SUMMARY
Although selective binding of 53BP1 to dimethylated histone H4 lysine 20 (H4K20me2) at DNA double strand breaks (DSBs) is a necessary and pivotal determinant of non-homologous end joining (NHEJ)-directed repair, the enzymes that generate H4K20me2 at DSBs were unclear. Here we determined that the PR-Set7 monomethyltransferase (H4K20me1) regulates de novo H4K20 methylation at DSBs. Rapid recruitment of PR-Set7 to DSBs was dependent on the NHEJ Ku70 protein and necessary for NHEJ-directed repair. PR-Set7 monomethyltransferase activity was required, but insufficient, for H4K20me2 and 53BP1 nucleation at DSBs. We determined that PR-Set7-mediated H4K20me1 facilitates Suv4-20 methyltransferase recruitment and catalysis to generate H4K20me2 necessary for 53BP1 binding. The orchestrated and concerted activities of PR-Set7 and Suv4-20 were required for proficient 53BP1 nucleation and DSB repair. This report identifies PR-Set7 as an essential component of NHEJ and implicates PR-Set7 as a central determinant of NHEJ-directed repair early in mammalian DSB repair pathway choice.
Keywords: PR-Set7/Set8/SetD8/KMT5a, Suv4-20/KMT5b/KMT5c, MMSET/NSD2/WHSC1, H4K20, histone methylation, 53BP1, Ku70, NHEJ
INTRODUCTION
p53 binding protein 1 (53BP1) is a critical component in mammalian DNA damage response (DDR) and recruitment of 53BP1 to DSBs is a necessary and pivotal determinant of NHEJ-directed repair (Panier and Boulton, 2014). Increasing evidence indicates that 53BP1 recruitment to chromatin near DSBs is dependent on proficient binding of the 53BP1 tandem Tudor domains to H4K20me2, which is required for competent 53BP1 nucleation at DSBs, mass accumulation of 53BP1 at irradiation-induced foci (IRIF), immunoglobulin class switch recombination (CSR) and NHEJ-directed repair (Bothmer et al., 2011; Botuyan et al., 2006; Bunting et al., 2010; Sanders et al., 2004). Consistent with this, recent reports demonstrate that 53BP1 binding to H4K20me2 is obstructed by acetylation of neighboring H4K16 and, conversely, H4K16 deacetylation robustly augments 53BP1-H4K20me2 binding resulting in enhanced 53BP1 nucleation at DSBs and 53BP1 IRIF (Hsiao and Mizzen, 2013; Tang et al., 2013). 53BP1 chromatin interaction is further strengthened by additional histone modification, including ubiquitinated H2AK15 that binds the 53BP1 ubiquitination-dependent recruitment (UDR) domain, but only in an H4K20me2-dependent manner (Fradet-Turcotte et al., 2013). While these findings indicate that H4K20me2 is an essential determinant of 53BP1 chromatin binding and NHEJ-directed repair, the upstream enzymatic mechanisms that facilitate H4K20me2 at DSBs were undetermined.
Since H4K20me2 is highly abundant, accounting for >80% of total H4 in human cells, the necessity for de novo H4K20me2 at DSBs was unclear (Pesavento et al., 2007). Due to the abundance of H4K20me2, it was postulated that the likely presence of pre-existing H4K20me2 near DSBs may suffice for 53BP1 binding, however, rigorous examination of this model is currently lacking (Sanders et al., 2004). A recent report seemed to support the model demonstrating that H4K20me2 near a DSB was only slightly elevated immediately after induction of the DSB but 53BP1 occupancy increased ~2-fold (Hsiao and Mizzen, 2013). At later time points following DSB induction, however, H4K20me2 and 53BP1 near the DSB were reported to be significantly elevated (~20-fold) (Pei et al., 2011). These results indicate that de novo H4K20me2 occurs at DSBs but the H4K20 methyltransferases responsible and their biological significance in DSB repair were unclear.
In this report we determined that the orchestrated and concerted activities of the PR-Set7 H4K20 monomethyltransferase (H4K20me1) and Suv4-20 H4K20me2 methyltransferases are required for de novo H4K20me2 near DSBs (Nishioka et al., 2002; Schotta et al., 2004). We found that the reported rapid recruitment of PR-Set7 to DSBs is dependent on the NHEJ Ku70 protein, which interacts with PR-Set7 in cells (Oda et al., 2010). Consistent with a role in NHEJ-directed repair, depletion of PR-Set7 and H4K20me1 severely impaired 53BP1 recruitment and NHEJ repair activity. Although 53BP1 was reported to bind H4K20me1 in vitro, we found that PR-Set7-mediated H4K20me1 is insufficient for 53BP1 nucleation in cells (Botuyan et al., 2006; Oda et al., 2010). We determined that PR-Set7-mediated H4K20me1 functions to facilitate de novo H4K20me2 by promoting Suv4-20 recruitment and catalysis, consistent with reports demonstrating that H4K20me1 is the preferred substrate for Suv4-20 catalysis (Schotta et al., 2008; Southall et al., 2014; Wu et al., 2013). Suv4-20-mediated H4K20me2 was necessary for 53BP1 nucleation near DSBs, consistent with reports demonstrating that Suv4-20 depletion impairs 53BP1 IRIF (Hsiao and Mizzen, 2013; Yang et al., 2008). This study reveals a progressive PR-Set7-dependent pathway required for proficient de novo H4K20me2 at DSBs, 53BP1 nucleation and DSB repair. Furthermore, this study identifies PR-Set7 as an essential factor of NHEJ that likely promotes NHEJ-directed repair early during mammalian DSB repair pathway selection.
