presented in Fig. 6
| CTRL | RGD | |||
|---|---|---|---|---|
| Patlak | Ki (ml min−1 100 g−1) | Ki (ml min−1 100 g−1) | ||
| 300–700 s | 0.26 | 0.59 | ||
| 700–1,296 | 0.11 | −0.083 | ||
| Tofts | Ki (ml min−1 100 g−1) | Ve (ml 100 g−1) | Ki (ml min−1 100 g−1) | Ve (ml 100 g−1) |
| 0–270 s | 0.83 | 4.6 | 8.43 | 2.9 |
| 0–1,296 s | 0.90 | 5.9 | 1.98 | 7.8 |
Tofts
The extended Tofts model was found to describe the CTRL nanoemulsion data well (Fig. 6c), indicating a backflow of the nanoemulsion which was expected from the Patlak plot. When fitting the Tofts model to the RGD nanoemulsion data unrealistic values for some of the fitted parameters were obtained (results not shown). Although obviously this model does not account for the observed fluctuations, when fixing Vp to 2.1 as was found for the CTRL nanoemulsion (under the assumption that Vp was the same in the two groups as the same types of tumor and mice were used), a reasonable fit was obtained (Fig. 6c). The nature of the Tofts model does not allow fitting of different subsequent time intervals, as we did using the Patlak approach. However, to improve the fit and obtain more quantitative information on initial targeting, we fitted the Tofts model to the first 4.5 min post injection data in which the fluctuation in the RGD nanoemulsion DCE-MRI data had not manifested itself yet (Fig. 6d). In this case the Tofts model fitted the RGD data very well. It was found that Ki was much larger for RGD nanoemulsion than for CTRL nanoemulsion, demonstrating targeting to occur very rapidly in this initial phase (Table 2). Ve was relatively small for both agents (Table 2). As the RGD nanoemulsion is mainly associated with endothelial cells, which constitute a small portion of the total voxel volume, this was a reasonable observation. It also indicated that the CTRL nanoemulsion once extravasated from the vasculature is not able to effectively diffuse throughout the tumor inter-stitium, but rather accumulates in a small volume close to the vasculature. This observation was confirmed by the histology and previous work [26].
Power spectra
To further assess the apparent periodicity in the accumulation rate of the RGD nanoemulsion, we conducted a Fourier transformation of the normalized signal intensity curves. From the Fourier transformations the powers for every frequency were calculated. The obtained power spectra show that the powers at 2.4 and 3.2 mHz, respectively corresponding to oscillations with periods of 6.9 and 5.2 min, were significantly higher in the RGD data than in the CTRL data (Fig. 7). Hence, this is a strong indication for the presence of oscillations in the accumulation rate of the RGD nanoemulsions, which were absent in case of the CTRL nanoemulsion.
Fig. 7.

Power spectra obtained from the normalized signal intensity DCE-MRI curves. Error bars represent standard errors of the mean (RGD n = 4, CTRL n = 4)
In vitro incubation experiment
To study in vitro αvβ3-integrin targeting dynamics, HU- VEC were incubated for 4 min with nanoemulsion containing medium, followed by a pause where the cells were exposed to nanoemulsion free medium, and incubated again for 4 min with nanoemulsion containing medium. In case of the CTRL nanoemulsion, we did not observe a clear effect of the length of the pause on total cellular internalization. In case of the RGD nanoemulsion we saw a marked increase in total cellular uptake with increasing pause between the first and second nanoemulsion incubation (Fig. 8a). The apparent slight decrease in cellular uptake from the 12 min pause to the 30 min pause samples for both CTRL and RGD, may be due to intracellular fluorophore quenching, disintegration, or excretion.
Fig. 8.
