Abstract
The molecular chaperone network plays a critical role in the formation and propagation of self-replicating yeast prions. Not only do individual prions differ in their requirements for certain chaperones, but structural variants of the same prion can also display distinct dependences on the chaperone machinery, specifically Hsp104. The AAA+ ATPase Hsp104 is a disaggregase required for the maintenance of most known yeast prions. As a key component in the propagation of prions, understanding how Hsp104 differs in its interaction with specific variants is crucial to understanding how prion variants may be selected or evolve. Here, we investigate two novel mutations in Hsp104, hsp104-G254D, and hsp104-G730D, which allow us to elucidate some mechanistic features of Hsp104 disaggregation and its requirement for activity in propagating specific prion variants. Both Hsp104 mutants propagate the [PSI+] prion to some extent, but show a high rate of prion loss. Both Hsp104-G254D and Hsp104-G730D display reduced biochemical activity, yet differ in their ability to efficiently resolubilize disordered, heat-aggregated substrates. Additionally, both mutants impair weak [PSI+] propagation, but are capable of propagating the less stable strong [PSI+] variant to some extent. One of the Hsp104 mutants also has the ability to propagate one variant of the [RNQ+] prion. Thus, our data suggest that changes in Hsp104 activity limit substrate disaggregation in a manner that depends more on the stability of the substrate than the nature of the aggregated species.
Keywords: Hsp104, chaperone biology, yeast prions, [PSI+], [RNQ+], prion variants
Introduction
Prions are self-templating, amyloidogenic protein aggregates. In mammals, prions are associated with several neurodegenerative diseases, including scrapie in sheep, chronic wasting disease in deer and elk, and Creutzfeldt-Jacob disease in humans.1,2 In the budding yeast Saccharomyces cerevisiae, prions are non-toxic, epigenetic elements of inheritance, but they do share many characteristics with mammalian prions and other disease-related amyloidogenic proteins.3-5 The yeast prion [PSI+] results from a self-propagating aggregated state of the translation termination factor Sup35.6,7 Aggregation of Sup35 results in partial loss-of-function that leads to an increase in global nonsense suppression.8 Consequently, the ability to generate [PSI+] cells, coupled with the reversible or metastable nature of this prion mechanism, has been proposed to be advantageous due to the ability to alter phenotypes through the [PSI+]-dependent translation of normally silent regions of the genome.9,10 Interestingly, the appearance of the [PSI+] prion appears to be regulated by the presence of another yeast prion, called [RNQ+] (or [PIN+]) which is the aggregated form of the Rnq1 protein.11-15 Though the soluble protein Rnq1 (in [rnq−] cells) has no known function, the prion state of Rnq1, [RNQ+], functions to promote the de novo induction of [PSI+], and is often required for the aggregation of other amyloidogenic proteins in yeast.16
Although the prion conformation is self-templating, the maintenance of prions in yeast relies on the molecular chaperone network to produce aggregates that can be transmitted from mother to daughter cells. The AAA+ ATPase chaperone Hsp104 is a disaggregase required for the propagation of both [PSI+] and [RNQ+], as well as all other recognized yeast prions.11,17-19 Hsp104, in concert with Hsp70 and Hsp40 chaperones, also functions to resolubilize proteins that aggregate as a consequence of various environmental stresses.20,21 Hsp104 has five distinct domains and is functional as a hexamer that contains a central pore used for threading substrates.22-24 The N-terminal domain is proposed to play a role in substrate recognition and may also be a site for interaction of co-chaperones, though the N-terminal domain is not required for either thermotolerance or prion propagation.25 The two ATP-binding domains (NBD1 and NBD2), connected by a coiled-coil linker domain, both bind and hydrolyze ATP to power the disaggregation mechanism.26,27 Lastly, the function of the fifth modular domain, the C-terminal domain, is still mostly unclear, though some data have implicated this domain as having a role in substrate interaction and processing.28,29
The proposed role of Hsp104 in prion propagation is to fragment prion aggregates in order to generate the smaller heritable species, or propagons.30-32 Consequently, deletion of Hsp104 eliminates both [PSI+] and [RNQ+], and several mutants in each domain of Hsp104 have been characterized that affect [PSI+] and [RNQ+] propagation.12,17,33-36 Interestingly, [PSI+] is especially sensitive to changes in the Hsp104 system as the overexpression of Hsp104 eliminates [PSI+] to generate [psi−] cells, whereas Hsp104 overexpression does not eliminate [RNQ+].12,17,37,38 Hsp104 has also been shown to catalyze amyloid formation of prion proteins in vitro.37,39,40 Furthermore, maintenance of prions by Hsp104 is aided by Hsp70 and Hsp40 co-chaperones, and overexpression or deletion of these chaperones can affect prion propagation.41-47
