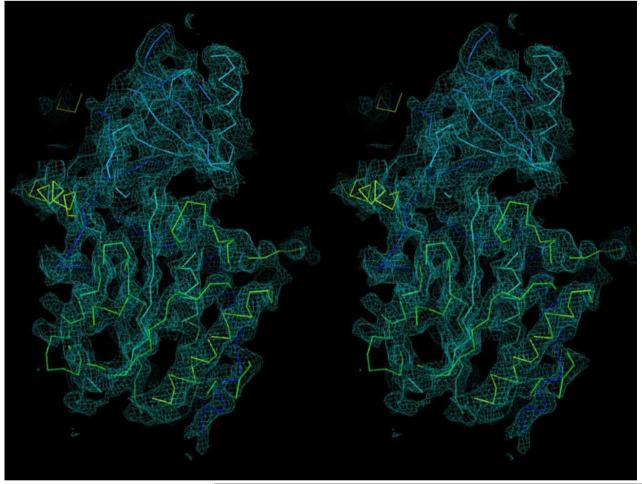
Abstract
The γ-secretase complex, comprising presenilin 1 (PS1), Pen-2, Aph-1, and Nicastrin, is a membrane-embedded protease that controls a number of important cellular functions through substrate cleavage. Aberrant cleavage of the amyloid precursor protein results in aggregation of β-amyloid peptide, which accumulates in the brain and consequently causes Alzheimer’s disease. Here we report the three-dimensional structure of an intact human γ-secretase complex at 4.5 Å resolution, determined by cryo-EM single-particle analysis. The γ-secretase complex comprises a horseshoe-shaped transmembrane domain, which contains 19 transmembrane segments (TMs), and a large extracellular domain (ECD) from Nicastrin, which sits right above the hollow space formed by the TM horseshoe. Intriguingly, Nicastrin ECD is structurally similar to a large family of peptidases exemplified by the glutamate carboxypeptidase PSMA. This structure serves as an important basis for understanding the functional mechanisms of the γ-secretase complex.
γ-Secretase is a membrane-embedded aspartyl protease that cleaves a large number of transmembrane substrate proteins within their membrane-spanning regions, with the cleavage products serving as signaling molecules1,2. This process is known as regulated intramembrane proteolysis (RIP)3. Two extensively studied substrates of γ-secretase are the amyloid precursor protein (APP) and the Notch receptor2. Successive cleavages of APP give rise to several β-amyloid peptides (Aβ), each with different length4. Aberrant accumulation of an aggregation-prone 42-residue Aβ (Aβ42) over a 40-residue product (Aβ40) leads to formation of Aβ plaques in the brain, triggering the development and pathogenesis of Alzheimer’s disease2. Cleavage of the Notch receptor results in the release and translocation of its intracellular domain into the nucleus2. Abnormal Notch signaling is linked to developmental defects and several types of cancer2.
The γ-secretase complex consists of four components: presenilin, Pen-2, Aph-1, and Nicastrin, each containing at least one predicted transmembrane segment (TM)5,6. Together, these proteins have a molecular weight of approximately 170 kDa, whereas the Nicastrin ECD has an additional 30-70 kDa of glycosylation7. Presenilin is the catalytic component and contains nine TMs8-11. Association with Pen-2 facilitates an autocatalytic cleavage of presenilin between TM6 and TM7, producing two fragments known as N-terminal fragment (NTF) and C-terminal fragment (CTF)12,13. Aph-1 and Nicastrin assemble into a stable subcomplex14,15, which then interacts with the CTF of presenilin16,17. Nicastrin contains a large extracellular domain that is thought to be responsible for substrate recruitment18,19. The central role of presenilin in the γ-secretase complex is evidenced by the identification of over 150 missense mutations2, each derived from an Alzheimer’s disease patient.
Despite advances on the functional aspects of γ-secretase, structural characterization has been extremely slow, due in large part to the daunting challenges of expression and purification of the intact γ-secretase. The limited structural information on γ-secretase is restricted to low-resolution images derived from electron microscopy (EM) analysis20-24, an NMR structure of the CTF of presenilin25, and a crystal structure of an archaeal homolog of presenilin26. Consequently, there is little mechanistic understanding of the γ-secretase functions.
During the past several years, we have mounted a rigorous effort to prepare homogeneous, active human γ-secretase for structural investigation. We attempted cryo-EM single particle reconstruction by exploiting technological advances in direct electron detection and statistical image processing27,28. Recent applications of this rapidly developing technology include near 3-Å resolution structures of a mitochondrial ribosome large subunit29, the 12-fold symmetric F420-reducing [NiFe] hydrogenase30, and the 4-fold symmetric TRPV1 complex31. Despite these advances, near-atomic resolution reconstruction remains challenging for smaller, non-symmetric proteins such as human γ-secretase. In this study, we report a three-dimensional structure of this membrane-embedded complex with an overall resolution of 4.5 Å, which reveals its domain architecture, secondary structural elements, TM arrangement, and ECD fold, and yields important functional insights.
Preparation of the γ-secretase complex
The human Aph-1 is encoded by two genes Aph1A and Aph1B, of which Aph1A appears to be more important32. Similarly, human presenilin has two forms: presenelin-1 (PS1) and presenilin-2 (PS2), and PS1 contains the vast majority of disease-derived mutations33. Due to these considerations, we focused our effort on the human γ-secretase that comprises PS1, Pen-2, Aph-1aL (referred to as Aph-1 hereafter), and Nicastrin. We initially assembled a systematic effort to examine the expression levels of the individual components, select subcomplexes, as well as the intact γ-secretase complex in four different expression systems: bacteria, yeast, insect cells, and mammalian cells. We succeeded in transient co-expression of all four components of the human γ-secretase complex in mammalian HEK293F cells. The coding sequences of PS1, Pen-2, Aph-1, and Nicastrin were individually cloned into our custom-designed pMLink plasmid, in which expression of each of the four γ-secretase components was under a separate promotor control (Extended Data Fig. 1a,b). The resulting pMLink plasmid was transfected into HEK293F cells (Fig. 1a).
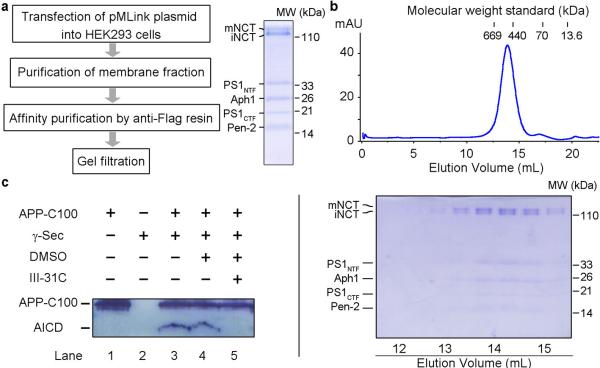
Figure 1. Expression and purification of active human γ-secretase.
a, A schematic diagram of the protocol for the expression and purification of the intact human γ-secretase complex. pMLink is our custom-designed vector for simultaneous co-expression of multiple proteins in mammalian cells. b, A representative gel filtration chromatography of human γ-secretase. The peak fractions were visualized on SDS-PAGE by Coomassie staining. PS1 had been completely auto-proteolyzed into NTF and CTF, whereas Nicastrin (NCT) existed in two forms: immature (iNCT) and mature (mNCT), reflecting differences in glycosylation. c, The purified γ-secretase was proteolytically active against the APP substrate C100. Cleavage of the substrate APP-C100 was blocked by the specific inhibitor III-31C.