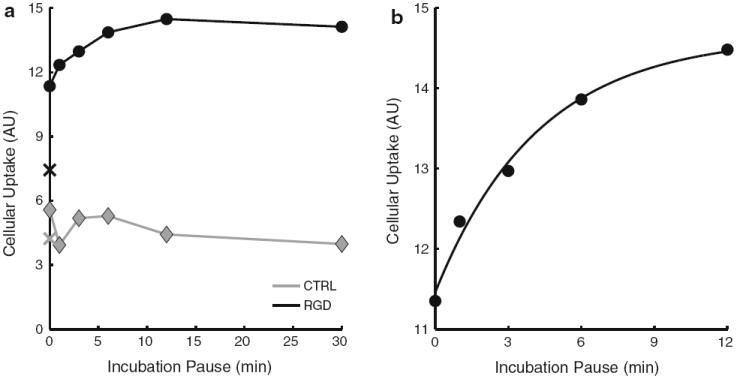
In vitro uptake of nanoemulsions by HUVEC as measured with flow cytometry. a Cellular uptake is plotted as a function of the pause between the first and second 4 min nanoemulsion incubations, for CTRL-NE in grey and for RGD-NE in black. The cellular uptake after 4 min of nanoemulsion incubation only is plotted as an X. Error bars were omitted as they are hidden behind the data points. b Monoexponential fit of y = a (b – e−ct) through the RGD nanoemulsion data of the samples with 0–12 min pause (R2 = 0.99, c = 0.24 min−1, corresponding to a half-life of 2.9 min)
Interestingly, in case of the RGD nanoemulsion, the maximum internalization observed (for a pause of 12 min), was about twice the amount as observed after only 4 min of incubation. However, the cells incubated for 8 min (the sample with 0 min pause between the first and second nanoemulsion incubation) only internalized approximately 50 % more than cells incubated for 4 min only (Fig. 8a). This demonstrated that the amount of internalization occurring during the second 4 min nanoemulsion incubation is substantially influenced by the length of the pause. When the RGD nanoemulsion data of the samples with 0-12 min pause was fitted with a mono-exponential y = a (b – e−ct) we obtained an R2 of 0.99, demonstrating a very good fit (Fig. 8b). The rate constant of this curve was 0.24 min−1, corresponding to a half-life of approximately 2.9 min.
Discussion
Intravital microscopy, DCE-MRI and compartment modeling were successfully used to study in vivo dynamics of αvβ3-integrin targeted RGD nanoemulsions and non-tar- geted CTRL nanoemulsions in xenografted tumors in mice. In accordance with previous studies, the RGD nanoemulsion exhibited a strong preference for the tumor rim where angiogenesis is most prevalent [4, 36]. In line with our previous work [26], we observed in vivo targeting of the RGD nanoemulsion within minutes after injection, which has been demonstrated for RGD conjugated liposomes as well [37]. Importantly, our approach allowed for quantifi-cation of nanoparticle targeting rates and provided the new insight that ligand targeting towards endothelial cell surface receptors can introduce a periodicity in nanoparticle accumulation rates.
Quantitative compartment modeling of the RGD nanoemulsion kinetics over the complete experimental time frame was complicated by the periodicity. We circumvented this problem by analyzing the first 4.5 min post injection where the periodicity had not manifested itself yet. In these first minutes post injection, Ki was much larger for the RGD nanoemulsion than for the CTRL nanoemulsion, demonstrating efficient and rapid targeting in those first minutes post injection to occur and that we were able to quantify this. In line with histology and IVM [26], the Tofts model indicated a relatively small extra-vascular space of distribution (Ve) for both nanoemulsions, corroborating that this model describes the nanoemulsion dynamics in the first minutes post injection well. In another study, dynamic imaging (first imaging time point 30 min post injection, 30 min temporal resolution,) after injection of αvβ3-integrin specific nanoparticles also showed a faster accumulation in the target tissue for targeted than for non-targeted particles, which was in line with our findings [38]. However, the much higher temporal resolution in the current DCE-MRI approach allowed for the observation of a periodicity in the targeting kinetics upon injection.
In an attempt to understand the observed periodicity in the RGD nanoemulsion accumulation we sought an explanation in integrin dynamics. αvβ3-integrin is a recycling receptor; in vitro studies show that it internalizes into intracellular vesicles and is rapidly recycled back to the plasma membrane with, depending on the intracellular route, a half-life of either 3 or 10 min [22, 24] ready to interact with the environment again.