Yeast prions exist as an array of self-propagating structures, known as prion variants (analogous to “prion strains” in mammalian prion nomenclature). Although formed from the same protein sequence, prion variants maintain distinct structures having varying stabilities and causing different cellular phenotypes, much like mammalian prion strains result in variation in disease pathology.48-52 Prion variants have been described for both [PSI+] and [RNQ+]. [PSI+] variants are characterized by the amount of nonsense suppression they confer, which is related to the ability of the aggregate to template monomeric Sup35.53-55 Weak variants of [PSI+] maintain an increased pool of monomeric Sup35 relative to strong [PSI+] variants. As such, weak [PSI+] variants show less nonsense suppression (more folded, soluble Sup35 means more translation termination). Additionally, distinct Sup35 amyloid structures have been generated using denatured, recombinant protein (usually using just the prion-forming domain, also called “NM”) by simply changing the temperature at which amyloid fibers are formed.49,50 From structural studies of in vitro formed amyloid, Sup35NM aggregates with a shorter protected amyloid core are less stable and give rise to a strong [PSI+] phenotype when [psi−] cells are infected with these aggregates, while fibers with longer, more stable cores tend to give rise to weak [PSI+] phenotypes upon infection.49,50,56 Recent data suggest that the longer, more stable core associated with the weaker [PSI+] variants is more refractory to Hsp104 activity or interaction.57,58 In contrast, the shorter core of strong variants is more labile to Hsp104 disaggregation, resulting in the generation of more propagons in strong [PSI+] cells, and thus increased conversion of monomeric Sup35.57 As such, strong [PSI+] variants present a stronger nonsense suppression phenotype as compared with weak [PSI+] variants, and are better able to propagate when challenged with fluctuations in Hsp104 activity.
Variants of [RNQ+] were first identified in vivo and were characterized by their differential ability to facilitate formation of the [PSI+] prion.15,59 These variants were named low, medium, high, and very high [PIN+] to indicate their rate of [PSI+] induction.15 These [PIN+]/[RNQ+] variants were further described by their patterns of fluorescence when Rnq1 was tagged with GFP.60 Two distinct phenotypes were observed: cells that contained multiple, small fluorescent foci termed multi-dot (m.d.), and cells that contained a single, large fluorescent focus termed single-dot (s.d.). [RNQ+] variants that induced [PSI+] at low, medium, high, and very high rates were all found within the microscopic s.d. pattern, while only one m.d. [RNQ+] variant was characterized, and it induced [PSI+] at a high rate. Further investigation of these established [RNQ+] variants showed that a distinct region of the Rnq1 protein was required for propagation of certain variants, suggesting that [RNQ+] variants, like [PSI+], may also differ in their core region.61 Similar to Sup35, Rnq1 fibers display distinct characteristics when generated under various conditions in vitro,62 though structural studies to elucidate the amyloid core of these [RNQ+] variants have not yet been done. Additionally, much less is understood about the interaction between [RNQ+] and Hsp104 and how changes in Hsp104 activity affect propagation of specific [RNQ+] variants.
Here, we characterize 2 novel mutations in Hsp104 that we identified as causing a defect in [PSI+] propagation. We show that these mutations decrease select activities of Hsp104, resulting in diminished disaggregation activity of specific substrates. Finally, we find that defects in [PSI+] and [RNQ+] propagation are specific for certain prion variants, thereby providing further evidence for the hypothesis that Hsp104 may interact more efficiently with less stable [PSI+] variants than more stable variants, and that this may also hold true for variants of [RNQ+].
Results
Point mutations in HSP104 cause a defect in the maintenance of [PSI+]
We performed a mutagenesis screen in order to identify factors required for the propagation of a specific variant of the [PSI+] prion. To identify such factors, we used a colony color based phenotypic assay commonly used to track [PSI+] prion propagation. In this colorimetric assay, a premature stop codon in the ADE1 gene, ade1–14, blocks the completion of the adenine biosynthesis pathway and results in the accumulation of a red-pigmented intermediate. Disruption of the pathway at this point also results in the inability of these cells to grow on media lacking adenine. Cellular changes that afford an increase in nonsense suppression of the premature stop codon in ade1–14 can result in production of sufficient Ade1 protein to generate light pink or white colonies and allow for growth on media lacking adenine. Such an increase in nonsense suppression can be generated by reduced function of the translation termination complex (eRF1 and eRF3, or Sup45 and Sup35, respectively).8 This can occur either by mutation of either termination factor or by conversion of wild type Sup35 into a prion state. When Sup35 is monomeric and fully functional in [psi-] cells, the premature stop codon in ade1–14 is recognized, resulting in colonies that are red in color on rich media and cannot grow on media lacking adenine. When Sup35 is aggregated in [PSI+] cells, however, the nonsense codon is suppressed and the [PSI+] cells appear white on rich media and can grow on media lacking adenine.