RESULTS
Rapid and focal recruitment of PR-Set7 to DSBs is necessary for proficient DSB repair
Prolonged PR-Set7 depletion results in the expected ablation of H4K20me1, but also reduced H4K20me2, coincident with phenotypes indicative of defective DSB repair, including elevated and sustained activation of canonical DDR proteins and unrepaired DSBs (Hartlerode et al., 2012; Houston et al., 2008; Oda et al., 2009). To exclude the possibility that defective DSB repair was an indirect consequence of reduced H4K20me2, U2OS cells were transduced with a control shRNA or shRNA targeting the 3’ UTR of endogenous PR-Set7 for 4 days to deplete PR-Set7 and reduce global H4K20me1, but not H4K20me2, prior to treatment with sub-lethal doses of ionizing radiation (IR) (Figure S1A and S1B). Short-term PR-Set7 depletion resulted in the rapid and significant decline of viable U2OS cells following low level IR exposure compared to control cells (Figure 1A). Since the non-irradiated PR-Set7 shRNA U2OS cells displayed no measurable changes in cell viability compared to control, the enhanced radiosensitivity of cells following short-term PR-Set7 depletion was not due to proliferative or cell cycle perturbations observed following prolonged PR-Set7 depletion.
Figure 1. Recruitment of PR-Set7 to DSBs is necessary for NHEJ-directed repair.
(A) U2OS cells transduced with control or PR-Set7 shRNA were irradiated (2 Gy) 48 hours post-transduction. Population doublings (PD) were calculated by monitoring cell viability by trypan blue exclusion 48 hours following IR (y-axis). Values represent the mean +/− SD from 5 independent biological replicates. The Student’s t-test was used to determine statistical significance as indicated.
(B) Early passage human foreskin fibroblast cells transduced with control or PR-Set7 shRNA were irradiated with the indicated doses. Whole cell lysates were collected 1 hour post-irradiation for Western analysis using the indicated antibodies.
(C) Illustration of the DR-GFP transgene and locations of the qPCR amplicons.
(D) ChIP-qPCR analysis of wild type (WT) DR-GFP mESCs in the absence (−DSB) or presence (+DSB) of I-SceI using the indicated antibodies (x-axis) and qPCR amplicons. Values represent the mean fold enrichment relative to histone H3 +/− SD from 5 independent biological replicates.
(E) Control or PR-Set7 shRNA U2OS cells containing integrated I-SceI-GFP reporters were used to measure NHEJ (EJ5-GFP), single strand annealing (SSA-GFP) and HDR (HDR-GFP) activity by flow cytometry. Values represent the mean GFP+ cells +/− SD from 6 independent biological replicates. The Student’s t-test was used to determine statistical significance between control and PR-Set7 shRNA as indicated: p<0.0001 (***), p=0.0012 (**), p=0.0024 (**).
See also Figure S1.
While these results demonstrate that PR-Set7 is necessary for proficient DSB repair, it remained unclear whether PR-Set7 functions in DDR and/or directly in DSB repair. To assess the role of PR-Set7 in DDR checkpoint activation, early passage diploid human foreskin fibroblasts (HFFs) transduced with control or PR-Set7 shRNA were subjected to increasing doses of IR (Figure 1B). Western analysis revealed that the dose-dependent activation of the canonical DDR checkpoint proteins, ATM, ATR, p53 and H2A.X, were nearly identical between control and PR-Set7 depleted cells indicating that PR-Set7 is dispensable for the initial activation of these DDR proteins. Conversely, a potential direct role for PR-Set7 in DSB repair was inferred following low energy laser-irradiation of GFP-PR-Set7 expressing HeLa cells (Kong et al., 2009). Fluorescence microscopy revealed that GFP-PR-Set7 rapidly mobilized and remained at laser-induced DSBs for >30 minutes in contrast to GFP-null control, consistent with a previous report (Figure S1C and S1D) (Oda et al., 2010).
These cytological observations suggested that PR-Set7 is directly recruited to DSBs. To test this hypothesis at a defined DSB in cells, murine embryonic stem cells (mESCs) containing an integrated DR-GFP transgene were electroporated with an I-SceI expression plasmid to create a genome-specific induced DSB (iDSB) prior to chromatin immunoprecipitations (ChIPs) and qPCR (Figure 1C) (Donoho et al., 1998). ChIP-qPCR analysis confirmed the iDSB by significant H2A.X phosphorylation (γH2A.X) throughout the transgene compared to the negative control GAPDH (Figures 1D and S1E). Significant enrichments of PR-Set7 and methylated H4K20 relative to H3 control were observed near the iDSB (−1 kb and −2 kb) but none were detected more distal to the iDSB at +5 kb (Figures 1D, S1F and S1G). Identical results were obtained in U2OS DR-GFP cells (Figure S1H). The NHEJ Ku70 protein displayed similar enrichment patterns indicating that PR-Set7 recruitment and de novo H4K20 methylation are focally localized to a region near the iDSB.
U2OS cells containing different integrated I-SceI reporter transgenes were utilized to delineate the role of PR-Set7 in NHEJ (EJ5-GFP), single strand annealing (SSA-GFP) and homology-directed repair (HDR-GFP) (Gunn and Stark, 2012). Following induction of the DSB in control and PR-Set7 depleted cells, proficient iDSB repair was assessed by flow cytometry of GFP+ cells. Compared to control, PR-Set7 depletion significantly reduced iDSB repair by NHEJ and significantly increased repair by SSA and HDR (Figure 1E). These findings indicate that PR-Set7 recruitment to DSBs is necessary for NHEJ-directed repair.