To facilitate purification, we used a range of different affinity tags to label the N- or C-termini of the four individual components. The best outcome was achieved with a FLAG tag at the N-terminus of Pen-2. The γ-secretase-containing membrane fractions of HEK293F cells, extracted by the detergent CHAPSO, was purified over an anti-FLAG affinity resin and further fractionated on a size exclusion column (Fig. 1a,b). The resulting γ-secretase complex exhibited excellent solution behavior and could be easily visualized on SDS-PAGE by Coomassie blue staining, free of any major contaminating protein. Importantly, the NTF and CTF were clearly visible, suggesting completion of PS1 autoproteolysis in the presence of the other three components. By contrast, expression of PS1 alone yielded the intact, uncleaved protein (Extended Data Fig. 1c).
Presence of the NTF and CTF is indicative of an active γ-secretase complex. To examine this scenario, we reconstituted a γ-secretase activity assay using the substrate APP-C100, which contains the C-terminal 100 amino acids of APP34. Incubation of γ-secretase with the substrate in a 1:10 molar ratio led to generation of APP intracellular domain (AICD) (Fig. 1c). The presenilin-specific inhibitor III-31C35, but not DMSO, blocked the cleavage of APP-C100. The level of γ-secretase activity is similar to what had been reported18. The same conclusion was obtained for γ-secretase in the presence of amphipol A8-35 under the same buffer condition as that employed in later cryo-EM analysis (Extended Data Fig. 1d). We concluded that the human γ-secretase was in an active conformation. Nonetheless, there is a possibility that, given sample manipulation, the EM structure described below may not represent the fully active conformation.
Cryo-EM analysis of γ-secretase
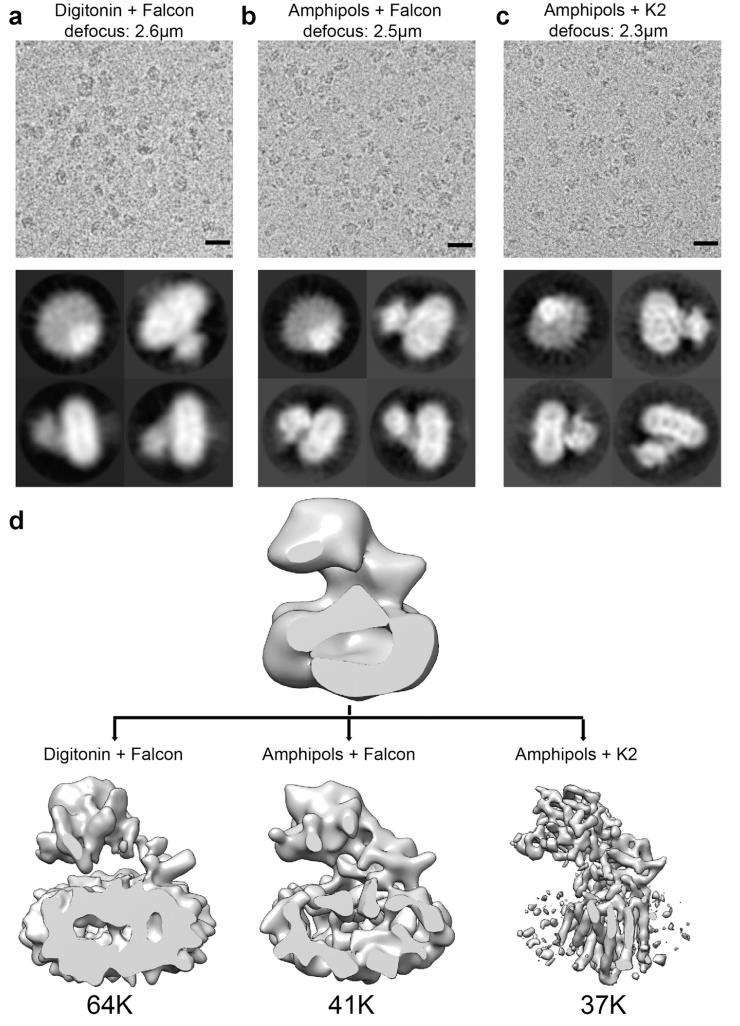
Initial attempts where we imaged γ-secretase in digitonin using an FEI Falcon-II direct-electron detector yielded a 3D reconstruction with a large disc-shaped “body” and a protruding “head”, which could accommodate the TMs and extracellular domains of γ-secretase, respectively (Extended Data Fig. 2a,d). However, despite sharp contrast in the individual particles, this reconstruction showed few internal features. The TMs were not clearly resolved, and the strongest density appeared at the periphery of the disc-shaped body, which likely derived from the detergent digitonin. These results concurred with relatively poor accuracies in the alignment of the particles as estimated in the employed statistical refinement procedure36, and suggested that the disordered nature of the detergent molecules and the small size of the complex precluded correct alignment of the particles.
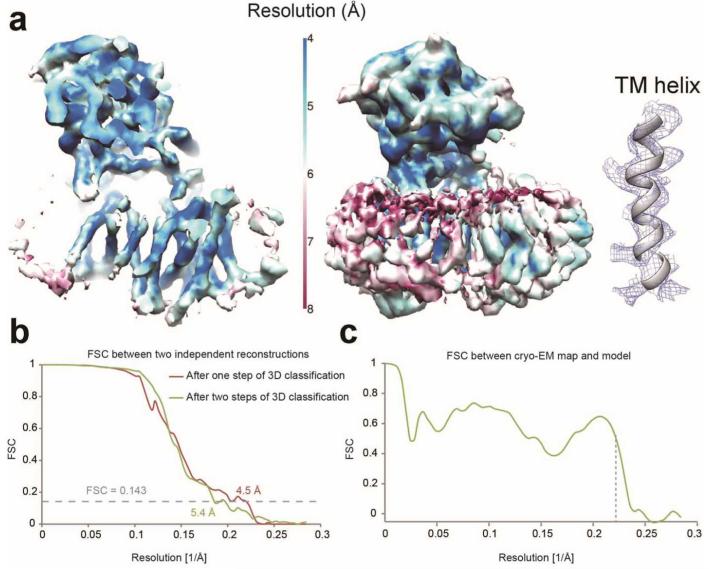
To minimize the effect of the disordered detergent on refinement, we replaced digitonin with amphipol A8-35 (Extended Data Fig. 2b,d). In addition, we also imaged these samples using a Gatan K2 Summit direct-electron detector in single-electron counting mode to achieve higher signal-to-noise ratios at the lower spatial frequencies, which are crucial for particle alignment. Combined with statistical image classification and movie processing27, this approach yielded a markedly improved map with an overall resolution of 4.5 Å (Fig. 2a; Extended Data Figs. 2c,d & 3).
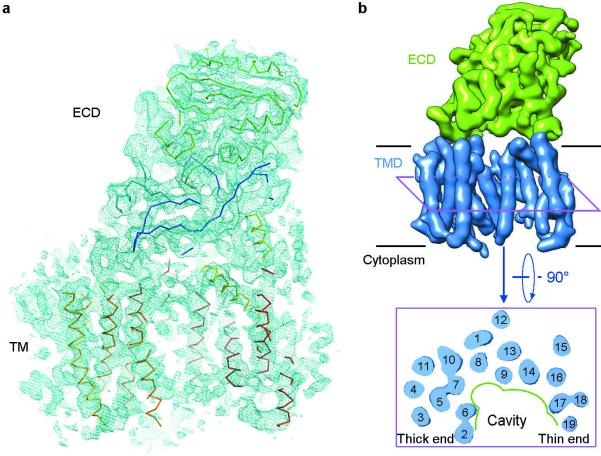
Figure 2. Overall structure of the human γ-secretase complex.
a, An overall density map for the entire human γ-secretase complex. α-Carbon traces are shown for some of the TMs and the ECD. The 5.4-Å map was used in both panels a and b. b, Overall structure of the human γ-secretase complex. Structure of the γ-secretase is viewed from within the plane of lipid membrane (upper panel). The 19 TMs from the four components of γ-secretase are colored blue, whereas the ECD of Nicastrin is shown in green. A cut-through section of the 19 TMs in γ-secretase is shown in the lower panel. The TMs form a horseshoe-shaped structure, with more TMs concentrated at the thick end. The TMs are numbered. Figs. 2a, 3, and 4b were prepared using PyMol49, and Figs. 2b and 4a were made in Chimera50.