In our in vitro targeting kinetics experiment, the cellular uptake of RGD nanoemulsion increased significantly with increasing incubation pause (time between the first and second nanoemulsion incubation), whereas no obvious effect of incubation pause on cellular uptake of CTRL nanoemulsion was observed. In other words, the amount of RGD nanoemulsion internalization occurring in the second 4 min of nanoemulsion incubation substantially increased with the length of the pause. The dependency of the total RGD nanoemulsion cellular uptake on incubation pause was described well by a mono-exponential function with a rate constant corresponding to a half-life of 2.9 min, nearly equaling the reported αvβ3-integrin short recycling half-life of 3 min [22]. This strongly indicated a role for integrin recycling in our in vitro observation; during the first 4 min integrin is extensively bound and internalized, and plausibly, during the incubation pause recycled back to the cell membrane.
In an in vivo situation, this could imply that integrin is extensively bound in the first minutes upon injection, followed by a period during which integrin is internalized and recycled and thus unavailable for circulating nanoparticles. Therefore, in the DCE-MRI experiment at any time point post injection, given that the RGD functionalized nanoparticles are still circulating at sufficient high concentrations, the amount of available luminal αvβ3-integrin is lower than upon injection. This implies that the initial accumulation rate of RGD contrast agent will be higher than the accumulation rate at any later time point, which was indeed observed. Furthermore, when a Fourier analysis of the DCE-curves was performed, the periods of the detected frequencies were 6.9 and 5.2 min, which is within the range of the reported αvβ3-integrin recycling half-lives of 3 and 10 min [22, 24].
Taken together this leads us to hypothesize that the observed periodicity was introduced by integrin binding, internalization and recycling kinetics. The bolus injection of RGD nanoemulsion will result in the majority of αvβ3-integrin being bound and internalized within minutes upon injection. Plausibly, the majority of these integrins will also reappear at a similar time point post injection. Hence, this binding and recycling of a pool of αvβ3-integrin may cause the periodicity observed after the initial binding of available integrin. A schematic representation of the hypothesized events introducing the periodicity is presented in Fig. 9.
Fig. 9.
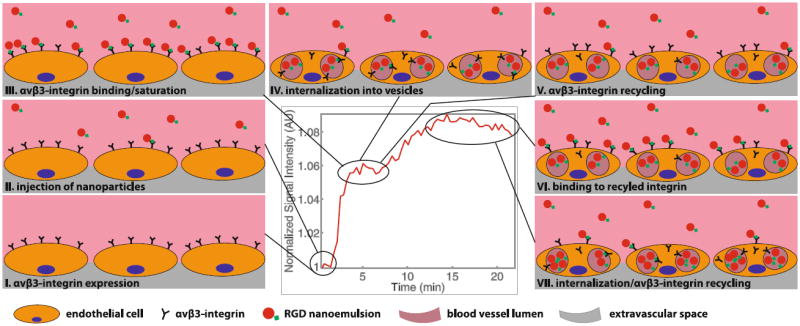
Graphical representation of the hypothesized chronological events leading to the observed periodicity. I, II Show the situation before and just after injection. III Depicts the binding/saturation which leads to the first signal increase. IV Shows the internalization during the first interval where no signal increase is observed and V shows the recycling of integrin back to the cell membrane which is occurring towards the end of the first interval with constant signal intensity. VI Shows the binding which has occurred after the second signal increase and VII shows internalization/recycling occurring again towards the end of the measurement
In vitro, RGD-conjugated nanoparticles are known to be internalized by αvβ3-integrin expressing cells [4, 26]. Moreover, indications for cellular uptake of RGD conjugated particles by αvβ3-integrin expressing cells in vivo can be found in literature as well [39, 40]. Whether this indeed occurred in our study as well was not evident from the histological analysis. As nanoemulsions retain their structure due to interaction with liquid water, and the histology involved freezing, cutting, drying and many washing procedures, this plausibly led to the removal of a significant part of the nanoemulsion from the histological sections. In IVM experiments however, our RGD nanoemulsion was found close to or in direct contact with cell nuclei (Fig. 1e), providing a strong indication that RGD nanoemulsion is internalized upon integrin binding in vivo as well, supporting our hypothesis.