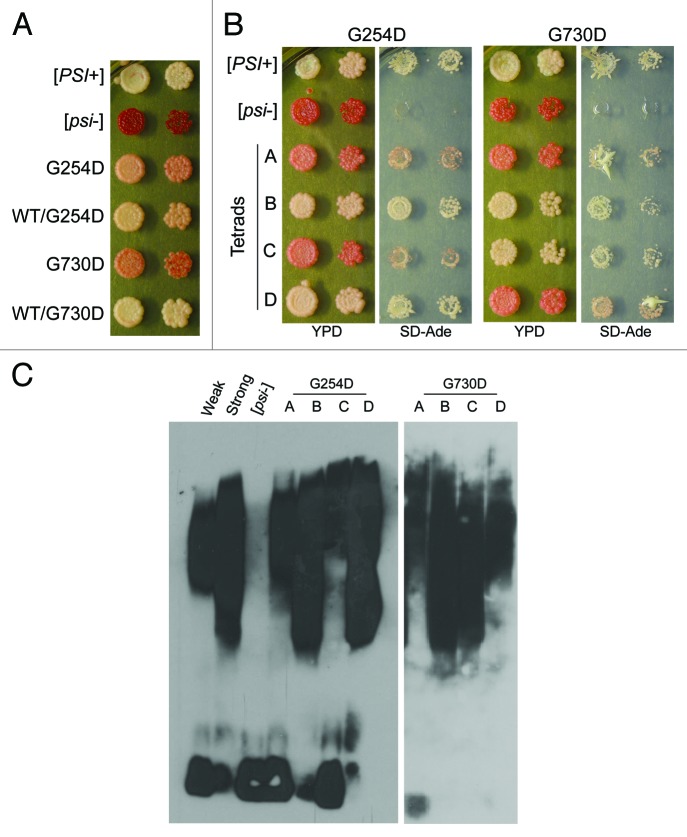
In addition, structural variants of the [PSI+] prion present a range of color phenotypes indicative of the amount of soluble Sup35 characteristic of those variants. For example, because of the relative increase in soluble Sup35, weak [PSI+] strains harboring ade1–14 are pink in color on rich media. In our screen, we expected to find mutants that affected [PSI+] propagation by a change in color phenotype from the light pink indicative of strong [PSI+] in our parent 74-D964 strain. Interestingly, in addition to mutations that completely inhibited [PSI+] propagation, we found a subset of mutations that partially inhibited [PSI+] inheritance. These mutants displayed a sectoring color phenotype on rich media (Fig. 1A). Color sectoring within a [PSI+] colony results when a fraction of the budding daughter cells lose the prion, thereby producing sections of the colony that are [psi−] and phenotypically red. We identified 2 point mutations in the chaperone Hsp104 that demonstrated an interesting effect on the inheritance of [PSI+] unlike the curing phenotype that is often characterized. These mutants, which we sequenced to identify as hsp104-G730D and hsp104-G254D, presented varying levels of sectoring and [PSI+] loss (Fig. 1A). We recreated both mutants in an unmutagenized [PSI+] 74-D694 strain by double homologous recombination to verify the phenotypic effects and used these strains for all subsequent analyses.
