Abstract
The zinc-finger DNA-binding domain (DBD) of poly (ADP-ribose) polymerase (PARP, EC 2.4.2.30) specifically recognizes DNA strand breaks induced by various DNA-damaging agents in eukaryotes. This, in turn, triggers the synthesis of polymers of ADP-ribose linked to nuclear proteins during DNA repair. The 46 kDa DBD of human PARP, and several derivatives thereof mutated in its first or second zinc-finger, were overproduced in Escherichia coli, in CV-1 monkey cells or in human fibroblasts to study their DNA-binding properties, the trans-dominant inhibition of resident PARP activity, and the consequences on DNA repair, respectively. A positive correlation was found between the in vitro DNA-binding capacity of the recombinant DBD polypeptides and their inhibitory effect on PARP activity stimulated by the alkylating agent N-methyl-N'-nitro-N-nitrosoguanidine (MNNG). Furthermore, overproduced wild-type DBD blocked unscheduled DNA synthesis induced in living cells by MNNG treatment, but not that induced by UV irradiation. These results define a critical role for the second zinc-finger of PARP for DNA single-stranded break binding and furthermore underscore the importance for PARP to act as a critical regulatory component in the repair of DNA damage induced by alkylating agents.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Althaus F. R. Poly ADP-ribosylation: a histone shuttle mechanism in DNA excision repair. J Cell Sci. 1992 Aug;102(Pt 4):663–670. doi: 10.1242/jcs.102.4.663. [DOI] [PubMed] [Google Scholar]
- Althaus F. R., Richter C. ADP-ribosylation of proteins. Enzymology and biological significance. Mol Biol Biochem Biophys. 1987;37:1–237. [PubMed] [Google Scholar]
- Alvarez-Gonzalez R., Jacobson M. K. Characterization of polymers of adenosine diphosphate ribose generated in vitro and in vivo. Biochemistry. 1987 Jun 2;26(11):3218–3224. doi: 10.1021/bi00385a042. [DOI] [PubMed] [Google Scholar]