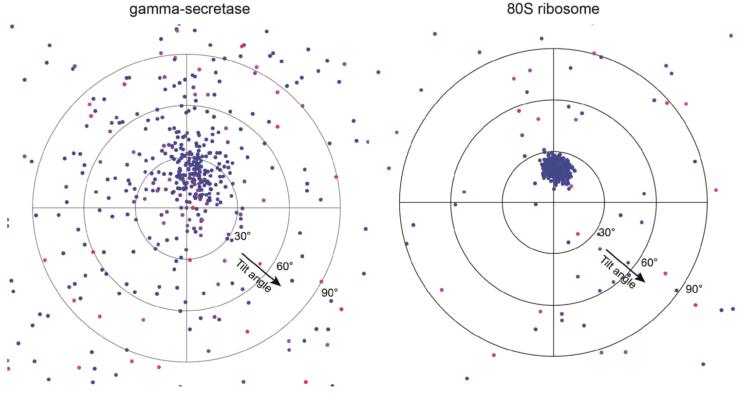
At this resolution, 19 TMs were identified, the β-strands in the Nicastrin ECD were well-resolved, and side-chain densities started to show for portions of the Nicastrin ECD and some of the TMs (Fig. 2a). Densities for some of the linker sequences between neighbouring TMs were improved by further image classification, which led to a map with an overall resolution of 5.4 Å from a subset of the particles (Extended Data Fig. 3b). The overall correctness of the density map and its handedness were confirmed by the tilt-pair test37 (Extended Data Fig. 4).
Overall structure of the γ-secretase
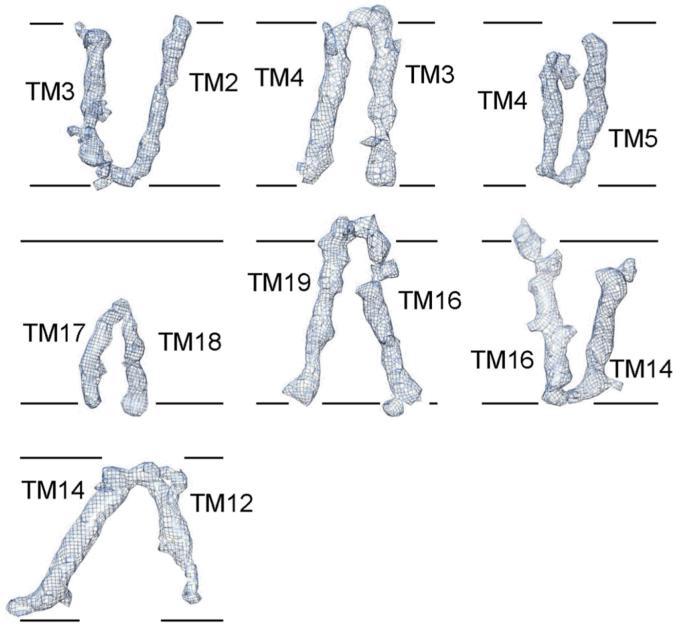
The 19 TMs are organized into a horseshoe-shaped structure (Fig. 2b, upper panel). In contrast to the density for the TMs, the density for the connecting sequences between neighbouring TMs is weak or absent, likely reflecting the disordered nature in these hydrophilic loops. Nonetheless, at least seven TMs are connected by strong density (Extended Data Fig. 5), suggesting their order of linkage in γ-secretase. For ease of discussion, we numbered the 19 TMs (Fig. 2b, lower panel). These TMs exhibit quite different lengths, with two connected TMs (TM17 and TM18) going half-way into the membrane from the cytoplasmic side (Fig. 2b, Extended Data Fig. 5). Two bent TMs (TM6 and TM7) are placed on the concave side of the horseshoe, facing the hollow center. The large, empty pocket appears to be poised for binding to some structural element – perhaps the substrate protein.
The distribution of the 19 TMs is uneven, with considerably more TMs concentrated on one end of the horseshoe-shaped structure (referred to as the “thick” end) than the other end (“thin” end). In the thin end, there are no more than two layers of TMs when viewed perpendicular to the membrane (Fig. 2b, lower panel). By contrast, the thick end has at least three layers of TMs. The archaeal homologue of PS1, mmPSH, exhibits a relatively complex membrane topology, with three layers of TMs in an inactive conformation26. Assuming all TMs in the γ-secretase have been identified in the current EM maps, this analysis suggests that PS1 might be located within the thick end of the TM horseshoe.
There is a large region of well-defined density outside the membrane-spanning region, and the density vastly exceeds that of the sequences from γ-secretase on the intracellular side. Among the four components of γ-secretase, Nicastrin is the only one that has a sizable ECD, and most of the extracellular density is thus attributable to Nicastrin (Fig. 2). Intriguingly, Nicastrin ECD is located right above the hollow center of the TM horseshoe and interacts closely with the extracellular loops of several TMs on both ends of the horseshoe (Fig. 2b). This organization is consistent with the reported function of substrate recruitment for Nicastrin18.
Our human γ-secretase contains four full-length proteins: PS1 (residues 1-467), Pen-2 (residues 1-101), Aph-1 (residues 1-265), and Nicastrin (residues 1-709). With glycosylation of Nicastrin, the predicted molecular weight of the intact γ-secretase is approximately 230-kDa22. The observed density accounts for approximately half of the total molecular weight of γ-secretase, with the 19 TMs accommodating about 500 residues and Nicastrin ECD containing about 650 residues. The lack of obvious density for the other sequences likely reflects their flexible nature, including the 30-70 kDa of oligosaccharides on glycosylated residues in the Nicastrin ECD. Only 43 of the 181 residues predicted to be on the cytoplasmic side of PS1 are hydrophobic, representing 24 percent of the total sequences and unlikely to be sufficient for formation of a stable structural core. In addition, the extracellular sequences for Pen-2 (residues 1-19 and 78-101) are predicted to be hydrophilic and flexible. These residues are missing in the current maps.
Structure of Nicastrin ECD
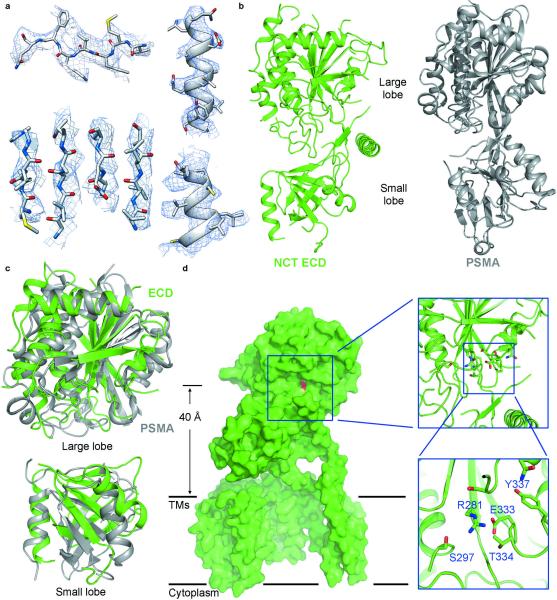
Nicastrin ECD was previously predicted to conform to the aminopeptidase superfamily fold38. The relatively high-resolution features in the density for Nicastrin ECD (Fig. 2a, Fig. 3a, Extended Data Fig. 6) prompted us to pursue a model for its domain architecture. To facilitate this task, we searched for sequences in the Protein Data Bank (PDB) that are homologous to those of Nicastrin ECD. One of the matches was the glutamate carboxyl peptidase PSMA (PDB code 2XEF39) (Extended Data Fig. 7), confirming the earlier prediction38. Of the 218 aligned amino acids between PSMA and Nicastrin, 52 are identical and 80 are similar. Visual inspection of the extracellular EM density revealed an excellent match to the structure of PSMA39. The conserved topology between these two structures allowed tracing of approximately 400 residues with side chains and 20 residues as poly-Ala sequences in the Nicastrin ECD (Fig. 3b). The remaining unassigned EM density is relatively poor and can accommodate about 200 residues. The modelled part of Nicastrin ECD resembles a dumbbell, with a large lobe and a small lobe (Fig. 3b), which can be superimposed with those of PSMA with root-mean-squared deviations (RMSD) of about 2.6 and 3.6 Å over 231 and 111 aligned Cα atoms, respectively (Fig. 3c).