De novo H4K20 methylation and 53BP1 nucleation are dependent on PR-Set7 monomethyltransferase activity
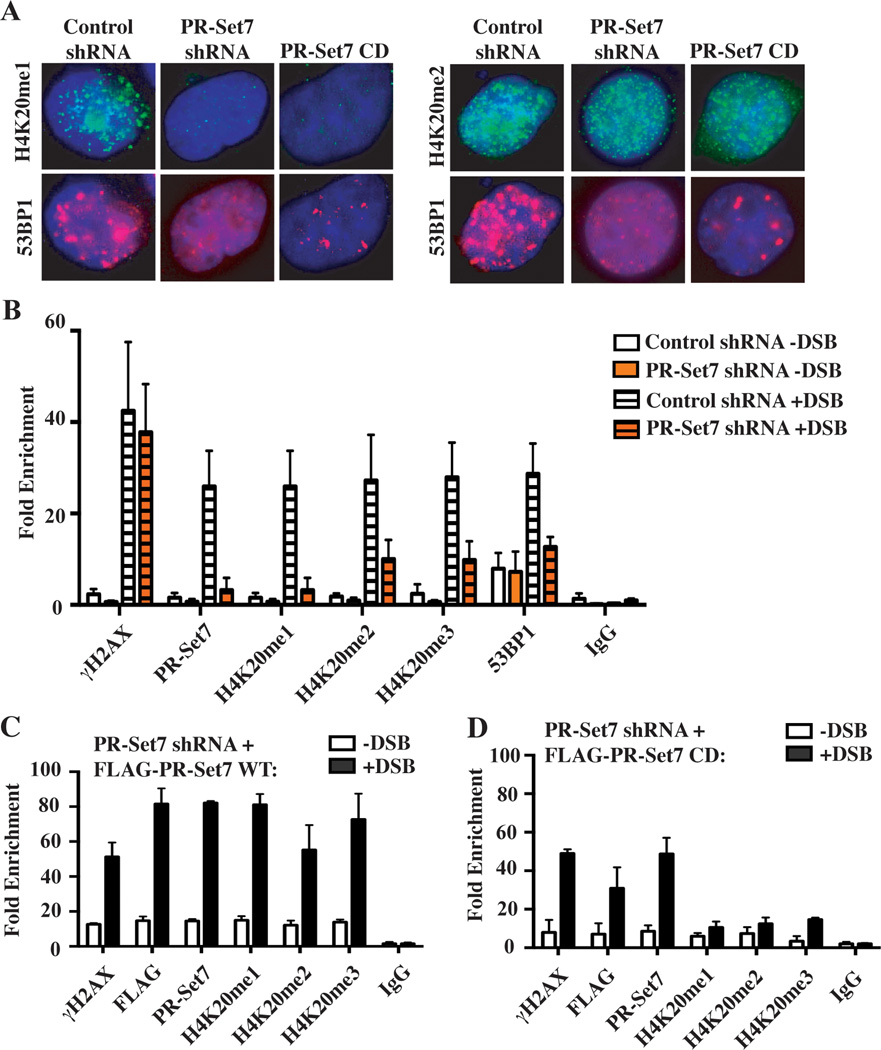
PR-Set7 may function in repair pathway choice by facilitating H4K20me2 near DSBs necessary for binding of 53BP1, a pivotal determinant of NHEJ-directed repair (Panier and Boulton, 2014). Consistent with this, several reports demonstrated that prolonged PR-Set7 depletion reduced global H4K20me2 levels and impaired 53BP1 IRIF (Botuyan et al., 2006; Hartlerode et al., 2012; Oda et al., 2010). Although short-term PR-Set7 depleted HeLa cells retained wild type H4K20me2 levels, we unexpectedly observed impaired 53BP1 IRIF (Figures 2A and S1B). To determine whether this defect was due to loss of PR-Set7 catalytic activity, HeLa cells were transfected with a dominant negative PR-Set7 catalytically dead (CD) point mutant (Sims and Rice, 2008). These cells also displayed impaired 53BP1 IRIF, consistent with previous reports, indicating that PR-Set7 monomethyltransferase activity, but not PR-Set7 per se, is required for proficient 53BP1 IRIF (Figure 2A) (Hartlerode et al., 2012; Houston et al., 2008; Oda et al., 2010).
Figure 2. 53BP1 recruitment requires PR-Set7 monomethyltransferase-dependent de novo H4K20 methylations.
(A) Representative immunofluorescence microscopy images of H4K20me1, H4K20me2 (green) and 53BP1 (red) in irradiated HeLa cells expressing control shRNA, PR-Set7-specific shRNA or a PR-Set7 catalytically dead (CD) mutant. Cells were counterstained with DAPI (blue).
(B) ChIP-qPCR analysis near the iDSB (−1 kb) in U2OS DR-GFP cells transduced with control or PR-Set7 shRNA in the absence (−DSB) or presence (+DSB) of I-SceI using the indicated antibodies (x-axis). Values represent the mean fold enrichment relative to histone H3 +/− SD from 5 independent biological replicates.
(C) ChIP-qPCR analysis in PR-Set7 depleted U2OS DR-GFP cells complemented with FLAG-PR-Set7 WT (C) or FLAG-PR-Set7 CD (D) as described in Figure 2B. Values represent the mean fold enrichment relative to histone H3 +/− SD from 5 independent biological replicates. See also Figure S2.
The results suggested that PR-Set7 recruitment is necessary for de novo H4K20 methylation and 53BP1 nucleation near DSBs. To test this, U2OS DR-GFP cells were transduced with a control or PR-Set7 shRNA to deplete PR-Set7 prior to I-SceI transfection (Bennardo et al., 2009). ChIP-qPCR demonstrated that PR-Set7 depletion did not affect γH2A.X enrichment but resulted in the ablation of H4K20me1 and significant reductions in H4K20me2/3 and 53BP1 occupancy near the iDSB compared to control, as hypothesized (Figures 2B and S2A). Complementation experiments were performed in PR-Set7-depleted U2OS DR-GFP cells transfected with either wild type FLAG-PR-Set7 (WT) or -PR-Set7 CD (Figure S2B). ChIP-qPCR analysis demonstrated that while recruitment of PR-Set7 WT restored de novo H4K20 methylations near the iDSB, recruitment of PR-Set7 CD did not (Figures 2C, 2D, S2C and S2D). To determine whether PR-Set7 recruitment is sufficient for de novo H4K20 methylations and 53BP1 nucleation at loci regardless of DNA damage, HEK293-TK22 cells containing an integrated 5xGAL4-UAS-TK-Luc reporter transgene were transfected with a GAL4-DBD-control, -PR-Set7 WT or -PR-Set7 CD plasmid (Figure S3A) (Ishizuka and Lazar, 2003). ChIP-qPCR analysis demonstrated that specific localization of PR-Set7 WT to the undamaged transgene resulted in significantly increased H4K20 methylations and 53BP1 occupancy (Figures 3A and S3B). Although H4K20 methylation enrichments were ~2–3x lower compared to iDSBs, 53BP1 occupancy relative to H4K20 methylation was similar to iDSBs (Figures 2B and 3A). In contrast, H4K20 methylations and 53BP1 occupancy were not observed following PR-Set7 CD localization. The collective results demonstrate that PR-Set7 recruitment to an iDSB or an undamaged locus is necessary and sufficient to induce de novo H4K20 methylations and 53BP1 nucleation in a PR-Set7 monomethyltransferase-dependent manner.