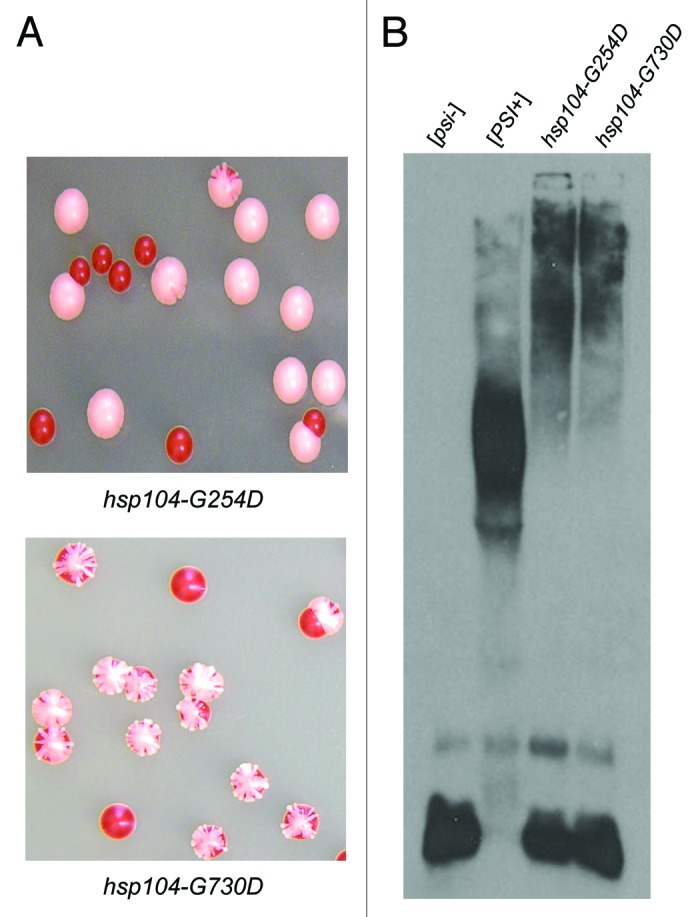
Figure 1.hsp104-G254D and hsp104-G730D strains show inefficient [PSI+] propagation. (A) hsp104-G254D and hsp104-G730D cells display a sectoring [PSI+] phenotype as a result of inefficient [PSI+] inheritance. (B) Lysates of sectoring hsp104-G254D and hsp104-G730D cells with strong [PSI+] cells ([PSI+]) and [psi−] cells ([psi−]) as controls were analyzed by SDD-AGE analysis followed by western blot and blotting for Sup35. These results were reproduced at least three times. An example of the shift is shown. The general loss of the lower aggregate species and the decrease in aggregated Sup35 is reproducible. The appearance of monomeric Sup35 on SDD-AGE western blots is more variable, even with controls, for unknown reasons.
In order to determine how the mutants affected the biochemical properties of Sup35 in [PSI+] cells, we first performed SDD-AGE (semi-denaturing with detergent agarose gel-electrophoresis)63 and analyzed the SDS-resistant Sup35 species. In hsp104-G254D and hsp104-G730D cell lysates, we found that the size of the Sup35 aggregate distribution was increased, as was the amount of Sup35 monomer (Fig. 1B). From these data, one could hypothesize that these Hsp104 mutants are unable to efficiently fragment Sup35 aggregates, resulting in larger aggregates that cannot be as easily passed on to daughter cells. In addition, less propagons would also result in decreased monomer addition and a larger pool of monomeric Sup35. Alternatively, the mutants could be propagating a weak variant of [PSI+] which would be predicted to show the same change in aggregate pattern.
Strong [PSI+] propagons are maintained in both hsp104-G254D and hsp104-G730D
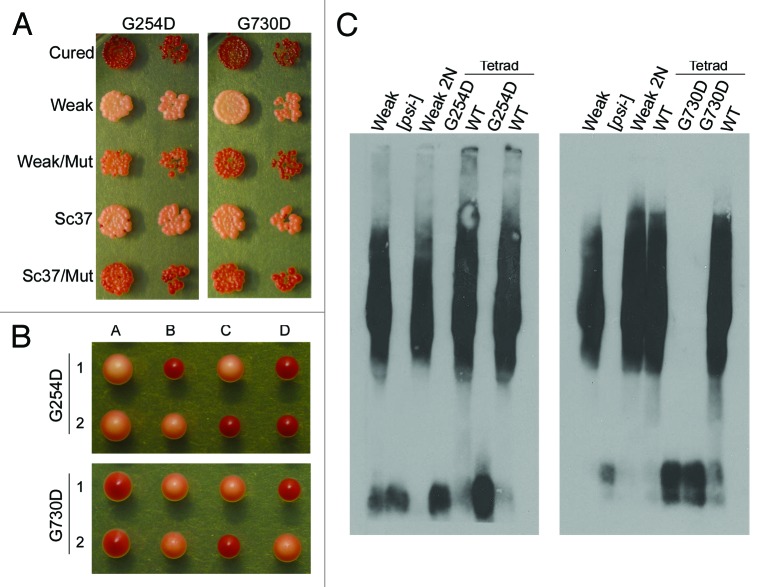
The SDD-AGE analysis of the mutant strains (Fig. 1B) showed an increase in both high-molecular weight species and Sup35 monomer, which is characteristic of weak [PSI+] variants.30 However, the original [PSI+] variant in the screen was strong [PSI+]. Thus, we used a genetic test and phenotypic test to analyze whether the mutants appeared to be propagating a different variant of [PSI+] or if the variant propagated in the sectoring colonies still appeared to be strong [PSI+]. As the mutants are recessive, we could readily analyze the properties of the variant by mating the sectoring hsp104-G254D and hsp104-G730D cells to wild type [psi−] cells, sporulating diploids, and analyzing resultant haploid progeny phenotypically and biochemically. Mating [PSI+] hsp104-G254D and [PSI+] hsp104-G730D cells to wild type [psi−] cells resulted in diploids that were light pink, and more similar to the strong [PSI+] parent than the mutant haploids (Fig. 2A). Sporulation of the diploids resulted in tetrads with 2 stable [PSI+] haploids and 2 sectoring/ weaker [PSI+] haploids. For both heterozygous diploids, the 2 haploid progeny that expressed wild type HSP104 resembled strong [PSI+] phenotypically, by both color and growth on media lacking adenine (Fig. 2B). Finally, we performed SDD-AGE analysis on cell lysates from full tetrads obtained from the heterozygous diploids and compared the aggregate distribution of the wild type HSP104 haploids to lysates from control cells containing the parental strong [PSI+] variant. The biochemical analyses correlated with the colorimetric phenotypes: all the wild type haploids propagated Sup35 aggregates that resembled those from control strong [PSI+] cell lysates (Fig. 2C). In sum, our phenotypic and biochemical analyses suggest that the underlying structure of Sup35 aggregates is retained in [PSI+] cells harboring the hsp104-G254D and hsp104-G730D mutations; however, when these mutations are present as the only copy of Hsp104 in the cell they are unable to efficiently propagate the prion to maintain the strong [PSI+] phenotype.