Figure 3. Structure of the extracellular domain (ECD) of Nicastrin.
a, Representative cryo-EM density for β-strands (left panels) and α-helices (right panels) of the Nicastrin ECD. The 4.5-Å map was used here. b, The overall structure of Nicastrin ECD closely resembles that of the glutamate carboxyl peptidase PSMA39. The atomic model of Nicastrin ECD is shown in rainbow cartoon, with the N- and C-termini colored blue and red, respectively. The structure of PSMA is displayed in the right panel for comparison. c, Structural comparison between Nicastrin (green) and PSMA39 (grey) for the large lobe (upper panel) and the small lobe (lower panel). d, Identification of a putative substrate-binding site in Nicastrin ECD. A surface groove on Nicastrin ECD, located 40 Å above the lipid membrane, faces the hollow center of the TM horseshoe. Glu333, which is thought to play an important role in substrate recruitment18,19, resides in the groove.
Discussion
The structural homology between Nicastrin ECD and the peptidase PSMA may not support the possibility that Nicastrin could serve as an active protease in cells. PSMA is a zinc metallo-protease and the majority of the residues that coordinate the two zinc ions in PSMA have been replaced in Nicastrin (Extended Data Fig. 7). Moreover, we have been unable to detect any protease activity for Nicastrin ECD in vitro using a variety of potential substrate proteins under diverse conditions. Nonetheless, the fact that Nicastrin ECD shares a conserved fold as PSMA and other peptidases supports the notion that Nicastrin may be involved in substrate recruitment18,19. Nicastrin ECD appears to contain a surface groove approximately 40 Å above the lipid membrane, facing the hollow center of the TM horseshoe (Fig. 3d). This surface groove could be a putative substrate-binding site. Because the active site of PS1 is predicted to be located approximately 20 Å below the surface of the lipid membrane26, the putative substrate-binding site is at least 60 Å away from the catalytic Asp residues in PS1. Assuming the N-terminus of the BACE cleavage product APP-C99 is recognized by this surface groove18, a distance of 60 Å can be conveniently spanned by the primary cleavage products of APP-C99 – Aβ40 and Aβ42. Supporting this analysis, Glu333, which was thought to be responsible for substrate binding18,19, resides at the center of the groove (Fig. 3d). The residue in PSMA that corresponds to Glu333 of Nicastrin is directly involved in zinc binding39.
The lack of side chain features in the density for the 19 TMs does not allow assignment of the four components. The weak density for the loops connecting neighbouring TMs further complicates the assignment task. Nonetheless, we suggest a speculative assignment, in which all 9 TMs of PS1 are located within the thick end of the TM horseshoe (Extended Data Fig. 8a). The PS1 homologue mmPSH contains three layers of TMs26. Based on the current EM density, the thick end is the only place in the horseshoe structure with three layers of TMs. The putative assignment of TM1 from PS1 was facilitated by the bent nature of TM1 in mmPSH26. Pen-2 was shown to be in close proximity of the CTF of PS140 and directly bind TM4 of PS141,42, and Aph-1 and Nicastrin were thought to interact with the CTF of PS116,17; these features are recapitulated in our model. In this speculative model, TM6 and TM7 of PS1, which harbor the catalytic Asp residues, and TM9, which contains the substrate recognition sequence, face the hollow center of the TM horseshoe (Extended Data Fig. 8a). This analysis suggests the location of substrate cleavage by γ-secretase. The two TMs of Pen-2 are likely inserted between TM7 and TM8 on the cytoplasmic side, leading to a major conformational rearrangement of the TMs in PS1 compared to those in mmPSH26 and opening of the putative substrate entry site (Extended Data Fig. 8b). This analysis might explain why PS1 autoproteolysis only occurs in the presence of Pen-2. Despite the charm of this model, we cannot rule out the opposing possibility where PS1 is placed in the thin end (Extended Data Fig. 8c). After all, it remains to be seen whether mmPSH represents a sound structural model for PS1 or whether all TMs in the γ-secretase have been identified.
Although the overall resolution of our structure is 4.5 Å, the resolution for the TMs is lower and thus does not allow modeling of specific side chains. Compared to the detergent choice of digitonin, amphipol was clearly better in the cryo-EM analysis of γ-secretase and helped improve the quality of image reconstruction. The use of amphipol was previously reported in at least two cryo-EM studies of membrane proteins31,43. As a new class of surfactants designed to improve the solubility of membrane proteins44, amphipols may prove to be an important tool for future EM-based investigation of many other membrane proteins.
Recent structural investigations of intramembrane proteases such as the prokaryotic homologues of rhomboid45-47, S2P48, and presenilin/SPP26 have provided tantalizing hints about functional mechanisms of these membrane-embedded signaling proteases. In this study, we report the first cryo-EM density map of human γ-secretase in which individual β-strands are clearly separated in Nicastrin ECD and 19 TMs form a horseshoe-like structure. Our observed structural features are different from those that were derived from previous low-resolution EM studies of γ-secretase20-24. Our structure marks an important step towards elucidating the molecular mechanisms of this key enzyme whose aberrant activity engenders the Alzheimer’s disease.