Additionally, it has been demonstrated that upon internalization into endosomes, either RGD-integrin mediated [23] or not [41], the r1 of Gd based agents decreases, resulting in a decrease in MRI signal, which is explained by a reduced water access to the Gd [42]. Furthermore, it has also been observed that cellular internalization of Gd-labeled RGD-conjugated liposomes can lead to an increase in r2 of the Gd as an effect of increased magnetic susceptibility due to clustering inside endosomes [23]. Although not related to actual Gd concentration, and not accounted for in conventional compartment models, both these effects will introduce a decrease in the observed DCE-MRI signal intensity. One indication for these effects to play a role is the slight decrease in the signal intensity curve of the RGD nanoemulsion from 3.5 to 5.5 min and after 13 min post injection. Another indication for these effects to play a role is the negative Ki in the second part of the Patlak plot of the RGD nanoemulsion. As the RGD nanoemulsion concentration in the plasma is only slightly decreasing during the measurement, it is unlikely that the contrast agent is leaving the tissue and thus a decrease in r1 and/or an increase in r2 upon cellular internalization could have occurred.
To better understand these initial targeting kinetics and periodicity, a higher temporal resolution, which may for example be achieved at the cost of spatial resolution, with more advanced imaging sequences, better scanners, or higher relaxivity contrast agent, will be important. Investigation of the accumulation with optical or radiotracer techniques to rule out the effect of possible contrast quenching may also proof useful. Possibly new compartment models can be created incorporating parameters such as in vivo affinity constants of the RGD-nanoemulsion for the integrin and rates describing integrin trafficking, however to date the required knowledge to do so is lacking. Including IVM and the dorsal window chamber mouse model in our study allowed us to collect low noise population based VIFs at high temporal resolution. Although in mice experimentally challenging, obtaining individual input functions may allow for a more accurate quantification of the investigated parameters as well.
Although additional data are needed to test our hypothesis, a recently published paper does provide an indication that targeting of MRI contrast agents to endo-thelial cell receptors may indeed introduce a periodicity in DCE-MRI curves [43]. In that study DCE-MRI was performed on tumor bearing mice using Gd labeled nanoparticles targeted towards CD13 through conjugation with NGR peptides [43]. CD13, like αvβ3-integrin, is a receptor upregulated on angiogenic endothelial cells [44]. Although not specifically addressed in the study, the DCE-MRI curve of the CD13 targeted agent also displays a fast initial rise, followed by a periodicity, both of which were absent in case of the non-targeted control [43]. As for RGD functionalized nanoparticles, NGR-conjugated nanoparticles are known to be internalized into endosomes by cells expressing CD13 in vitro [45, 46]. Furthermore, CD13 is likely recycled back to the cell membrane after internalization as well [45, 47]. Hence, this periodicity may thus also be explained by our hypothesis.
In conclusion, we developed a useful experimental setup for studying in vivo nanoparticle targeting dynamics, which allowed us to monitor nanoparticle accumulation kinetics and quantify targeting rates. It provided us with the new insight that ligand targeting of nanoparticles can introduce a periodicity in nanoparticle accumulation. Herein we hypothesize that this periodicity is introduced by receptor internalization and recycling kinetics. The presented approach can both become a valuable tool to increase our understanding of in vivo nanoparticle targeting and optimize targeted nanoparticle design as well as provide more quantitative information on in vivo receptor levels and perhaps kinetics.
Supplementary Material
Acknowledgments
We gratefully thank K. Sæterbø for implanting the dorsal window chambers in mice and culturing cells, S. Andersen, K. Pettersen and S. Moestue for providing and taking care of the mice with subcutaneous xenografts used in the in vivo MRI experiments, and B. Ytterhus and L. Dyrnes for preparation of the histological tumor sections. The work was supported by the Norwegian Cancer Society and Medical Imaging Laboratory (MI-lab, NTNU, Norway) and the National Institute of Health National Cancer Institute Grant R01 CA155432 (W.J.M.M.).
Footnotes
Electronic supplementary material The online version of this article (doi:10.1007/s10456-013-9380-7) contains supplementary material, which is available to authorized users.
Contributor Information
Sjoerd Hak, Email: sjoerd.hak@ntnu.no, MI Lab, The Norwegian University of Science and Technology, Trondheim, Norway; Department of Circulation and Medical Imaging, The Norwegian University of Science and Technology, Trondheim, Norway; MR Senteret, Institutt for sirkulasjon og bildediagnostikk, Postboks 8905, 7491 Trondheim, Norway.