- Belt P. B., Groeneveld H., Teubel W. J., van de Putte P., Backendorf C. Construction and properties of an Epstein-Barr-virus-derived cDNA expression vector for human cells. Gene. 1989 Dec 14;84(2):407–417. doi: 10.1016/0378-1119(89)90515-5. [DOI] [PubMed] [Google Scholar]
- Benjamin R. C., Gill D. M. Poly(ADP-ribose) synthesis in vitro programmed by damaged DNA. A comparison of DNA molecules containing different types of strand breaks. J Biol Chem. 1980 Nov 10;255(21):10502–10508. [PubMed] [Google Scholar]
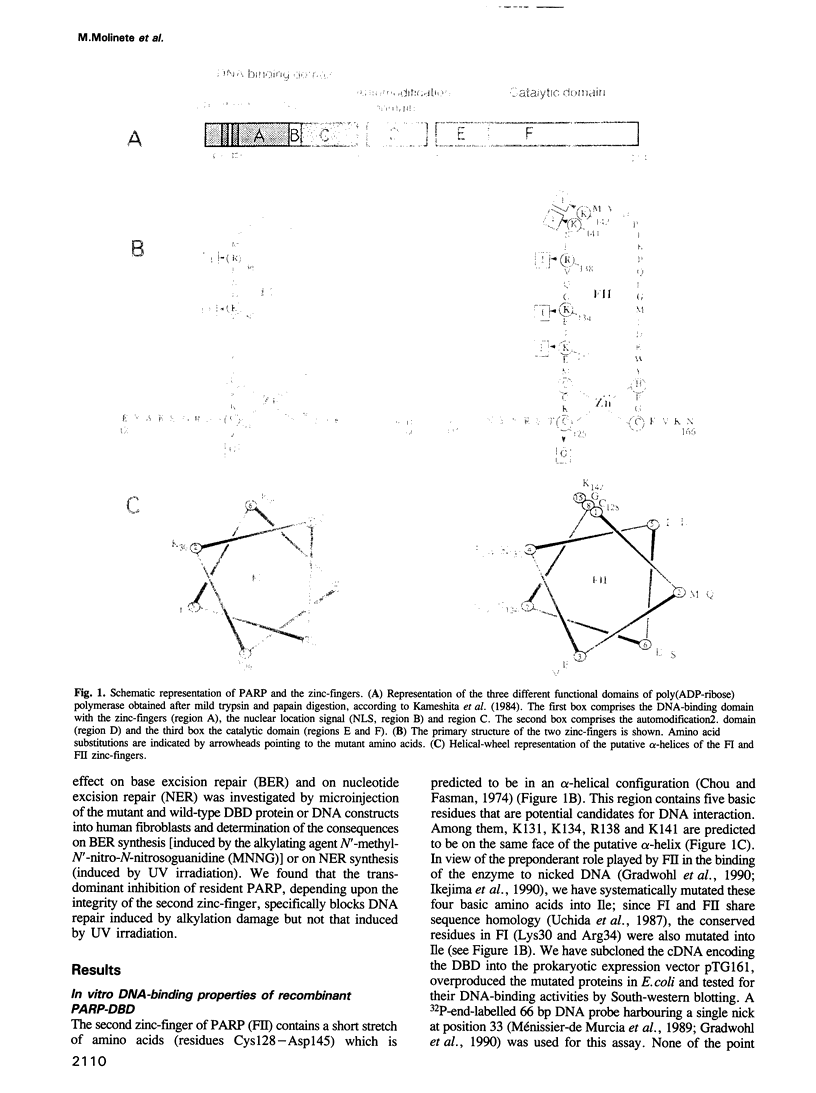
- Chou P. Y., Fasman G. D. Prediction of protein conformation. Biochemistry. 1974 Jan 15;13(2):222–245. doi: 10.1021/bi00699a002. [DOI] [PubMed] [Google Scholar]
- Cleaver J. E., Bodell W. J., Morgan W. F., Zelle B. Differences in the regulation by poly(ADP-ribose) of repair of DNA damage from alkylating agents and ultraviolet light according to cell type. J Biol Chem. 1983 Aug 10;258(15):9059–9068. [PubMed] [Google Scholar]
- Courtney M., Buchwalder A., Tessier L. H., Jaye M., Benavente A., Balland A., Kohli V., Lathe R., Tolstoshev P., Lecocq J. P. High-level production of biologically active human alpha 1-antitrypsin in Escherichia coli. Proc Natl Acad Sci U S A. 1984 Feb;81(3):669–673. doi: 10.1073/pnas.81.3.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Weerd-Kastelein E. A., Keijzer W., Bootsma D. Genetic heterogeneity of xeroderma pigmentosum demonstrated by somatic cell hybridization. Nat New Biol. 1972 Jul 19;238(81):80–83. doi: 10.1038/newbio238080a0. [DOI] [PubMed] [Google Scholar]