Figure 2.hsp104-G254D and hsp104-G730D cells propagate the original strong [PSI+] variant. (A) hsp104-G254D and hsp104-G730D cells (G254D and G730D, respectively) and diploids from the mating of hsp104-G254D and hsp104-G730D to wild type [psi−] cells (WT/G254D and WT/G730D) were spotted on rich media (YPD) and media lacking adenine (SD-Ade). Strong [PSI+] and [psi−] cells are spotted for comparison. The second spot in each row is a 5-fold dilution of the first spot. (B) Full tetrads from the sporulation of the diploids in (A) were spotted for each mutant as indicated. Each haploid progeny in the tetrad is labeled, A–D. Strong [PSI+] and [psi−] are spotted for color comparison. The second spot in each row is a 5-fold dilution of the first spot. (C) Representative tetrads from (B) were analyzed by SDD-AGE analysis and western blot. The letters A–D correspond to the haploids with the same label in (B) and weak [PSI+] (Weak), strong [PSI+] (Strong) and [psi−] were analyzed for comparison. SDD-AGE analysis was performed four times with haploids from both hsp104-G254D and hsp104-G730D cells.
Hsp104 missense mutant proteins are defective in ATP hydrolysis and efficient hexamer formation
As the increased size of the Sup35 aggregates in hsp104-G254D and hsp104-G730D cells could suggest a fragmentation defect of Hsp104, we next investigated whether these mutations affected ATP hydrolysis. We purified recombinant wild type Hsp104, Hsp104-G254D, and Hsp104-G730D proteins from E. coli cells. We then tested the ability of Hsp104-G254D and Hsp104-G730D to hydrolyze ATP by monitoring release of inorganic phosphate using the Malachite Green assay.64 The G254 residue is in NBD1 and G730 is in NBD2 and as such, may be involved in ATP binding or hydrolysis. As compared with wild type Hsp104, both mutants (G730D and G254D) had significantly reduced ATPase activity (Table 1). Surprisingly, these results contradict previous results in the literature, which demonstrate that mutants of Hsp104 that display similarly low levels of ATP hydrolysis often cure [PSI+].17,33
Table 1. Hsp104 mutants display defects in ATP hydrolysis under physiological salt conditions.
| Physiological* | Low† | |
|---|---|---|
| WT | 0.225 ± 0.0397 | 0.732 ± 0.0543 |
| G254D | 0.0325 ± 0.00177 | 0.211 ± 0.00177 |
| G730D | 0.00583 ± 0.00439 | 0.677 ± 0.0321 |
Numbers represent the average initial rate (nmol·µg−1·min−1) of ATP hydrolysis for WT Hsp104, Hsp104-G254D, and Hsp104-G730D. ATPase assays were measured by the Malachite Green assay. Rates were calculated from three separate experiments from 2 different preparations of purified protein. *Buffer contains 150 mM NaCl. †Buffer contains only 15 mM NaCl.
As ATP hydrolysis is dependent on the ability of Hsp104 to form hexamers,64 we next investigated whether these mutants were capable of oligomerizing in the presence of nucleotide.65 Using glycerol gradient fractionation in the presence of ATP, we found that Hsp104-G254D and Hsp104-G730D do form hexamers, though appear to do so less efficiently than wild type, as there was a slight shift in the peak of Hsp104-G254D and Hsp104-G730D toward the top of the gradient (Fig. 3). In the absence of ATP, wild type Hsp104 and the mutant proteins do not form hexamers (Fig. 3). Thus, the reduced ATP hydrolysis of both Hsp104-G254D and Hsp104-G730D may correlate with their inefficient hexamerization ability.