Jana Cebulla, Department of Circulation and Medical Imaging, The Norwegian University of Science and Technology, Trondheim, Norway; MR Senteret, Institutt for sirkulasjon og bildediagnostikk, Postboks 8905, 7491 Trondheim, Norway.
Else Marie Huuse, MI Lab, The Norwegian University of Science and Technology, Trondheim, Norway; Department of Circulation and Medical Imaging, The Norwegian University of Science and Technology, Trondheim, Norway; MR Senteret, Institutt for sirkulasjon og bildediagnostikk, Postboks 8905, 7491 Trondheim, Norway.
Catharina de L. Davies, Department of Physics, The Norwegian University of Science and Technology, Trondheim, Norway Høgskoleringen 5, 7591 Trondheim, Norway.
Willem J. M. Mulder, Translational and Molecular Imaging Institute, Icahn School of Medicine at Mount Sinai, New York, NY, USA Mount Sinai School of Medicine, One Gustave L. Levy Place, Box 1234, New York, NY 10029, USA.
Henrik B.W. Larsson, Department of Circulation and Medical Imaging, The Norwegian University of Science and Technology, Trondheim, Norway Functional Imaging Unit, Diagnostic Department, Glostrup University Hospital, Glostrup, Denmark; Ndr. Ringvej 57, 2600 Glostrup, Denmark.
Olav Haraldseth, MI Lab, The Norwegian University of Science and Technology, Trondheim, Norway; Department of Circulation and Medical Imaging, The Norwegian University of Science and Technology, Trondheim, Norway; MR Senteret, Institutt for sirkulasjon og bildediagnostikk, Postboks 8905, 7491 Trondheim, Norway; Department of Medical Imaging, St. Olav’s University Hospital, Trondheim, Norway.
References
- 1.Lobatto ME, Fuster V, Fayad ZA, Mulder WJ. Perspectives and opportunities for nanomedicine in the management of atherosclerosis. Nat Rev Drug Discov. 2011;10(11):835–852. doi: 10.1038/nrd3578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Choi KY, Liu G, Lee S, Chen X. Theranostic nanoplatforms for simultaneous cancer imaging and therapy: current approaches and future perspectives. Nanoscale. 2012;4(2):330–342. doi: 10.1039/c1nr11277e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lanza GM, Wallace KD, Scott MJ, Cacheris WP, Abendschein DR, Christy DH, Sharkey AM, Miller JG, Gaffney PJ, Wickline SA. A novel site-targeted ultrasonic contrast agent with broad biomedical application. Circulation. 1996;94(12):3334–3340. doi: 10.1161/01.cir.94.12.3334. [DOI] [PubMed] [Google Scholar]
- 4.Mulder WJ, van der Schaft DW, Hautvast PA, Strijkers GJ, Koning GA, Storm G, Mayo KH, Griffioen AW, Nicolay K. Early in vivo assessment of angiostatic therapy efficacy by molecular MRI. FASEB J. 2007;21(2):378–383. doi: 10.1096/fj.06-6791com. [DOI] [PubMed] [Google Scholar]
- 5.Medarova Z, Pham W, Farrar C, Petkova V, Moore A. In vivo imaging of siRNA delivery and silencing in tumors. Nat Med. 2007;13(3):372–377. doi: 10.1038/nm1486. [DOI] [PubMed] [Google Scholar]
- 6.Winter PM, Caruthers SD, Kassner A, Harris TD, Chinen LK, Allen JS, Lacy EK, Zhang H, Robertson JD, Wickline SA, Lanza GM. Molecular imaging of angiogenesis in nascent Vx-2 rabbit tumors using a novel alpha(nu)beta3-targeted nanoparticle and 1.5 tesla magnetic resonance imaging. Cancer Res. 2003;63(18):5838–5843. [PubMed] [Google Scholar]
- 7.Nel AE, Madler L, Velegol D, Xia T, Hoek EM, Somasundaran P, Klaessig F, Castranova V, Thompson M. Understanding biophysicochemical interactions at the nano-bio interface. Nat Mater. 2009;8(7):543–557. doi: 10.1038/nmat2442. [DOI] [PubMed] [Google Scholar]