- Gradwohl G., Ménissier de Murcia J. M., Molinete M., Simonin F., Koken M., Hoeijmakers J. H., de Murcia G. The second zinc-finger domain of poly(ADP-ribose) polymerase determines specificity for single-stranded breaks in DNA. Proc Natl Acad Sci U S A. 1990 Apr;87(8):2990–2994. doi: 10.1073/pnas.87.8.2990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J. C., Svoboda D. L., Reardon J. T., Sancar A. Human nucleotide excision nuclease removes thymine dimers from DNA by incising the 22nd phosphodiester bond 5' and the 6th phosphodiester bond 3' to the photodimer. Proc Natl Acad Sci U S A. 1992 Apr 15;89(8):3664–3668. doi: 10.1073/pnas.89.8.3664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikejima M., Noguchi S., Yamashita R., Ogura T., Sugimura T., Gill D. M., Miwa M. The zinc fingers of human poly(ADP-ribose) polymerase are differentially required for the recognition of DNA breaks and nicks and the consequent enzyme activation. Other structures recognize intact DNA. J Biol Chem. 1990 Dec 15;265(35):21907–21913. [PubMed] [Google Scholar]
- Jacobson E. L., Antol K. M., Juarez-Salinas H., Jacobson M. K. Poly(ADP-ribose) metabolism in ultraviolet irradiated human fibroblasts. J Biol Chem. 1983 Jan 10;258(1):103–107. [PubMed] [Google Scholar]
- James M. R., Lehmann A. R. Role of poly(adenosine diphosphate ribose) in deoxyribonucleic acid repair in human fibroblasts. Biochemistry. 1982 Aug 17;21(17):4007–4013. doi: 10.1021/bi00260a016. [DOI] [PubMed] [Google Scholar]
- Kameshita I., Matsuda Z., Taniguchi T., Shizuta Y. Poly (ADP-Ribose) synthetase. Separation and identification of three proteolytic fragments as the substrate-binding domain, the DNA-binding domain, and the automodification domain. J Biol Chem. 1984 Apr 25;259(8):4770–4776. [PubMed] [Google Scholar]
- Kawamitsu H., Hoshino H., Okada H., Miwa M., Momoi H., Sugimura T. Monoclonal antibodies to poly(adenosine diphosphate ribose) recognize different structures. Biochemistry. 1984 Jul 31;23(16):3771–3777. doi: 10.1021/bi00311a032. [DOI] [PubMed] [Google Scholar]