Method
Construction of pMLink vector for co-expression of multiple proteins
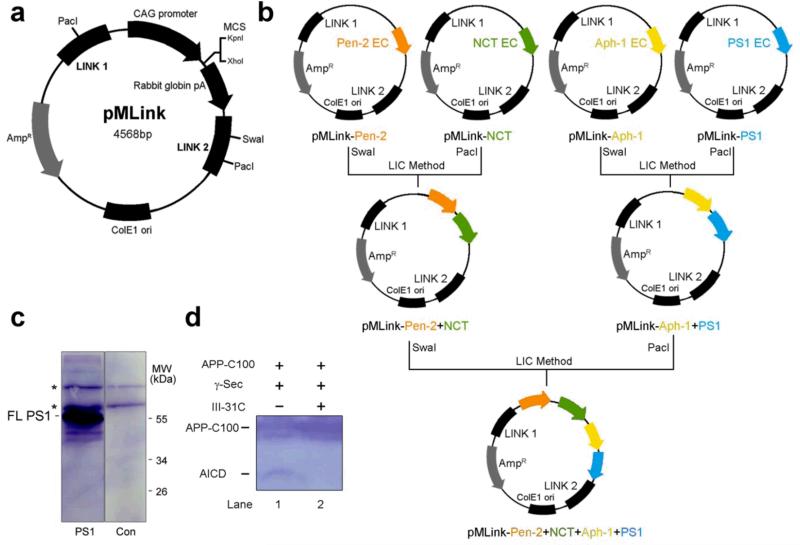
To construct the pMLink vector, the plasmid pCAG51 was digested by SalI and BamHI. The ~2000-bp product that contains the AmpR gene and the ColE1 sequence was ligated with the annealing product of two primers (Forward: 5′-tcgagCTTAATTAACAACACCATTTGctcgagacagatctGTAACAACACCATTTAAATGGAGTGGTTACAAATGGAGTGGTTAATTAAG-3′; Reverse: 5′-gatcgCTTAATTAACCACTCCATTTGTAACCACTCCATTTAAATGGTGTTGTTACagatctgtctcgagCAAATGGTGTTGTTAATTAAG-3′). Then the resulting plasmid was digested by XhoI and BglII, and ligated with the ~2500-bp fragment of pCAG that was released by SalI and BamHI digestion. The final empty pMLink vector harbors two LINK sequences52,53, one upstream (LINK1) and the other downstream (LINK2) of the expression cassette of CAG, which contains a promoter, multiple cloning sites, and a transcriptional terminator ploy-A sequence. LINK1 (5′-CTTAATTAACAACACCATTTG-3′) contains a Pac I restriction enzyme cleavage site and LINK2 (5′-GTAACAACACCATTTAAATGGAGTGGTTACAAATGGAGTGGTTAATTAA G-3′) a SwaI and a PacI sites53. The LINK sequences allow insertion of a PacI-digested fragment from one plasmid at the SwaI site of a second plasmid by ligation-independent cloning (LIC) method52,53. In the first step, the coding DNA sequences for human PS1, Pen-2, Aph1aL, and Nicastrin were individually cloned into the pMLink vector at the multiple cloning sites. Pen-2 contains an N-terminal FLAG tag, Aph1aL has a C-terminal HA tag, and Nicastrin carries a C-terminal V5-His6 tag. Then, pMLink-Pen-2 and pMLink-Nicastrin were combined by the LIC method to generate pMLink-Pen-2-Nicastrin, whereas pMLink-Aph-1 and pMLink-PS1 gave rise to pMLink-Aph-1-PS1. In the final step, the two plasmids pMLink-Pen-2-Nicastrin and pMLink-Aph-1-PS1 were combined to generate the final plasmid pMLink-Pen-2-Nicastrin-Aph-1-PS1.
Transient expression of the human γ-secretase complex
The coding DNA sequences for the human Pen-2, Aph1aL, PS1, and Nicastrin (NCT) were sequentially cloned into the pMLink vector. Pen-2 contains an N-terminal FLAG tag, Aph1aL has a C-terminal HA tag, and NCT carries a C-terminal V5-His6 tag. HEK 293F cells (Invitrogen) were cultured in SMM 293T-I medium (Sino Biological Inc.) at 37 °C under 5% CO2 and 75% humidity in a Multitron-Pro shaker (Infors, 130 rpm). When the cell density reached 2×106 cells/ml, the pMLink plasmid containing all four components of the human γ-secretase complex was transfected into the cells. For one liter of cell culture, 1.5 mg of plasmid were pre-mixed with 4 μg of 25-kDa linear polyethylenimines (PEIs) (Polysciences) in 50 ml fresh medium for 15-30 minutes before transfection. Transfection was started by adding the mixture into cell culture and incubating for 30 minutes, followed by dilution of the cell culture to 1.5×106 cells/ml with fresh medium. Transfected cells were cultured for 60 hours before harvesting. Initial expression tests were examined by Western blot using monoclonal antibodies against the FLAG tag, HA tag, V5 tag and the NTF of PS1 (Merck Millipore).
Purification of the γ-secretase complex
Transfected cells were centrifuged at 800g and resuspended in a lysis buffer containing 25 mM HEPES, pH 7.4, 150 mM NaCl and protease inhibitor cocktails (Amresco). After sonication on ice, the suspension was centrifuged at 150,000g for 1 hour to pellet the cell membrane. Cell membrane was resuspended by the same lysis buffer mentioned above and supplemented with 1% CHAPSO. After incubation for 2 hours at 4 °C, the suspension was centrifuged at 150,000 g for 30 minutes and the supernatant was incubated with Anti-FLAG M2 affinity gel (Sigma) for 30 minutes at 4 °C. The resin was washed three times, each with 10 ml buffer containing 25 mM HEPES, pH 7.4, 150 mM NaCl, and 0.1% digitonin (Sigma). The γ-secretase complex was eluted with a buffer containing 25 mM HEPES, pH 7.4, 150 mM NaCl, 0.1% digitonin and 200 μg/ml FLAG peptide (Sigma). Protein solution was concentrated with a 10-kD cut-off Centricon (Millipore) and further purified by Superose-6 column (GE Healthcare). The peak fractions were concentrated to 1.5 mg/ml, and supplemented with amphipol A8-35 (Anatrace) to a final concentration of 4.5 mg/ml. After incubation at 4 °C for 4 hours, 60 mg Bio-Beads SM-2 (Bio-Rad) was added and incubated overnight. Free amphipol molecules were removed by Superose-6 column. Peak fractions were collected for cryo-EM studies. This expression and purification strategy gives a typical yield of 0.2 mg homogeneous γ-secretase complex for every liter of HEK293F cell culture.
γ-secretase activity assays
APP-C100 protein with a C-terminal Myc-His6 tag was overexpressed in E. coli and purified by Ni2+-NTA affinity resin. The elution buffer contained 1% (w/v) CHAPSO, 25 mM HEPES, pH 7.4, and 250 mM imidazole. The eluted materials were applied to gel filtration (Superdex-200, GE Healthcare) in 1% (w/v) CHAPSO, 25 mM HEPES, pH 7.4, and 150 mM NaCl. 1μM purified APP-C100 protein were mixed with purified γ-secretase in 0.2% (w/v) CHAPSO, 50 mM HEPES, pH 7.0, 0.1% (w/v) phosphatidylcholine, and 0.025% (w/v) phosphatidylethanolamine, and incubated at 37 °C for 12 hours. The cleavage product, AICD-Myc was detected by anti-Myc antibody (GE Healthcare). A γ-secretase inhibitor, N-[[[(2R,3S)-3-[[(1,1-dimethylethoxy)carbonyl]amino]-2-hydroxy-4-phenylbutyl](phenylmethyl) amino]carbonyl]-L-leucyl-L-valine methyl ester WPE-III-31-C (abbreviated as III-31C, Sigma-Aldrich), was dissolved in DMSO and used for inhibition assay with a final concentration of 1 μM.
Electron microscopy
Aliquots of 3 μl of purified γ-secretase at a concentration of ~4 μM were placed on glow-discharged holey carbon grids (Quantifoil Cu R1.2/1.3). Grids were blotted for 4 seconds and flash frozen in liquid ethane using an FEI Vitrobot. Grids were transferred to an FEI Tecnai Polara or a Titan Krios electron microscope that was operating at 300 kV. Images were recorded manually using an FEI Falcon-II detector on the Polara microscope at a calibrated magnification of 79,096 (yielding a pixel size of 1.77 Å), or using a K2 Summit detector in counting mode on the Titan Krios microscope at a calibrated magnification of 28,409 (yielding a pixel size of 1.76 Å). The K2 detector was mounted after a Gatan Imaging Filter (GIF), but the filter was not used to remove any inelastic scattering. However, to prevent any image distortions, GIF-tuning was carefully checked throughout data collection. A dose rate of 17 electrons/Å2 per second and an exposure time of 2 seconds were used on the Falcon, and a dose rate of ~2.5 electrons/Å2 per second and exposure time of 15 seconds on the K2. Defocus values in the final K2 data set ranged from 1.4-4.4 μm, and 15 video frames were recorded.