- 8.Choi HS, Frangioni JV. Nanoparticles for biomedical imaging: fundamentals of clinical translation. Mol imaging. 2010;9(6):291–310. [PMC free article] [PubMed] [Google Scholar]
- 9.Lammers T, Kiessling F, Hennink WE, Storm G. Drug targeting to tumors: principles, pitfalls and (pre-) clinical progress. J Control Release. 2012;161(2):175–187. doi: 10.1016/j.jconrel.2011.09.063. [DOI] [PubMed] [Google Scholar]
- 10.Alberts B, Johnson A, Lewis J, Raff M, Roberts K, Walter P. Molecular biology of the cell. 5. Garland Science; New York: 2007. [Google Scholar]
- 11.Ocak I, Baluk P, Barrett T, McDonald DM, Choyke P. The biologic basis of in vivo angiogenesis imaging. Front Biosci. 2007;12:3601–3616. doi: 10.2741/2337. [DOI] [PubMed] [Google Scholar]
- 12.O’Connor JP, Jackson A, Parker GJ, Roberts C, Jayson GC. Dynamic contrast-enhanced MRI in clinical trials of antivascular therapies. Nat Rev Clin oncol. 2012;9(3):167–177. doi: 10.1038/nrclinonc.2012.2. [DOI] [PubMed] [Google Scholar]
- 13.Larsson HB, Stubgaard M, Frederiksen JL, Jensen M, Henriksen O, Paulson OB. Quantitation of blood-brain barrier defect by magnetic resonance imaging and gadolinium-DTPA in patients with multiple sclerosis and brain tumors. Magn Reson Med. 1990;16(1):117–131. doi: 10.1002/mrm.1910160111. [DOI] [PubMed] [Google Scholar]
- 14.Patlak CS, Blasberg RG. Graphical evaluation of blood-to-brain transfer constants from multiple-time uptake data. Generalizations. J Cereb Blood Flow Metab. 1985;5(4):584–590. doi: 10.1038/jcbfm.1985.87. [DOI] [PubMed] [Google Scholar]
- 15.Tofts PS, Kermode AG. Measurement of the blood-brain barrier permeability and leakage space using dynamic MR imaging. 1. Fundamental concepts. Magn Reson Med. 1991;17(2):357–367. doi: 10.1002/mrm.1910170208. [DOI] [PubMed] [Google Scholar]
- 16.Backer MV, Backer JM. Imaging key biomarkers of tumor angiogenesis. Theranostics. 2012;2(5):502–515. doi: 10.7150/thno.3623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Folkman J. Role of angiogenesis in tumor growth and metastasis. Semin Oncol. 2002;29(6, Supplement 16):15–18. doi: 10.1016/S0093-7754(02)70065-1. [DOI] [PubMed] [Google Scholar]
- 18.McKeage MJ, Baguley BC. Disrupting established tumor blood vessels: an emerging therapeutic strategy for cancer. Cancer. 2010;116(8):1859–1871. doi: 10.1002/cncr.24975. [DOI] [PubMed] [Google Scholar]
- 19.Liu Z, Wang F, Chen X. Integrin alpha(v)beta(3)-targeted cancer therapy. Drug Dev Res. 2008;69(6):329–339. doi: 10.1002/ddr.20265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sipkins DA, Cheresh DA, Kazemi MR, Nevin LM, Bednarski MD, Li KC. Detection of tumor angiogenesis in vivo by alphaVbeta3-targeted magnetic resonance imaging. Nat Med. 1998;4(5):623–626. doi: 10.1038/nm0598-623. [DOI] [PubMed] [Google Scholar]
- 21.Tateishi U, Oka T, Inoue T. Radiolabeled RGD peptides as integrin alpha(v)beta3-targeted PET tracers. Curr Med Chem. 2012;19(20):3301–3309. doi: 10.2174/092986712801215937. [DOI] [PubMed] [Google Scholar]
- 22.Caswell PT, Norman JC. Integrin trafficking and the control of cell migration. Traffic. 2006;7(1):14–21. doi: 10.1111/j.1600-0854.2005.00362.x. [DOI] [PubMed] [Google Scholar]