- Küpper J. H., de Murcia G., Bürkle A. Inhibition of poly(ADP-ribosyl)ation by overexpressing the poly(ADP-ribose) polymerase DNA-binding domain in mammalian cells. J Biol Chem. 1990 Nov 5;265(31):18721–18724. [PubMed] [Google Scholar]
- Lamarre D., Talbot B., de Murcia G., Laplante C., Leduc Y., Mazen A., Poirier G. G. Structural and functional analysis of poly(ADP ribose) polymerase: an immunological study. Biochim Biophys Acta. 1988 Jul 13;950(2):147–160. doi: 10.1016/0167-4781(88)90007-3. [DOI] [PubMed] [Google Scholar]
- Lijinsky W. Interaction with nucleic acids of carcinogenic and mutagenic N-nitroso compounds. Prog Nucleic Acid Res Mol Biol. 1976;17:247–269. doi: 10.1016/s0079-6603(08)60072-0. [DOI] [PubMed] [Google Scholar]
- Luisi B. F., Xu W. X., Otwinowski Z., Freedman L. P., Yamamoto K. R., Sigler P. B. Crystallographic analysis of the interaction of the glucocorticoid receptor with DNA. Nature. 1991 Aug 8;352(6335):497–505. doi: 10.1038/352497a0. [DOI] [PubMed] [Google Scholar]
- Luisi B. DNA transcription. Zinc standard for economy. Nature. 1992 Apr 2;356(6368):379–380. doi: 10.1038/356379a0. [DOI] [PubMed] [Google Scholar]
- Marmorstein R., Carey M., Ptashne M., Harrison S. C. DNA recognition by GAL4: structure of a protein-DNA complex. Nature. 1992 Apr 2;356(6368):408–414. doi: 10.1038/356408a0. [DOI] [PubMed] [Google Scholar]
- Mazen A., Gradwohl G., de Murcia G. Zinc-binding proteins detected by protein blotting. Anal Biochem. 1988 Jul;172(1):39–42. doi: 10.1016/0003-2697(88)90408-3. [DOI] [PubMed] [Google Scholar]