Image processing
We used RELION (version 1.3-beta) for automated selection of 606,755 particles from 1,471 micrographs in the final K2 data set. Contrast transfer function parameters were estimated using CTFFIND354. All 2D and 3D classifications and refinements were performed using RELION36. We used reference-free 2D class averaging and 3D classification to discard false positives from the automated picking procedure, and selected 144,545 particles for a first 3D refinement. The program PRIME in the software package SIMPLE was used to generate an initial model55. In a subsequent 3D classification run with four classes, an angular sampling of 3.75° was combined with local angular searches around the refined orientations, and the refined model from the first refinement was used as a starting model. This yielded a subset of 37,310 particles, for which a reconstruction with improved density in the trans-membrane domain was obtained.
In order to correct for beam-induced movements, the 15 video frames for each micrograph were first aligned using whole-image motion correction28. Secondly, particle-based beam-induced movement correction was performed using statistical movie processing in RELION27. For these calculations, we used running averages of 7 movie frames, and a standard deviation of 1 pixel for the translational alignment. To further increase the accuracy of the per-particle movement correction, we used the beta version of RELION-1.3 to fit linear tracks through the optimal translations for all running averages, and included neighbouring particles on the micrograph in these fits. In addition, we employed a resolution and dose-dependent model for the radiation damage, where each frame is weighted with a different B-factor as estimated from single-frame reconstructions.
After beam-induced movement correction, the first refinement from 144,545 particles yielded a map with a resolution of 4.5 Å. FSCs calculated for separately masked domain indicated local resolutions of 4.5 and 5.1 Å for the ECD and TMD, respectively. The second refinement, after selecting 37,310 in an additional 3D classification step, yielded a map with improved density for some connecting helices in the trans-membrane domain at an overall resolution of 5.4 Å, and local resolutions of 4.7 and 5.6 Å for the ECD and TMD, respectively.
Reported resolutions are based on the gold-standard FSC=0.143 criterion56, and FSC curves were corrected for the effects of a soft mask on the FSC curve using high-resolution noise substitution56. Prior to visualization, all density maps were corrected for the modulation transfer function (MTF) of the detector, and then sharpened by applying a negative B-factor that was estimated using automated procedures37. A tilt-pair test37 was performed to validate the overall correctness of our map and to confirm its handedness (Extended Data Fig. 4). Local resolution variations were estimated using ResMap57.
Model building and refinement
The atomic model for Nicastrin ECD was built in COOT58 and refined with stereo-chemical and homology restraints in REFMAC59, using the modified versions of these programs for cryo-EM maps29. Throughout refinement, reference and secondary structure restraints were applied using the PSMA structure as a reference model60.
Extended Data
Extended Data Figure 1. Construction of the pMLink vector for co-expression of the four components of γ-secretase.
a, A schematic diagram of the empty pMLink plasmid. The empty pMLink plasmid was created by insertion of the LINK1 and LINK2 sequences52,53 into the pCAG vector51. The two LINK sequences allow insertion of a PacI digested fragment from one plasmid at the SwaI site of a second plasmid by ligation-independent cloning (LIC) method52,53. The four components of γ-secretase were individually cloned into the multiple cloning sites of the empty pMLink vector, generating four plasmids. b, Construction of the pMLink vector that contains all four components of γ-secretase. First, pMLink-Pen-2 and pMLink-Nicastrin were combined by the LIC method to generate pMLink-Pen-2-Nicastrin, whereas pMLink-Aph-1aL and pMLink-PS1 gave rise to pMLink-Aph-1aL-PS1. Then, the two plasmids pMLink-Pen-2-Nicastrin and pMLink-Aph-1aL-PS1 were combined to generate the final plasmid pMLink-Pen-2-Nicastrin-Aph-1aL-PS1. EC: expression cassette (containing promoter, target gene and poly A sequence). c, Expression of PS1 alone resulted in the production of the uncleaved PS1 protein. Transfection of pMLink-PS1 into HEK293F cells led to expression of the uncleaved, full-length PS1. Shown here is result of a Western blot using a monoclonal antibody against the NTF of PS1. d, The purified γ-secretase in amphipol A8-35 was proteolytically active against the APP substrate C100. Cleavage of the substrate APP-C100 was blocked by the specific inhibitor III-31C. The same γ-secretase in amphipol A8-35 under the same buffer was used for cryo-EM analysis.
Extended Data Figure 2. Cryo-EM analysis of the human γ-secretase complex.
a, Analysis of γ-secretase in digitonin imaged on back-thinned Falcon II. A representative electron micrograph (scale bar, 20 nm) and reference-free 2D class averages are shown in the upper and lower panels, respectively. b, Analysis of γ-secretase in amphipols imaged on back-thinned Falcon II. c, Analysis of γ-secretase in amphipols imaged on K2 Summit. d, Comparison of densities for the three methods of imaging described above. The fewest number of particles (37K) was used for the generation of higher-resolution images for samples in amphipols on the K2 Summit.
Extended Data Figure 3. Density and resolution of the human γ-secretase complex.
a, Resolution limit (color-coded) of the cryo-EM density for γ-secretase in amphipols imaged on K2 Summit. The left panel shows a cut-through view on the interior of the maps. The middle panel shows the entire maps. The low-resolution density (reddish color) likely represents that from amphipols. The right panel shows the best density for TM in the map, with some of the side chain features visible. The 4.5-Å EM map was used here. b, Gold-standard FSC curves for the density maps. The resolution limits reached 4.5 Å and 5.4 Å, respectively, after one (red) or two (green) steps of 3D classification. c, The FSC curve between the Nicastrin ECD atomic model and the corresponding part of the cryo-EM map.
Extended Data Figure 4. Tilt-pair validation of the correct handedness of γ-secretase.
Tilt-pair validation plots are shown for γ-secretase and 80S ribosome imaged on the same electron microscope at 0° and 20° tilt angles, under the same magnification, and using the same detector. The position of each dot represents the direction and the amount of tilting for a particle pair in polar coordinates. Blue dots correspond to in-plane tilt transformations; red dots to out-of-plane tilt transformations. For both samples blue dots cluster in the same region of the plot at a tilt angle of approximately 20°, which validates the structure and confirms that our γ-secretase map is in the same handedness as the 80S ribosome map. Note that the increased scatter of the points in the γ-secretase plot results from the significantly smaller molecular weight compared to the 80S ribosome, which results in much less accurate orientational assignments.
Extended Data Figure 5. Connecting density of the TMs.
Seven pairs of TMs are shown for which there is EM density connecting the two TMs. The orientation of these seven pairs of TMs reflects that in the intact γ-secretase complex, with extracellular space on the upper side and cytoplasm on the lower side. The 5.4-Å density map was used here. This figure was prepared using Chimera50.
Extended Data Figure 6. The density map for the ECD of Nicastrin in stereo view.
The EM density is colored cyan, whereas the built model is displayed in Cα trace. The 4.5-Å density map was used here. This figure was prepared using PyMol49.
Extended Data Figure 7. Sequence alignment between Nicastrin ECD and the glutamate carboxyl peptidase PSMA.
Identical amino acids between Nicastrin and PSMA are highlighted in red, and conserved residues are colored blue. PSMA39 residues that coordinate the first and second zinc atoms are identified by red squares and red circles, respectively, above the sequences. Residues in Nicastrin that are aligned to the proximity of zinc-binding sites in PSMA are indicated by blue triangles.