Figure 3. 53BP1 nucleation requires H4K20me1-dependent Suv4-20 recruitment and catalysis at DSBs.
(A) Illustration of the integrated 5xGAL4-UAS-TK-Luc transgene in HEK293-TK22 cells and location of qPCR amplicon. ChIP-qPCR analysis from cells transfected with GAL4-DBD-null, -PR-Set7 WT, -PR-Set7 CD, -Suv4-20H1 or -Suv4-20H2 using the indicated antibodies (x-axis). Values represent the mean fold enrichment relative to histone H3 +/− SD from 3 independent biological replicates.
(B) ChIP-qPCR analysis near the iDSB (−1 kb) in wild type (WT) DR-GFP mESCs and Suv4-20h1/h2 double knockout (DKO) DR-GFP mESCs as described in Figure 1D. Values represent the mean fold enrichment relative to histone H3 +/− SD from 3 independent biological replicates.
(C) ChIP-qPCR analysis near the iDSB (−1 kb) in control or PR-Set7 shRNA U2OS DR-GFP cells expressing FLAG-null, -Suv4-20H1 or -Suv4-20H2 as described in Figure 2B. Values represent the mean fold enrichment relative to histone H3 +/− SD from 3 independent biological replicates.
See also Figure S3.
Suv4-20 methyltransferases are required for H4K20me2 and 53BP1 nucleation at an iDSB
Based on the findings above, we hypothesized that PR-Set7-mediated H4K20me1 near DSBs facilitates subsequent H4K20me2 by a different methyltransferase. It was previously reported that the MMSET/NSD2/WHSC1 methyltransferase was required for H4K20me2 and 53BP1 recruitment (Hajdu et al., 2011; Pei et al., 2011). However, our results and recent independent reports demonstrate that MMSET does not regulate H4K20 methylation, MMSET is not recruited to DSBs, MMSET is dispensable for 53BP1 IRIF and MMSET is not required for CHK2 activation (Figure S4G–Q) (Hartlerode et al., 2012; Hsiao and Mizzen, 2013; Kuo et al., 2011).
In contrast to MMSET, the Suv4-20h1/h2 methyltransferases are responsible for the majority of H4K20me2/3 in mammalian cells and H4K20me1 is the preferred substrate for Suv4-20 catalysis (Schotta et al., 2004; Schotta et al., 2008; Southall et al., 2014; Wu et al., 2013). To test the hypothesis that the Suv4-20s are required for H4K20me2 and 53BP1 nucleation at an iDSB, the DR-GFP transgene was inserted at the hprt locus in Suv4-20h1/h2 double knock-out (DKO) mESCs (Donoho et al., 1998; Schotta et al., 2008). The DKO DR-GFP mESCs displayed similar levels of γH2A.X enrichment near the iDSB as control (Figures 1D, 3B and S3C). Consistent with the hypothesis, significantly reduced H4K20me2/3 and 53BP1 occupancy, but increased H4K20me1, were observed near the iDSB in the DKO DR-GFP mESCs compared to control. These results indicate that Suv4-20h1 and/or Suv4-20h2 are required for de novo H4K20me2 and 53BP1 nucleation near an iDSB, consistent with reports demonstrating impaired 53BP1 IRIF in Suv4-20 depleted human cells (Hsiao and Mizzen, 2013; Yang et al., 2008). The findings also demonstrate that the Suv4-20s are dispensable for PR-Set7 recruitment to DSBs, that PR-Set7 specifically monomethylates H4K20 near DSBs and that, although 53BP1 binds H4K20me1 in vitro, PR-Set7-mediated H4K20me1 is insufficient for 53BP1 nucleation in cells (Botuyan et al., 2006; Oda et al., 2010).
PR-Set7-mediated H4K20me1 is requisite for Suv4-20 catalysis and 53BP1 nucleation
We hypothesized that Suv4-20-dependent H4K20me2 and 53BP1 nucleation near DSBs required prior PR-Set7-mediated H4K20me1. To address this first, control or PR-Set7 depleted U2OS DR-GFP cells were transfected with FLAG-Suv4-20H1 or -Suv4-20H2 (Figure S3D). ChIP-qPCR analysis demonstrated equivalent enrichment of each Suv4-20 indicating that both are recruited to the iDSB in control cells (Figures 3C and S3E). PR-Set7 depletion resulted in an ~2–3x reduction in Suv4-20 occupancy near the iDSB indicating that Suv4-20 recruitment is dependent, in part, on PR-Set7 and/or H4K20me1 (Figures 3C and S3F). To evaluate the necessity of H4K20me1 for Suv4-20 catalysis and 53BP1 nucleation, HEK293-TK22 cells were transfected with GAL4-DBD-Suv4-20H1 or -Suv4-20H2 (Figure S3A). In contrast to PR-Set7 WT, ChIP-qPCR analysis demonstrated that Suv4-20 localization to the undamaged integrated transgene was insufficient for de novo H4K20 methylation and 53BP1 nucleation (Figures 3A and S3B). These findings demonstrate that PR-Set7-mediated H4K20me1 is required for proficient Suv4-20 recruitment and catalysis to generate de novo H4K20me2 near DSBs necessary for 53BP1 nucleation.