- Mazen A., Menissier-de Murcia J., Molinete M., Simonin F., Gradwohl G., Poirier G., de Murcia G. Poly(ADP-ribose)polymerase: a novel finger protein. Nucleic Acids Res. 1989 Jun 26;17(12):4689–4698. doi: 10.1093/nar/17.12.4689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ménissier-de Murcia J., Molinete M., Gradwohl G., Simonin F., de Murcia G. Zinc-binding domain of poly(ADP-ribose)polymerase participates in the recognition of single strand breaks on DNA. J Mol Biol. 1989 Nov 5;210(1):229–233. doi: 10.1016/0022-2836(89)90302-1. [DOI] [PubMed] [Google Scholar]
- Pavletich N. P., Pabo C. O. Zinc finger-DNA recognition: crystal structure of a Zif268-DNA complex at 2.1 A. Science. 1991 May 10;252(5007):809–817. doi: 10.1126/science.2028256. [DOI] [PubMed] [Google Scholar]
- Satoh M. S., Lindahl T. Role of poly(ADP-ribose) formation in DNA repair. Nature. 1992 Mar 26;356(6367):356–358. doi: 10.1038/356356a0. [DOI] [PubMed] [Google Scholar]
- Schreiber V., Molinete M., Boeuf H., de Murcia G., Ménissier-de Murcia J. The human poly(ADP-ribose) polymerase nuclear localization signal is a bipartite element functionally separate from DNA binding and catalytic activity. EMBO J. 1992 Sep;11(9):3263–3269. doi: 10.1002/j.1460-2075.1992.tb05404.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shall S. ADP-ribosylation of proteins: a ubiquitous cellular control mechanism. Biochem Soc Trans. 1989 Apr;17(2):317–322. doi: 10.1042/bst0170317. [DOI] [PubMed] [Google Scholar]
- Simonin F., Briand J. P., Muller S., de Murcia G. Detection of poly(ADP ribose) polymerase in crude extracts by activity-blot. Anal Biochem. 1991 Jun;195(2):226–231. doi: 10.1016/0003-2697(91)90321-j. [DOI] [PubMed] [Google Scholar]
- Simonin F., Ménissier-de Murcia J., Poch O., Muller S., Gradwohl G., Molinete M., Penning C., Keith G., de Murcia G. Expression and site-directed mutagenesis of the catalytic domain of human poly(ADP-ribose)polymerase in Escherichia coli. Lysine 893 is critical for activity. J Biol Chem. 1990 Nov 5;265(31):19249–19256. [PubMed] [Google Scholar]