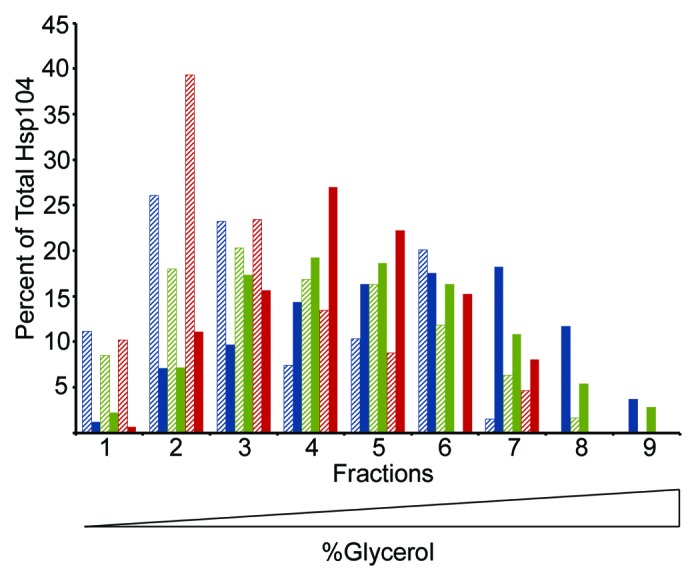
Figure 3. Hsp104-G254D and Hsp104-G730D mutant proteins form hexamers in vitro. Hsp104 (blue), Hsp104-G254D (green), and Hsp104-G730D (red) were incubated with (solid color) or without (hatched) 5 mM ATP for 10 min then subjected to ultracentrifugation through a linear (10–35%) glycerol gradient. Equal volume fractions were collected and the amount of Hsp104 protein in each fraction was analyzed by SDS-PAGE and western blot. Individual bands were quantified and the amount of Hsp104 in each fraction was plotted as a percent of the total Hsp104 protein. The graph shows the data from one assay. This assay was performed three times and all gave similar results.
We reasoned that if the majority of the mutant were in the hexameric state, we might observe wild type levels of ATPase activity from the mutants. Hsp104 has previously been shown to hexamerize even in the absence of ATP when incubated in a buffer containing low salt (<20 mM).64 Therefore, we again measured hydrolysis of ATP by the Malachite green assay, this time in a low salt (15 mM NaCl) buffer to promote hexamerization. In low salt buffer, we found that the NBD2 mutant, Hsp104-G730D, displayed ATPase activity similar to wild type (Table 1). Hsp104-G254D, however, did show an increase in ATP hydrolysis in low salt buffer, but not to the same extent as Hsp104 or Hsp104-G730D, suggesting the assembly defect of Hsp104-G254D may not be the only cause of the reduced rate of ATP hydrolysis.
Low activity mutations in HSP104 vary in their ability to disaggregate non-prion substrates
In addition to its role in prion propagation, Hsp104 is also necessary for cell survival following acute heat shock.21 With the aid of Hsp70 and Hsp40 co-chaperones, Hsp104 resolubilizes proteins that aggregate as a result of heat or other stresses and is crucial for cellular recovery from such stresses.20 Therefore, we tested the ability of Hsp104-G254D and Hsp104-G730D to promote thermotolerance. We first pre-treated cells at 37 °C to induce Hsp104 expression, and then heat-shocked HSP104, hsp104Δ, hsp104-G254D, and hsp104-G730D cells at 50 °C for various times as indicated, before plating on rich media to determine the relative viability and recovery. We found that both wild type HSP104 and hsp104-G254D were thermotolerant, but hsp104-G730D cells were not, and resembled the hsp104Δ strain (Fig. 4). Thus, despite having lower ATPase activity, Hsp104-G254D was more active in thermotolerance assays than Hsp104-G730D, suggesting that the mechanism of disaggregation is de-coupled from the ATP hydrolysis activity for these 2 mutants.
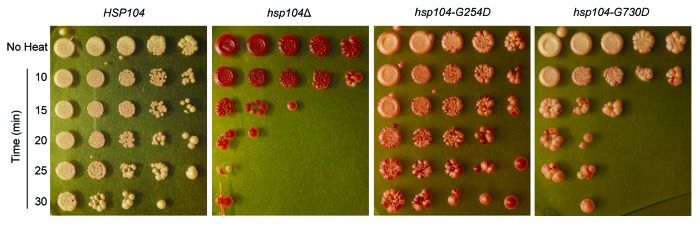
Figure 4.hsp104-G254D and hsp104-G730D display different levels of non-prion disaggregation. The thermotolerance of HSP104, hsp104Δ, hsp104-G254D, and hsp104-G730D cells was tested. Cells were first grown at 37 °C in liquid culture to induce HSP104 expression, heat-shocked at 50 °C for 10 to 30 min as indicated, and then spotted on rich media. Control cells (No Heat) were plated without heat shock. Thermotolerance assays were repeated four times and all showed similar results.