Proficient recruitment of PR-Set7 to DSBs is dependent on Ku70
A previous report demonstrated that cytological accumulation of ectopic PR-Set7 at laser-induced DSB foci required direct binding to PCNA (Oda et al., 2010). To determine the necessity of PCNA binding on PR-Set7 recruitment to an iDSB, PR-Set7 depleted U2OS DR-GFP cells were transfected with FLAG-PR-Set7 WT or -PR-Set7 PIPm2, which encodes mutations in the second PCNA-interacting protein (PIP) box of PR-Set7 that ablates PCNA binding (Figure S4B) (Abbas et al., 2010; Centore et al., 2010; Jorgensen et al., 2011). Despite the inability to bind PCNA, ChIP-qPCR analysis revealed significant enrichments of PR-Set7 PIPm2 and H4K20 methylations near the iDSB, although these values were proportionally lower compared to PR-Set7 WT (Figures 4A and S4A). These results demonstrate that PR-Set7 recruitment to an iDSB can occur independent of PCNA binding.
Figure 4. Ku70-dependent recruitment of PR-Set7 to DSBs.
(A) ChIP-qPCR analysis near the iDSB (−1 kb) in control and PR-Set7 shRNA U2OS DR-GFP cells expressing FLAG-PR-Set7 WT or -PR-Set7 PIPm2. Values represent the mean fold enrichment relative to histone H3 +/− SD from 3 independent biological replicates.
(B) FLAG-immunoprecipitations from nuclear lysates of HEK293 cells expressing FLAG-PR-Set7 or -GFP negative control followed by Western analysis using the indicated antibodies. 5% of the input material and 20% of the bound material was used.
(C) Autoradiography of SDS-PAGE fractionated proteins following S-Tag-immunoprecipitation of recombinant S-Tag-HIS-PR-Set7 incubated with in vitro transcribed and translated (TnT) 35S-null, -Ku70, -Ku80 or -PARP1 (top). Equivalent immunoprecipitation between samples was assessed by Western analysis for HIS (bottom).
(D) Representative fluorescence microscopy images of wild type (WT) or Ku70 depleted HeLa cells expressing GFP-PR-Set7 (green) 5 minutes after generation of laser-induced DSBs (yellow arrows).
(E) ChIP-qPCR analysis near the iDSB (−1 kb) in Ku70−/−, DNA-PKcs−/− (F) or XRCC4−/− (G) DR-GFP mESCs in the presence (+DSB) or absence (−DSB) of I-SceI using the indicated antibodies (x-axis). Values represent the mean fold enrichment relative to histone H3 +/− SD from 5 independent biological replicates.
(H) Proposed model. PR-Set7 interaction with canonical NHEJ factors, including Ku70/80 and DNA-PKcs, results in rapid recruitment of PR-Set7 and de novo H4K20me1 at DSBs (first panel). PR-Set7-mediated H4K20me1 is required for subsequent Suv4-20 recruitment and catalysis to generate H4K20me2 at the lesion (second panel). The orchestrated recruitment and concerted enzymatic activities of PR-Set7 and Suv4-20 are necessary for 53BP1 nucleation at the DSB and NHEJ-directed repair (third panel).
See also Figure S4.
To identify potential PR-Set7-interacting proteins necessary for PR-Set7 recruitment to DSBs, two independent tandem affinity purifications of ectopically expressed FLAG-HA-null and -PR-Set7 in HeLa cells were performed followed by comparative mass spectrometry analyses (Spektor and Rice, 2009). Identification of proteins unique to PR-Set7 included the NHEJ-associated factors Ku70, Ku80 and PARP1 (Table S1). Immunoprecipitation of FLAG-PR-Set7 or -GFP negative control expressed in HEK293 cells confirmed the specific interaction of PR-Set7 with endogenous Ku70, Ku80 and PARP1 (Figure 4B). In vitro assays demonstrated that recombinant PR-Set7 directly binds Ku70 and Ku80, but not PARP1 (Figure 4C).
These findings suggested that PR-Set7 recruitment to DSBs is dependent on certain NHEJ-associated factors. To determine the necessity of specific canonical NHEJ factors on PR-Set7 recruitment to an iDSB, ChIP-qPCR was performed in several DR-GFP mESC lines lacking individual NHEJ factors. In Ku70−/− DR-GFP mESCs, PR-Set7 recruitment and de novo H4K20 methylations near the iDSB were severely impaired compared to WT control (Figures 1D, 4E and S4C). Consistent with this, GFP-PR-Set7 mobilization to laser-induced DSBs was blunted in Ku70-depleted HeLa cells (Figure 4D). These results demonstrate that proficient PR-Set7 recruitment to DSBs is dependent on Ku70, however, PR-Set7 was found to be dispensable for Ku70 localization to DSBs (Figure S4F and S4G). In DNA-PKcs−/− DR-GFP mESCs, PR-Set7 recruitment and de novo H4K20 methylations near the iDSB were lower compared to WT control (Figures 1D, 4F and S4D). These results suggest that proficient PR-Set7 recruitment to DSBs requires DNA-PKcs, consistent with the interaction detected between PR-Set7 and endogenous DNA-PKcs in HEK293 cells (Figure 4B). However, the concurrent reduction of Ku70 near the iDSB in these cells alternatively suggests that the observed decreased PR-Set7 occupancy was Ku70-dependent. In XRCC4−/− DR-GFP mESCs, PR-Set7 and H4K20 methylations near the iDSB were comparable to WT control indicating that XRCC is dispensable for PR-Set7 recruitment (Figures 1D, 4G and S4E). These collective results demonstrate that proficient PR-Set7 recruitment to DSBs is dependent on Ku70.