- 23.Kok MB, Hak S, Mulder WJ, van der Schaft DW, Strijkers GJ, Nicolay K. Cellular compartmentalization of internalized paramagnetic liposomes strongly influences both T1 and T2 relaxivity. Magn Reson Med. 2009;61(5):1022–1032. doi: 10.1002/mrm.21910. [DOI] [PubMed] [Google Scholar]
- 24.Roberts M, Barry S, Woods A, van der Sluijs P, Norman J. PDGF-regulated rab4-dependent recycling of alphavbeta3 integrin from early endosomes is necessary for cell adhesion and spreading. Curr Biol. 2001;11(18):1392–1402. doi: 10.1016/s0960-9822(01)00442-0. [DOI] [PubMed] [Google Scholar]
- 25.Janssen APCA, Schiffelers RM, ten Hagen TLM, Koning GA, Schraa AJ, Kok RJ, Storm G, Molema G. Peptide-targeted PEG-liposomes in anti-angiogenic therapy. Int J Pharm. 2003;254(1):55–58. doi: 10.1016/S0378-5173(02)00682-8. [DOI] [PubMed] [Google Scholar]
- 26.Hak S, Helgesen E, Hektoen HH, Huuse EM, Jarzyna PA, Mulder WJ, Haraldseth O, Davies C de L. The effect of nanoparticle polyethylene glycol surface density on ligand-directed tumor targeting studied in vivo by dual modality imaging. ACS Nano. 2012;6(6):5648–5658. doi: 10.1021/nn301630n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Seymour LW. Passive tumor targeting of soluble macromolecules and drug conjugates. Crit Rev Ther Drug Carr Syst. 1992;9(2):135–187. [PubMed] [Google Scholar]
- 28.Jarzyna PA, Skajaa T, Gianella A, Cormode DP, Samber DD, Dickson SD, Chen W, Griffioen AW, Fayad ZA, Mulder WJ. Iron oxide core oil-in-water emulsions as a multifunc-tional nanoparticle platform for tumor targeting and imaging. Biomaterials. 2009;30(36):6947–6954. doi: 10.1016/j.biomaterials.2009.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Erikson A, Tufto I, Bjonnum AB, Bruland OS, Davies C de L. The impact of enzymatic degradation on the uptake of differently sized therapeutic molecules. Anticancer Res. 2008;28(6A):3557–3566. [PubMed] [Google Scholar]
- 30.Tofts PS. Modeling tracer kinetics in dynamic Gd-DTPAMR imaging. J Magn Reson Imaging. 1997;7(1):91–101. doi: 10.1002/jmri.1880070113. [DOI] [PubMed] [Google Scholar]
- 31.Larsson HB, Fritz-Hansen T, Rostrup E, Sondergaard L, Ring P, Henriksen O. Myocardial perfusion modeling using MRI. Magn Reson Med. 1996;35(5):716–726. doi: 10.1002/mrm.1910350513. [DOI] [PubMed] [Google Scholar]
- 32.Parker GJM, Padhani AR. Quantitative MRI of the brain. Wiley, Ltd; 2004. T1-W DCE-MRI: T1-weighted dynamic contrast-enhanced MRI; pp. 341–364. [DOI] [Google Scholar]
- 33.Larsson HB, Courivaud F, Rostrup E, Hansen AE. Measurement of brain perfusion, blood volume, and blood-brain barrier permeability, using dynamic contrast-enhanced T(1)-weighted MRI at 3 tesla. Magn Reson Med. 2009;62(5):1270–1281. doi: 10.1002/mrm.22136. [DOI] [PubMed] [Google Scholar]
- 34.Patlak CS, Blasberg RG, Fenstermacher JD. Graphical evaluation of blood-to-brain transfer constants from multiple-time uptake data. J Cereb Blood Flow Metab. 1983;3(1):1–7. doi: 10.1038/jcbfm.1983.1. [DOI] [PubMed] [Google Scholar]
- 35.Mulder WJ, Strijkers GJ, Habets JW, Bleeker EJ, van der Schaft DW, Storm G, Koning GA, Griffioen AW, Nicolay K. MR molecular imaging and fluorescence microscopy for identification of activated tumor endothelium using a bimodal lipidic nanoparticle. FASEB J. 2005;19(14):2008–2010. doi: 10.1096/fj.05-4145fje. [DOI] [PubMed] [Google Scholar]