Extended Data Figure 8. Speculative assignment of TMs and implications for γ-secretase function.
a, TMs 1-11 are putatively assigned to PS1 (blue) and Pen-2 (purple). This assignment is based on three assumptions: (1) PS1 is structurally homologous to mmPSH26 that contains three layers of TMs; (2) all TMs in the intact γ-secretase have been identified in the current EM structure; and (3) PS1 does not undergo drastic structural rearrangement upon binding to the other three components. Two perpendicular views and a cut-through section are shown. b, Structural comparison of PS1 (blue) with its archaeal homologue mmPSH (grey). PS1 and mmPSH share 23 percent sequence identity in the TM region. Structure of mmPSH represents the inactive conformation26. In this speculative comparison, the two TMs of Pen-2 are inserted between TM7 and TM8 of PS1, likely triggering marked conformational shifts in nearby TMs. c, A contrasting model of TM assignment in γ-secretase. Although less likely, we cannot rule out the possibility that PS1 is located at the thin end of the TM horseshoe. In this case, Aph-1 would be assigned to the thick end.
ACKNOWLEDGMENTS
We thank Mike Wolfe and Dennis Selkoe for support and encouragement, and Shaoxia Chen, Christos Savva, Jake Grimmett and Toby Darling for technical support. This work was funded by the Ministry of Science and Technology of China (2009CB918801 to YS), National Natural Science Foundation of China (projects 30888001, 31021002 and 31130002 to YS), a European Union Marie Curie Fellowship (to XCB), and the UK Medical Research Council (MC_UP_A025_1013, to SHWS).
Footnotes
Author Information
The modelled atomic coordinates of Nicastrin has been deposited in the Protein Data Bank with the accession code XXXX. In addition, the 4.5 Å and 5.4 Å EM maps have been deposited in EMDB with accession codes YYYY and ZZZZ, respectively.
The authors declare no competing financial interests.
References
- 1.Selkoe DJ, Wolfe MS. Presenilin: running with scissors in the membrane. Cell. 2007;131:215–221. doi: 10.1016/j.cell.2007.10.012. [DOI] [PubMed] [Google Scholar]
- 2.De Strooper B, Iwatsubo T, Wolfe MS. Presenilins and gamma-Secretase: Structure, Function, and Role in Alzheimer Disease. Cold Spring Harbor perspectives in medicine. 2012;2:a006304. doi: 10.1101/cshperspect.a006304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brown MS, Ye J, Rawson RB, Goldstein JL. Regulated intramembrane proteolysis: a control mechanism conserved from bacteria to humans. Cell. 2000;100:391–398. doi: 10.1016/s0092-8674(00)80675-3. [DOI] [PubMed] [Google Scholar]
- 4.Wolfe MS. Toward the structure of presenilin/gamma-secretase and presenilin homologs. Biochim Biophys Acta. 2013;1828:2886–97. doi: 10.1016/j.bbamem.2013.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.De Strooper B. Aph-1, Pen-2, and Nicastrin with Presenilin generate an active gamma-Secretase complex. Neuron. 2003;38:9–12. doi: 10.1016/s0896-6273(03)00205-8. [DOI] [PubMed] [Google Scholar]
- 6.Kimberly WT, et al. Gamma-secretase is a membrane protein complex comprised of presenilin, nicastrin, Aph-1, and Pen-2. Proc Natl Acad Sci U S A. 2003;100:6382–7. doi: 10.1073/pnas.1037392100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schedin-Weiss S, Winblad B, Tjernberg LO. The role of protein glycosylation in Alzheimer disease. FEBS J. 2014;281:46–62. doi: 10.1111/febs.12590. [DOI] [PubMed] [Google Scholar]
- 8.Wolfe MS, et al. Two transmembrane aspartates in presenilin-1 required for presenilin endoproteolysis and gamma-secretase activity. Nature. 1999;398:513–7. doi: 10.1038/19077. [DOI] [PubMed] [Google Scholar]
- 9.De Strooper B, et al. Deficiency of presenilin-1 inhibits the normal cleavage of amyloid precursor protein. Nature. 1998;391:387–90. doi: 10.1038/34910. [DOI] [PubMed] [Google Scholar]
- 10.Strooper B, et al. A presenilin-1-dependent gamma-secretase-like protease mediates release of Notch intracellular domain. Nature. 1999;398:518–22. doi: 10.1038/19083. [DOI] [PubMed] [Google Scholar]
- 11.Struhl G, Greenwald I. Presenilin is required for activity and nuclear access of Notch in Drosophila. Nature. 1999;398:522–5. doi: 10.1038/19091. [DOI] [PubMed] [Google Scholar]
- 12.Thinakaran G, et al. Endoproteolysis of presenilin 1 and accumulation of processed derivatives in vivo. Neuron. 1996;17:181–90. doi: 10.1016/s0896-6273(00)80291-3. [DOI] [PubMed] [Google Scholar]
- 13.Takasugi N, et al. The role of presenilin cofactors in the gamma-secretase complex. Nature. 2003;422:438–441. doi: 10.1038/nature01506. [DOI] [PubMed] [Google Scholar]
- 14.Gu Y, et al. APH-1 interacts with mature and immature forms of presenilins and nicastrin and may play a role in maturation of presenilin.nicastrin complexes. J Biol Chem. 2003;278:7374–80. doi: 10.1074/jbc.M209499200. [DOI] [PubMed] [Google Scholar]
- 15.LaVoie MJ, et al. Assembly of the gamma-secretase complex involves early formation of an intermediate subcomplex of Aph-1 and nicastrin. J Biol Chem. 2003;278:37213–22. doi: 10.1074/jbc.M303941200. [DOI] [PubMed] [Google Scholar]
- 16.Steiner H, Winkler E, Haass C. Chemical cross-linking provides a model of the gamma-secretase complex subunit architecture and evidence for close proximity of the C-terminal fragment of presenilin with APH-1. J Biol Chem. 2008;283:34677–86. doi: 10.1074/jbc.M709067200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kaether C, et al. The presenilin C-terminus is required for ER-retention, nicastrin-binding and gamma-secretase activity. Embo J. 2004;23:4738–48. doi: 10.1038/sj.emboj.7600478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shah S, et al. Nicastrin functions as a gamma-secretase-substrate receptor. Cell. 2005;122:435–47. doi: 10.1016/j.cell.2005.05.022. [DOI] [PubMed] [Google Scholar]
- 19.Dries DR, et al. Glu-333 of nicastrin directly participates in gamma-secretase activity. J Biol Chem. 2009;284:29714–24. doi: 10.1074/jbc.M109.038737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lazarov VK, et al. Electron microscopic structure of purified, active gamma-secretase reveals an aqueous intramembrane chamber and two pores. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:6889–94. doi: 10.1073/pnas.0602321103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ogura T, et al. Three-dimensional structure of the gamma-secretase complex. Biochemical and biophysical research communications. 2006;343:525–34. doi: 10.1016/j.bbrc.2006.02.158. [DOI] [PubMed] [Google Scholar]
- 22.Osenkowski P, et al. Cryoelectron microscopy structure of purified gamma-secretase at 12 A resolution. Journal of molecular biology. 2009;385:642–52. doi: 10.1016/j.jmb.2008.10.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Renzi F, et al. Structure of gamma-secretase and its trimeric pre-activation intermediate by single-particle electron microscopy. J Biol Chem. 2011;286:21440–9. doi: 10.1074/jbc.M110.193326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li Y, et al. Structural Interactions between Inhibitor and Substrate Docking Sites Give Insight into Mechanisms of Human PS1 Complexes. Structure. 2014;22:125–35. doi: 10.1016/j.str.2013.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sobhanifar S, et al. Structural investigation of the C-terminal catalytic fragment of presenilin 1. Proc Natl Acad Sci U S A. 2010;107:9644–9649. doi: 10.1073/pnas.1000778107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li X, et al. Structure of a presenilin family intramembrane aspartate protease. Nature. 2013;493:56–61. doi: 10.1038/nature11801. [DOI] [PubMed] [Google Scholar]