- Towbin H., Staehelin T., Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci U S A. 1979 Sep;76(9):4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchida K., Morita T., Sato T., Ogura T., Yamashita R., Noguchi S., Suzuki H., Nyunoya H., Miwa M., Sugimura T. Nucleotide sequence of a full-length cDNA for human fibroblast poly(ADP-ribose) polymerase. Biochem Biophys Res Commun. 1987 Oct 29;148(2):617–622. doi: 10.1016/0006-291x(87)90921-1. [DOI] [PubMed] [Google Scholar]
- Vermeulen W., Osseweijer P., de Jonge A. J., Hoeijmakers J. H. Transient correction of excision repair defects in fibroblasts of 9 xeroderma pigmentosum complementation groups by microinjection of crude human cell extracts. Mutat Res. 1986 May;165(3):199–206. doi: 10.1016/0167-8817(86)90055-6. [DOI] [PubMed] [Google Scholar]
- Zahradka P., Ebisuzaki K. Poly(ADP-ribose) polymerase is a zinc metalloenzyme. Eur J Biochem. 1984 Aug 1;142(3):503–509. doi: 10.1111/j.1432-1033.1984.tb08314.x. [DOI] [PubMed] [Google Scholar]
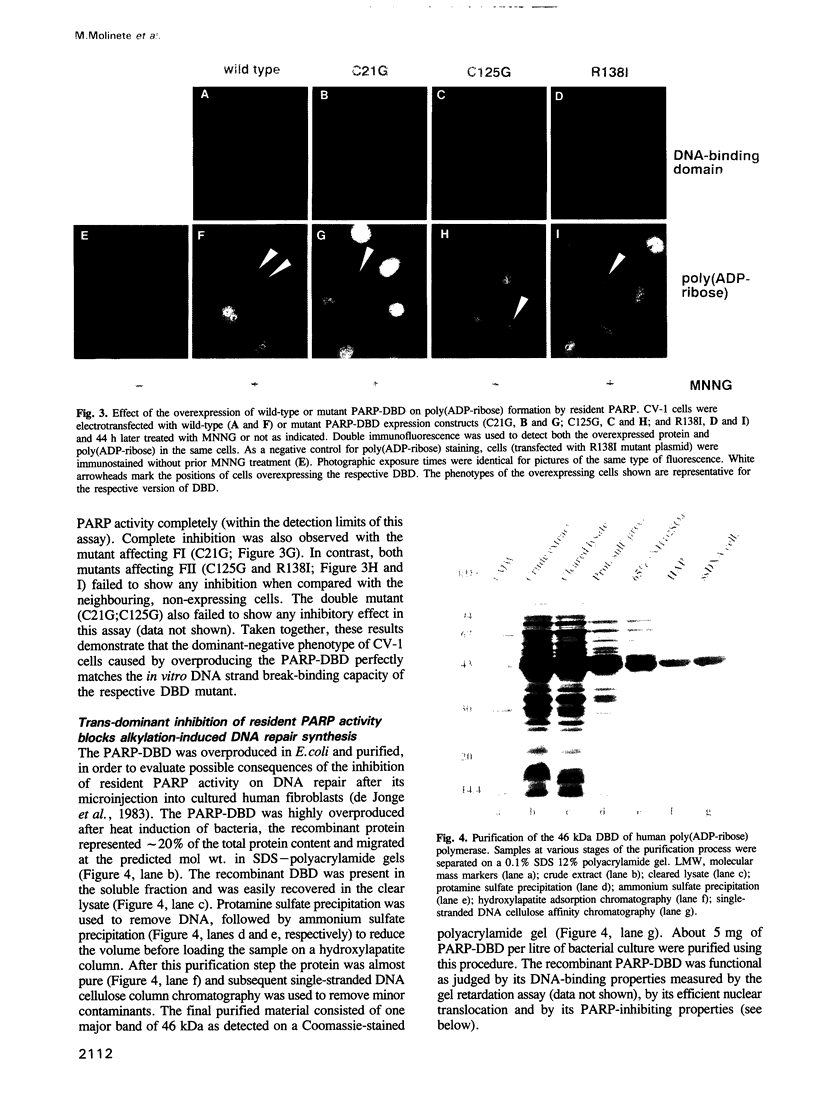
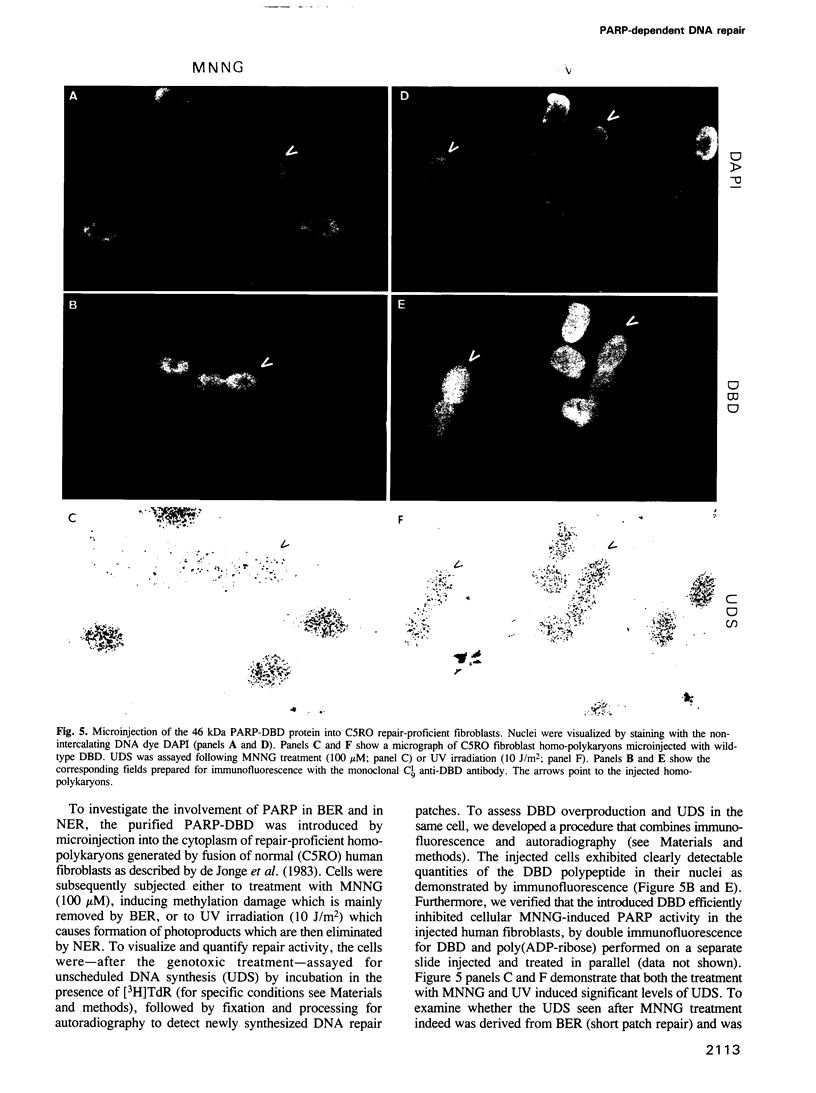
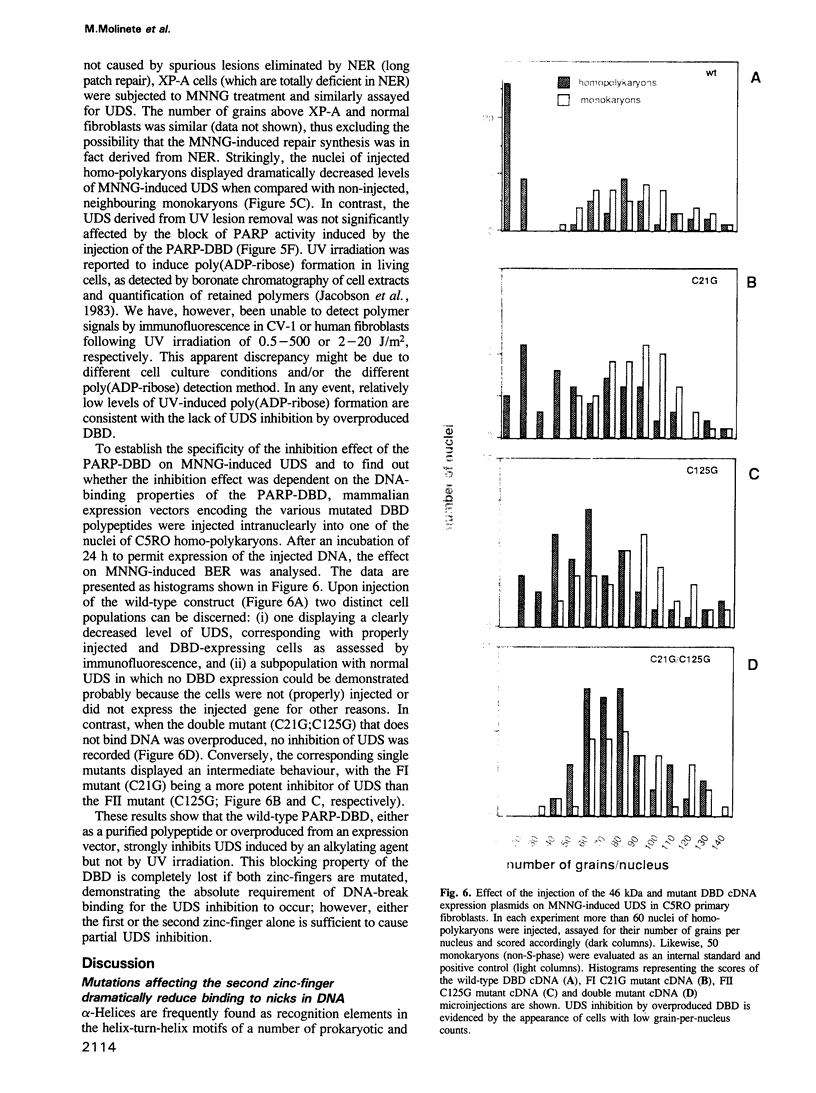
- de Jonge A. J., Vermeulen W., Klein B., Hoeijmakers J. H. Microinjection of human cell extracts corrects xeroderma pigmentosum defect. EMBO J. 1983;2(5):637–641. doi: 10.1002/j.1460-2075.1983.tb01477.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Murcia G., Huletsky A., Poirier G. G. Modulation of chromatin structure by poly(ADP-ribosyl)ation. Biochem Cell Biol. 1988 Jun;66(6):626–635. doi: 10.1139/o88-072. [DOI] [PubMed] [Google Scholar]
- de Murcia G., Ménissier-de Murcia J., Schreiber V. Poly(ADP-ribose) polymerase: molecular biological aspects. Bioessays. 1991 Sep;13(9):455–462. doi: 10.1002/bies.950130905. [DOI] [PubMed] [Google Scholar]