hsp104-G254D and hsp104-G730D do not propagate weak [PSI+]
Recently, it has been shown that some mutations in Hsp104 that reduce ATPase activity can impair the propagation of weak [PSI+] variants, but not strong [PSI+] variants.66 Accordingly, we next tested the ability of hsp104-G254D and hsp104-G730D to propagate weak [PSI+] variants. We mated hsp104-G254D [psi−] and hsp104-G730D [psi−] cells to 2 previously characterized weak [PSI+] variants and examined the diploids and haploid progeny phenotypically and biochemically.49,53 In both weak [PSI+] variants, the heterozygous diploids displayed decreased nonsense suppression as compared with the wild type weak [PSI+] parent (Fig. 5A). These data suggest that both hsp104-G254D and hsp104-G730D were inhibiting weak [PSI+] propagation, even in the presence of wild type HSP104. Heterozygous diploids expressing wild type HSP104 and hsp104Δ maintained weak [PSI+], suggesting that the decreased nonsense suppression observed in the mutant diploids is not just the result of reduced Hsp104 expression (data not shown). Resultant tetrads were either all red and [psi−], or segregated 2:2 weak [PSI+]:[psi−] (Fig. 5B and data not shown) where the [PSI+] progeny segregated with the wild type HSP104. Presumably, the reduced activity of Hsp104-G254D and Hsp104-G730D prevent weak [PSI+] propagation and also partially inhibit wild type Hsp104 from stably propagating weak variants. Next, we chose tetrads that maintained weak [PSI+] in the wild type haploids for SDD-AGE analysis and found that the variant propagated resembled the weak [PSI+] parent (Fig. 5C). Thus, hsp104-G254D and hsp104-G730D do not irreversibly decrease the ability of wild type HSP104 to propagate weak [PSI+] variants and do not appear to alter the prion variant structure by phenotypic assays.
Figure 5. Neither hsp104-G254D nor hsp104-G730D can propagate weak [PSI+]. (A) [psi-] hsp104-G254D and [psi-] hsp104-G730D cells were mated to either weak [PSI+] or the weak [PSI+] variant Sc37. Heterozygous HSP104 and hsp104 mutant diploids (Weak/mut and Sc37/mut) from these matings and haploid parents (Cured, Weak, Sc37) were spotted on YPD to assess phenotype. The second spot in each row is a 5-fold dilution of the first spot. (B) Diploids of the mutants crossed to weak [PSI+] (Weak/Mut in A) were sporulated and 16 tetrads dissected. Two representative tetrads on YPD are shown. Each haploid is labeled A–D. (C) Diploids of weak [PSI+] crossed to the mutants (Weak/Mut in A) and a representative tetrad from these heterozygous diploids were subjected to SDD-AGE and western blot analysis to determine Sup35 aggregate distribution. SDD-AGE analysis was performed twice using four distinct tetrads.
hsp104-G730D propagates a known variant of [RNQ+]
Like [PSI+], the [RNQ+] prion is also dependent on Hsp104 for its maintenance.12 To determine if the prion propagation defect observed in hsp104-G254D and hsp104-G730D cells was specific to [PSI+], we tested the ability of both mutants to propagate several established [RNQ+] variants. After mating [psi−] hsp104-G254D and [psi−] hsp104-G730D cells to the s.d. low, medium, high, very high, and m.d. high variants of [RNQ+]59 and sporulating the diploids, we performed SDD-AGE on the hsp104-G254D and hsp104-G730D haploids to determine the [RNQ+] state of the cells. Interestingly, we found one variant of [RNQ+], the m.d. high variant, that was propagated, at least to some extent, in hsp104-G730D cells (Fig. 6). None of the s.d. variants could be propagated by hsp104-G730D, and hsp104-G254D did not propagate any of the [RNQ+] variants tested (data not shown). Thus, similar to recent reports for [PSI+], these data may suggest that the m.d. [RNQ+] variant is less stable than the s.d. [RNQ+] variants and thus more refractory to changes in the activity or interaction with Hsp104.