- 27.Bai XC, Fernandez IS, McMullan G, Scheres SH. Ribosome structures to near-atomic resolution from thirty thousand cryo-EM particles. Elife. 2013;2:e00461. doi: 10.7554/eLife.00461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li X, et al. Electron counting and beam-induced motion correction enable near-atomic-resolution single-particle cryo-EM. Nat Methods. 2013;10:584–90. doi: 10.1038/nmeth.2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Amunts A, et al. Structure of the yeast mitochondrial large ribosomal subunit. Science. 2014;343:1485–9. doi: 10.1126/science.1249410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Allegretti M, Mills DJ, McMullan G, Kuhlbrandt W, Vonck J. Atomic model of the F420-reducing [NiFe] hydrogenase by electron cryo-microscopy using a direct electron detector. Elife. 2014;3:e01963. doi: 10.7554/eLife.01963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liao M, Cao E, Julius D, Cheng Y. Structure of the TRPV1 ion channel determined by electron cryo-microscopy. Nature. 2013;504:107–12. doi: 10.1038/nature12822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Serneels L, et al. Differential contribution of the three Aph1 genes to gamma-secretase activity in vivo. Proc Natl Acad Sci U S A. 2005;102:1719–24. doi: 10.1073/pnas.0408901102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tang YP, Gershon ES. Genetic studies in Alzheimer’s disease. Dialogues Clin Neurosci. 2003;5:17–26. doi: 10.31887/DCNS.2003.5.1/yptang. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li YM, et al. Presenilin 1 is linked with gamma-secretase activity in the detergent solubilized state. Proc Natl Acad Sci U S A. 2000;97:6138–43. doi: 10.1073/pnas.110126897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kornilova AY, Das C, Wolfe MS. Differential effects of inhibitors on the gamma-secretase complex. Mechanistic implications. J Biol Chem. 2003;278:16470–3. doi: 10.1074/jbc.C300019200. [DOI] [PubMed] [Google Scholar]
- 36.Scheres SH. RELION: Implementation of a Bayesian approach to cryo-EM structure determination. J Struct Biol. 2012;180:519–30. doi: 10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rosenthal PB, Henderson R. Optimal determination of particle orientation, absolute hand, and contrast loss in single-particle electron cryomicroscopy. J Mol Biol. 2003;333:721–45. doi: 10.1016/j.jmb.2003.07.013. [DOI] [PubMed] [Google Scholar]
- 38.Fagan R, Swindells M, Overington J, Weir M. Nicastrin, a presenilin-interacting protein, contains an aminopeptidase/transferrin receptor superfamily domain. Trends Biochem Sci. 2001;26:213–4. doi: 10.1016/s0968-0004(01)01789-3. [DOI] [PubMed] [Google Scholar]
- 39.Zhang AX, et al. A remote arene-binding site on prostate specific membrane antigen revealed by antibody-recruiting small molecules. J Am Chem Soc. 2010;132:12711–6. doi: 10.1021/ja104591m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bammens L, Chavez-Gutierrez L, Tolia A, Zwijsen A, De Strooper B. Functional and topological analysis of Pen-2, the fourth subunit of the gamma-secretase complex. J Biol Chem. 2011;286:12271–82. doi: 10.1074/jbc.M110.216978. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 41.Watanabe N, et al. Pen-2 is incorporated into the gamma-secretase complex through binding to transmembrane domain 4 of presenilin 1. J Biol Chem. 2005;280:41967–75. doi: 10.1074/jbc.M509066200. [DOI] [PubMed] [Google Scholar]
- 42.Kim SH, Sisodia SS. Evidence that the “NF” motif in transmembrane domain 4 of presenilin 1 is critical for binding with PEN-2. J Biol Chem. 2005;280:41953–66. doi: 10.1074/jbc.M509070200. [DOI] [PubMed] [Google Scholar]
- 43.Althoff T, Mills DJ, Popot JL, Kuhlbrandt W. Arrangement of electron transport chain components in bovine mitochondrial supercomplex I1III2IV1. EMBO J. 2011;30:4652–64. doi: 10.1038/emboj.2011.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tribet C, Audebert R, Popot JL. Amphipols: polymers that keep membrane proteins soluble in aqueous solutions. Proc Natl Acad Sci U S A. 1996;93:15047–50. doi: 10.1073/pnas.93.26.15047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang Y, Zhang Y, Ha Y. Crystal structure of a rhomboid family intramembrane protease. Nature. 2006;444:179–180. doi: 10.1038/nature05255. Epub 2006 Oct 11. [DOI] [PubMed] [Google Scholar]
- 46.Wu Z, et al. Structural analysis of a rhomboid family intramembrane protease reveals a gating mechanism for substrate entry. Nat Struct Mol Biol. 2006;13:1084–91. doi: 10.1038/nsmb1179. [DOI] [PubMed] [Google Scholar]
- 47.Ben-Shem A, Fass D, Bibi E. Structural basis for intramembrane proteolysis by rhomboid serine proteases. Proc Natl Acad Sci USA. 2007;104:462–466. doi: 10.1073/pnas.0609773104. Epub 2006 Dec 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Feng L, et al. Structure of a site-2 protease family intramembrane metalloprotease. Science. 2007;318:1608–1612. doi: 10.1126/science.1150755. [DOI] [PubMed] [Google Scholar]
- 49.DeLano WL. The PyMOL Molecular Graphics System. 2002 on World Wide Web http://www.pymol.org.
- 50.Pettersen EF, et al. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–12. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 51.Matsuda T, Cepko CL. Electroporation and RNA interference in the rodent retina in vivo and in vitro. Proc Natl Acad Sci U S A. 2004;101:16–22. doi: 10.1073/pnas.2235688100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Alexandrov A, et al. A facile method for high-throughput co-expression of protein pairs. Mol Cell Proteomics. 2004;3:934–8. doi: 10.1074/mcp.T400008-MCP200. [DOI] [PubMed] [Google Scholar]
- 53.Scheich C, Kummel D, Soumailakakis D, Heinemann U, Bussow K. Vectors for co-expression of an unrestricted number of proteins. Nucleic Acids Res. 2007;35:e43. doi: 10.1093/nar/gkm067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mindell JA, Grigorieff N. Accurate determination of local defocus and specimen tilt in electron microscopy. J Struct Biol. 2003;142:334–47. doi: 10.1016/s1047-8477(03)00069-8. [DOI] [PubMed] [Google Scholar]
- 55.Elmlund H, Elmlund D, Bengio S. PRIME: probabilistic initial 3D model generation for single-particle cryo-electron microscopy. Structure. 2013;21:1299–306. doi: 10.1016/j.str.2013.07.002. [DOI] [PubMed] [Google Scholar]
- 56.Chen S, et al. High-resolution noise substitution to measure overfitting and validate resolution in 3D structure determination by single particle electron cryomicroscopy. Ultramicroscopy. 2013;135:24–35. doi: 10.1016/j.ultramic.2013.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kucukelbir A, Sigworth FJ, Tagare HD. Quantifying the local resolution of cryo-EM density maps. Nat Methods. 2014;11:63–5. doi: 10.1038/nmeth.2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 59.Murshudov GN, et al. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr D Biol Crystallogr. 2011;67:355–67. doi: 10.1107/S0907444911001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nicholls RA, Long F, Murshudov GN. Low-resolution refinement tools in REFMAC5. Acta Crystallogr D Biol Crystallogr. 2012;68:404–17. doi: 10.1107/S090744491105606X. [DOI] [PMC free article] [PubMed] [Google Scholar]