- 36.Jarzyna PA, Deddens LH, Kann BH, Ramachandran S, Calcagno C, Chen W, Gianella A, Dijkhuizen RM, Griffioen AW, Fayad ZA, Mulder WJ. Tumor angiogenesis phenotyping by nanoparticle-facilitated magnetic resonance and near-infrared fluorescence molecular imaging. Neoplasia. 2012;14(10):964–973. doi: 10.1593/neo.121148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schiffelers RM, Koning GA, ten Hagen TL, Fens MH, Schraa AJ, Janssen AP, Kok RJ, Molema G, Storm G. Anti-tumor efficacy of tumor vasculature-targeted liposomal doxorubicin. J Control Release. 2003;91(1–2):115–122. doi: 10.1016/s0168-3659(03)00240-2. [DOI] [PubMed] [Google Scholar]
- 38.Neubauer AM, Sim H, Winter PM, Caruthers SD, Williams TA, Robertson JD, Sept D, Lanza GM, Wickline SA. Nanoparticle pharmacokinetic profiling in vivo using magnetic resonance imaging. Magn Reson Med. 2008;60(6):1353–1361. doi: 10.1002/mrm.21795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Montet X, Montet-Abou K, Reynolds F, Weissleder R, Josephson L. Nanoparticle imaging of integrins on tumor cells. Neoplasia. 2006;8(3):214–222. doi: 10.1593/neo.05769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.van de Ven AL, Kim P, Haley O, Fakhoury JR, Adriani G, Schmulen J, Moloney P, Hussain F, Ferrari M, Liu X, Yun SH, Decuzzi P. Rapid tumoritropic accumulation of systemically injected plateloid particles and their biodistribution. J Control Release. 2012;158(1):148–155. doi: 10.1016/j.jconrel.2011.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Terreno E, Geninatti Crich S, Belfiore S, Biancone L, Cabella C, Esposito G, Manazza AD, Aime S. Effect of the intracel-lular localization of a Gd-based imaging probe on the relaxation enhancement of water protons. Magn Reson Med. 2006;55(3):491–497. doi: 10.1002/mrm.20793. [DOI] [PubMed] [Google Scholar]
- 42.Strijkers GJ, Hak S, Kok MB, Springer CS, Jr, Nicolay K. Three-compartment T1 relaxation model for intracellular paramagnetic contrast agents. Magn Reson Med. 2009;61(5):1049–1058. doi: 10.1002/mrm.21919. [DOI] [PubMed] [Google Scholar]
- 43.Oostendorp M, Douma K, Hackeng TM, van Zandvoort MA, Post MJ, Backes WH. Pharmacokinetics of contrast agents targeted to the tumor vasculature in molecular magnetic resonance imaging. Contrast Media Mol Imaging. 2010;5(1):9–17. doi: 10.1002/cmmi.361. [DOI] [PubMed] [Google Scholar]
- 44.Pasqualini R, Koivunen E, Kain R, Lahdenranta J, Sakamoto M, Stryhn A, Ashmun RA, Shapiro LH, Arap W, Ruoslahti E. Aminopeptidase N is a receptor for tumor-homing peptides and a target for inhibiting angiogenesis. Cancer Res. 2000;60(3):722–727. [PMC free article] [PubMed] [Google Scholar]
- 45.Wang X, Wang Y, Chen X, Wang J, Zhang X, Zhang Q. NGR-modified micelles enhance their interaction with CD13-overexpressing tumor and endothelial cells. J Control Release. 2009;139(1):56–62. doi: 10.1016/j.jconrel.2009.05.030. [DOI] [PubMed] [Google Scholar]
- 46.Negussie AH, Miller JL, Reddy G, Drake SK, Wood BJ, Dreher MR. Synthesis and in vitro evaluation of cyclic NGR peptide targeted thermally sensitive liposome. J Control Release. 2010;143(2):265–273. doi: 10.1016/j.jconrel.2009.12.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lakhotia S, Bauer KD, Papoutsakis ET. Fluid-mechanical forces in agitated bioreactors reduce the CD13 and CD33 surface protein content of HL60 cells. Biotechnol Bioeng. 1993;41(9):868–877. doi: 10.1002/bit.260410906. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