DISCUSSION
This study reveals that recruitment of PR-Set7 to DSBs is required to induce de novo H4K20 methylation, 53BP1 nucleation and NHEJ-directed repair (Figure 4H). PR-Set7 recruitment to laser-induced DSBs was previously reported to require direct binding of PR-Set7 to PCNA (Oda et al., 2010). The observed accumulation and retention of PR-Set7 at laser-induced DSBs, however, was inconsistent with reports demonstrating that PCNA binding results in the rapid degradation of PR-Set7 (Abbas et al., 2010; Centore et al., 2010; Jorgensen et al., 2011; Oda et al., 2010). Our results demonstrate that loss of PCNA binding had little effect on PR-Set7 recruitment to an iDSB and, alternatively, that proficient PR-Set7 recruitment to an iDSBs and laser-induced DSBs was dependent on the canonical NHEJ Ku70 protein, suggesting a role for PR-Set7 in NHEJ. Consistent with this, NHEJ-directed repair was severely impaired in PR-Set7 depleted cells concurrent with the accumulation of unrepaired DSBs (Houston et al., 2008; Oda et al., 2009). These results identify PR-Set7 as an essential component of NHEJ-directed repair necessary for the maintenance of genomic stability.
Based on our findings and previous reports, we propose that PR-Set7 is a primary early determinant of DSB repair pathway choice that promotes NHEJ-directed repair. The observed rapid recruitment of PR-Set7 to DSBs and the PR-Set7-dependent de novo H4K20me2 required for subsequent 53BP1 localization to DSBs supports an early direct mechanism for PR-Set7 in promoting NHEJ-directed repair (Oda et al., 2010). This is further supported by the findings that PR-Set7 recruitment to an undamaged locus induced de novo H4K20me2 and 53BP1 nucleation, implicating PR-Set7 as a principal initiating factor of NHEJ-directed repair. It was previously reported that PR-Set7 protein levels are dynamically regulated during cell cycle progression where PR-Set7 is abundant in G1-phase, when NHEJ is the dominant DSB repair pathway, but depleted in S-phase, when HDR dominates (Wu et al., 2010). These findings suggest that the regulation of PR-Set7 abundance is a central determinant of DSB repair pathway choice where the presence of PR-Set7 directly promotes NHEJ-directed repair and absence of PR-Set7 indirectly promotes HDR by impeding NHEJ-directed repair. Consistent with this, PR-Set7 depletion severely impaired NHEJ-directed repair and, concurrently, significantly enhanced HDR. The necessity for PR-Set7 degradation during initiation of fundamental HDR-associated programs, including DNA replication and UV damage repair, further supports the model that PR-Set7 regulates DSB repair pathway choice (Abbas et al., 2010; Centore et al., 2010; Jorgensen et al., 2011).
PR-Set7 monomethyltransferase activity was found to be necessary, but insufficient, for de novo H4K20me2 and 53BP1 nucleation. We determined that PR-Set7-mediated H4K20me1 functions to facilitate subsequent Suv4-20 recruitment and catalysis required to generate H4K20me2 and nucleate 53BP1 at an iDSB and an undamaged locus. Previous reports suggested that Suv4-20 and H4K20me2 were dispensable, but that H4K20me1 was sufficient, for 53BP1 recruitment since 53BP1 also binds H4K20me1 in vitro and Suv4-20h1/h2 DKO primary mouse embryonic fibroblasts (pMEFs), that have reduced global H4K20me2 but elevated H4K20me1, displayed only minor 53BP1 IRIF defects (Botuyan et al., 2006; Hartlerode et al., 2012; Oda et al., 2010; Schotta et al., 2008). In contrast to DKO pMEFs, Suv4-20 depleted HeLa cells, which also have reduced global H4K20me2 and elevated H4K20me1, displayed significant 53BP1 IRIF defects (Hsiao and Mizzen, 2013; Yang et al., 2008). While the reasons for these differences remain unclear, a report demonstrating that Suv4-20h1/h2 DKO B cells displayed significantly impaired CSR supports the necessity of Suv4-20 and H4K20me2 for 53BP1 localization and function (Bothmer et al., 2011; Schotta et al., 2008). Consistent with these reports, our results directly demonstrate that PR-Set7-mediated H4K20me1 alone is insufficient, and that Suv4-20-mediated H4K20me2 is required, for 53BP1 nucleation at an iDSB. The collective results of this study indicate that the orchestrated and concerted activities of PR-Set7 and Suv4-20 are requisite rate-limiting steps for proficient 53BP1 localization and NHEJ-directed repair. Since H4K20me2-dependent binding of 53BP1 to chromatin is augmented by additional histone modifications, including H4K16 deacetylation and H2AK15 ubiquitination, we propose that the de novo generation of a DSB-specific “histone code” facilitates the selective recruitment and retention of 53BP1 at DSBs necessary for proficient NHEJ-directed repair (Fradet-Turcotte et al., 2013; Hsiao and Mizzen, 2013; Tang et al., 2013).
EXPERIMENTAL PROCEDURES
U2OS, HFF, HEK293, mESCs and HeLa cells were cultured as previously described (Houston et al., 2008; Sims and Rice, 2008). Glycofect (Techulon) was used to transfect U2OS and Lipofectamine 2000 (Invitrogen) was used to transfect HEK293 and HeLa according to manufacturer’s instructions. The mESCs were electroporated as previously described (Donoho et al., 1998). Lentivirus was produced and cells transduced as previously reported (Sims and Rice, 2008). Approximately 106 U2OS or 5×106 mES DR-GFP cells were used for each ChIP as previously described (Congdon et al., 2010). ChIPs were performed 24 or 36 hrs following I-SceI transfection in U2OS or mES DR-GFP cells, respectively, when γH2AX enrichment was determined to be maximal at the iDSB. Other ChIP antibodies used in the studies were first titrated at these time points to optimize enrichment levels near the iDSB in control cells and compensate for possible differences in avidity between different antibodies. An iQ5 iCycler (Bio Rad) was used for qPCR with data presented as fold enrichment (%IP/%Input)/(%IP H3/%Input H3) and statistical significance determined by the Student’s t-test. Western analysis, immunoprecipitations, comparative mass spectrometry analysis and in vitro binding assays were performed as previously described (Spektor and Rice, 2009). Laser irradiation and immunofluorescence studies were performed as previously reported (Kong et al., 2009; Wu et al., 2010). Detailed descriptions of the reagents and methods used can be found in Supplemental Information.
Supplementary Material
HIGHLIGHTS.
Rapid PR-Set7 recruitment to DSBs is dependent on Ku70 and required for NHEJ repair
De novo H4K20 methylations at DSBs require PR-Set7 monomethyltransferase activity
PR-Set7-mediated H4K20me1 is necessary but insufficient for 53BP1 nucleation
H4K20me1 facilitates Suv4-20-mediated H4K20me2 required for 53BP1 binding at DSBs
ACKNOWLEDGEMENTS
This project was supported by the NIH (GM075094, J.C.R.; CA100710, K.Y.), an NIH Training Grant (5T32CA009320, C.T.T.), an NCI Cancer Center Support Grant (P30CA014089), the American Cancer Society (RSG117619, J.C.R.; PF0627301, C.T.T.), the Margaret E. Early Research and Pew Charitable Trusts (J.C.R.) and Deutsche Forschungsgemeinschaft (SFB1064, SFB684, SPP1356, G.S.). We thank Danny Reinberg (NYU, HHMI) for PR-Set7 plasmids, Mitch Lazar (UPenn) for HEK293-TK22 cells, Maria Jasin (MSKCC) for DR-GFP mESCs and Jeremy Stark (City of Hope) for U2OS DR-GFP cells and insightful discussions.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
SUPPLEMENTAL INFORMATION
Supplemental Information includes six figures and detailed Experimental Procedures and can be found with this article at X.
REFERENCES
- Abbas T, Shibata E, Park J, Jha S, Karnani N, Dutta A. CRL4(Cdt2) regulates cell proliferation and histone gene expression by targeting PR-Set7/Set8 for degradation. Mol Cell. 2010;40:9–21. doi: 10.1016/j.molcel.2010.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennardo N, Gunn A, Cheng A, Hasty P, Stark JM. Limiting the persistence of a chromosome break diminishes its mutagenic potential. PLoS Genet. 2009;5:e1000683. doi: 10.1371/journal.pgen.1000683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bothmer A, Robbiani DF, Di Virgilio M, Bunting SF, Klein IA, Feldhahn N, Barlow J, Chen HT, Bosque D, Callen E, et al. Regulation of DNA end joining, resection, and immunoglobulin class switch recombination by 53BP1. Mol Cell. 2011;42:319–329. doi: 10.1016/j.molcel.2011.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botuyan MV, Lee J, Ward IM, Kim JE, Thompson JR, Chen J, Mer G. Structural basis for the methylation state-specific recognition of histone H4-K20 by 53BP1 and Crb2 in DNA repair. Cell. 2006;127:1361–1373. doi: 10.1016/j.cell.2006.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunting SF, Callen E, Wong N, Chen HT, Polato F, Gunn A, Bothmer A, Feldhahn N, Fernandez-Capetillo O, Cao L, et al. 53BP1 inhibits homologous recombination in Brca1-deficient cells by blocking resection of DNA breaks. Cell. 2010;141:243–254. doi: 10.1016/j.cell.2010.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Centore RC, Havens CG, Manning AL, Li JM, Flynn RL, Tse A, Jin J, Dyson NJ, Walter JC, Zou L. CRL4(Cdt2)-mediated destruction of the histone methyltransferase Set8 prevents premature chromatin compaction in S phase. Mol Cell. 2010;40:22–33. doi: 10.1016/j.molcel.2010.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Congdon LM, Houston SI, Veerappan CS, Spektor TM, Rice JC. PR-Set7-mediated monomethylation of histone H4 lysine 20 at specific genomic regions induces transcriptional repression. J Cell Biochem. 2010;110:609–619. doi: 10.1002/jcb.22570. [DOI] [PubMed] [Google Scholar]
- Donoho G, Jasin M, Berg P. Analysis of gene targeting and intrachromosomal homologous recombination stimulated by genomic double-strand breaks in mouse embryonic stem cells. Mol Cell Biol. 1998;18:4070–4078. doi: 10.1128/mcb.18.7.4070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fradet-Turcotte A, Canny MD, Escribano-Diaz C, Orthwein A, Leung CC, Huang H, Landry MC, Kitevski-LeBlanc J, Noordermeer SM, Sicheri F, et al. 53BP1 is a reader of the DNA-damage-induced H2A Lys 15 ubiquitin mark. Nature. 2013;499:50–54. doi: 10.1038/nature12318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunn A, Stark JM. I-SceI-based assays to examine distinct repair outcomes of mammalian chromosomal double strand breaks. Methods in molecular biology. 2012;920:379–391. doi: 10.1007/978-1-61779-998-3_27. [DOI] [PubMed] [Google Scholar]
- Hajdu I, Ciccia A, Lewis SM, Elledge SJ. Wolf-Hirschhorn syndrome candidate 1 is involved in the cellular response to DNA damage. Proc Natl Acad Sci U S A. 2011;108:13130–13134. doi: 10.1073/pnas.1110081108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartlerode AJ, Guan Y, Rajendran A, Ura K, Schotta G, Xie A, Shah JV, Scully R. Impact of histone H4 lysine 20 methylation on 53BP1 responses to chromosomal double strand breaks. PLoS One. 2012;7:e49211. doi: 10.1371/journal.pone.0049211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houston SI, McManus KJ, Adams MM, Sims JK, Carpenter PB, Hendzel MJ, Rice JC. Catalytic function of the PR-Set7 histone H4 lysine 20 monomethyltransferase is essential for mitotic entry and genomic stability. J Biol Chem. 2008;283:19478–19488. doi: 10.1074/jbc.M710579200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsiao KY, Mizzen CA. Histone H4 deacetylation facilitates 53BP1 DNA damage signaling and double-strand break repair. Journal of molecular cell biology. 2013;5:157–165. doi: 10.1093/jmcb/mjs066. [DOI] [PubMed] [Google Scholar]
- Ishizuka T, Lazar MA. The N-CoR/histone deacetylase 3 complex is required for repression by thyroid hormone receptor. Mol Cell Biol. 2003;23:5122–5131. doi: 10.1128/MCB.23.15.5122-5131.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen S, Eskildsen M, Fugger K, Hansen L, Larsen MS, Kousholt AN, Syljuasen RG, Trelle MB, Jensen ON, Helin K, et al. SET8 is degraded via PCNA-coupled CRL4(CDT2) ubiquitylation in S phase and after UV irradiation. The Journal of cell biology. 2011;192:43–54. doi: 10.1083/jcb.201009076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong X, Mohanty SK, Stephens J, Heale JT, Gomez-Godinez V, Shi LZ, Kim JS, Yokomori K, Berns MW. Comparative analysis of different laser systems to study cellular responses to DNA damage in mammalian cells. Nucleic acids research. 2009;37:e68. doi: 10.1093/nar/gkp221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo AJ, Cheung P, Chen K, Zee BM, Kioi M, Lauring J, Xi Y, Park BH, Shi X, Garcia BA, et al. NSD2 links dimethylation of histone H3 at lysine 36 to oncogenic programming. Mol Cell. 2011;44:609–620. doi: 10.1016/j.molcel.2011.08.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishioka K, Rice JC, Sarma K, Erdjument-Bromage H, Werner J, Wang Y, Chuikov S, Valenzuela P, Tempst P, Steward R, et al. PR-Set7 is a nucleosome-specific methyltransferase that modifies lysine 20 of histone H4 and is associated with silent chromatin. Mol Cell. 2002;9:1201–1213. doi: 10.1016/s1097-2765(02)00548-8. [DOI] [PubMed] [Google Scholar]
- Oda H, Hubner MR, Beck DB, Vermeulen M, Hurwitz J, Spector DL, Reinberg D. Regulation of the Histone H4 Monomethylase PR-Set7 by CRL4(Cdt2)-Mediated PCNA-Dependent Degradation during DNA Damage. Mol Cell. 2010;40:364–376. doi: 10.1016/j.molcel.2010.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oda H, Okamoto I, Murphy N, Chu J, Price SM, Shen MM, Torres-Padilla ME, Heard E, Reinberg D. Monomethylation of histone H4-lysine 20 is involved in chromosome structure and stability and is essential for mouse development. Mol Cell Biol. 2009;29:2278–2295. doi: 10.1128/MCB.01768-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panier S, Boulton SJ. Double-strand break repair: 53BP1 comes into focus. Nature reviews Molecular cell biology. 2014;15:7–18. doi: 10.1038/nrm3719. [DOI] [PubMed] [Google Scholar]
- Pei H, Zhang L, Luo K, Qin Y, Chesi M, Fei F, Bergsagel PL, Wang L, You Z, Lou Z. MMSET regulates histone H4K20 methylation and 53BP1 accumulation at DNA damage sites. Nature. 2011;470:124–128. doi: 10.1038/nature09658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pesavento JJ, Yang H, Kelleher NL, Mizzen CA. Certain and Progressive Methylation of Histone H4 at Lysine 20 During the Cell Cycle. Mol Cell Biol. 2007 doi: 10.1128/MCB.01517-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders SL, Portoso M, Mata J, Bahler J, Allshire RC, Kouzarides T. Methylation of histone H4 lysine 20 controls recruitment of Crb2 to sites of DNA damage. Cell. 2004;119:603–614. doi: 10.1016/j.cell.2004.11.009. [DOI] [PubMed] [Google Scholar]
- Schotta G, Lachner M, Sarma K, Ebert A, Sengupta R, Reuter G, Reinberg D, Jenuwein T. A silencing pathway to induce H3-K9 and H4-K20 trimethylation at constitutive heterochromatin. Genes Dev. 2004;18:1251–1262. doi: 10.1101/gad.300704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schotta G, Sengupta R, Kubicek S, Malin S, Kauer M, Callen E, Celeste A, Pagani M, Opravil S, De La Rosa-Velazquez IA, et al. A chromatin-wide transition to H4K20 monomethylation impairs genome integrity and programmed DNA rearrangements in the mouse. Genes Dev. 2008;22:2048–2061. doi: 10.1101/gad.476008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sims JK, Rice JC. PR-Set7 establishes a repressive trans-tail histone code that regulates differentiation. Mol Cell Biol. 2008;28:4459–4468. doi: 10.1128/MCB.00410-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southall SM, Cronin NB, Wilson JR. A novel route to product specificity in the Suv4-20 family of histone H4K20 methyltransferases. Nucleic acids research. 2014;42:661–671. doi: 10.1093/nar/gkt776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spektor TM, Rice JC. Identification and characterization of posttranslational modification-specific binding proteins in vivo by mammalian tethered catalysis. Proc Natl Acad Sci U S A. 2009;106:14808–14813. doi: 10.1073/pnas.0907799106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang J, Cho NW, Cui G, Manion EM, Shanbhag NM, Botuyan MV, Mer G, Greenberg RA. Acetylation limits 53BP1 association with damaged chromatin to promote homologous recombination. Nature structural & molecular biology. 2013;20:317–325. doi: 10.1038/nsmb.2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Siarheyeva A, Zeng H, Lam R, Dong A, Wu XH, Li Y, Schapira M, Vedadi M, Min J. Crystal structures of the human histone H4K20 methyltransferases SUV420H1 and SUV420H2. FEBS letters. 2013;587:3859–3868. doi: 10.1016/j.febslet.2013.10.020. [DOI] [PubMed] [Google Scholar]
- Wu S, Wang W, Kong X, Congdon LM, Yokomori K, Kirschner MW, Rice JC. Dynamic regulation of the PR-Set7 histone methyltransferase is required for normal cell cycle progression. Genes Dev. 2010;24:2531–2542. doi: 10.1101/gad.1984210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Pesavento JJ, Starnes TW, Cryderman DE, Wallrath LL, Kelleher NL, Mizzen CA. Preferential dimethylation of histone H4-lysine 20 by Suv4-20. J Biol Chem. 2008;283:12085–12092. doi: 10.1074/jbc.M707974200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.