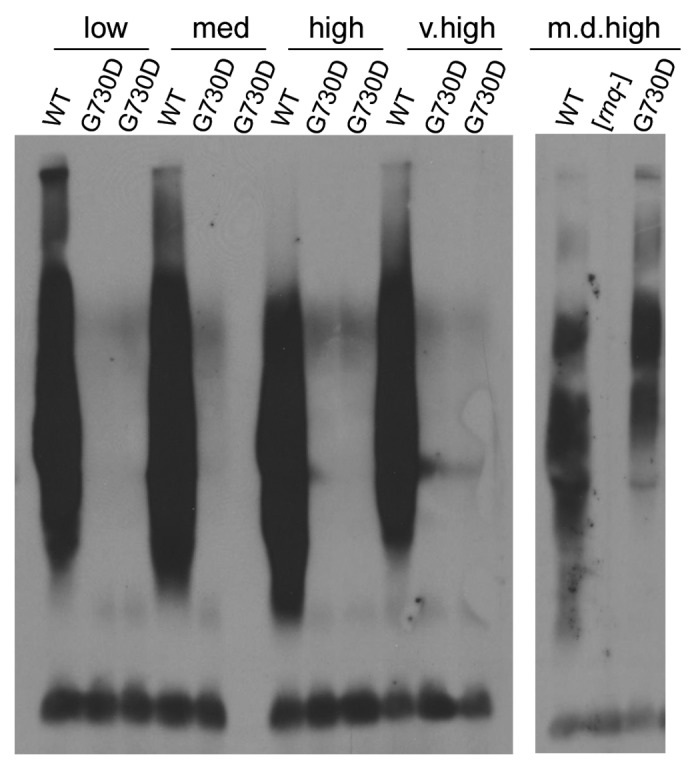
Figure 6.hsp104-G730D can propagate multi-dot high [RNQ+]. hsp104-G730D cells were mated to s.d. low (low), medium (med), high (high), very high (v.high) and m.d. high (m.d. high) [RNQ+] and then sporulated. The hsp104-G730D haploid progeny from four separate tetrads of each variant mating, along with the unmated [RNQ+] variants as controls (WT), were analyzed by SDD-AGE and western blot.
Discussion
Here, we identify 2 novel mutations in Hsp104, Hsp104-G254D, and Hsp104-G730D, which affect Hsp104 activity in a substrate- and prion variant-specific manner. Cells carrying the NBD1 mutant, hsp104-G254D, or the NBD2 mutant, hsp104-G730D, display high mitotic loss of the strong variant of [PSI+]. Phenotypically, hsp104-G730D displays a high degree of sectoring while hsp104-G254D exhibits fewer sectoring colonies but more completely red [psi−] colonies. Both mutants showed marked reductions in ATP hydrolysis activity under physiological conditions, though a second copy of either mutant did not alter the sectoring [PSI+] phenotype (data not shown). Despite similar biochemical activities, the mutants differed in their ability to disaggregate a broad range of substrates, though both are unable to propagate weak variants of [PSI+]. The hsp104-G254D mutant is active in thermotolerance, but is defective in the propagation of all tested variants of [RNQ+]. On the other hand, Hsp104-G730D expressing strains are unable to resolve disordered aggregates but, curiously, can propagate at least one variant of the [RNQ+] prion to some extent.
Manipulation of the chaperone network has long been known to regulate prion propagation and formation.17,20,27,67 Hsp104 is a general prion regulator;26,68,69 deletion of Hsp104 eliminates all of the known yeast prions and dependence on Hsp104 is often one of the criteria used when identifying new yeast prions.18,70-72 Therefore, we were not surprised when several candidates from our mutagenesis screen for factors involved in [PSI+] propagation were identified as mutations in Hsp104. Several interdependent features of Hsp104 contribute to its overall function in substrate disaggregation, and understanding the effect these features have on each other is not simple. For example, as an AAA+ ATPase, hydrolysis of ATP provides the primary energy for the disaggregase ability of Hsp104 and yet we identified 2 mutations, Hsp104-G254D in NBD1 and Hsp104-G730D in NBD2, that still disaggregate substrates despite having significantly lower rates of ATP hydrolysis. Interestingly, reducing the ATPase activity of Hsp104 was previously shown to enhance substrate disaggregation.73,74 However, the increased disaggregation ability extended only to disordered substrates; decreased ATPase activity instead impaired prion remodeling.66,73 On the other hand, a highly characterized mutation in the Walker B motif in NBD1, E285A/Q, hydrolyzes ATP at a rate much higher than wild type (300–500%), but fails to support either thermotolerance or prion propagation.74 Therefore, ATP hydrolysis is not always a reliable indicator for substrate disaggregation ability.
One hypothesis proposed suggested that disordered substrates are less stable and thus require less overall force by Hsp104 to be disaggregated.66 High temperature and other stresses result in the increased exposure of hydrophobic regions in proteins, leading to the formation of unstable, heterogeneous protein aggregates. Unlike these stress-induced aggregates, prions are highly ordered, stable protein aggregates that are typically resistant to denaturation by detergents and high temperatures. Mutations in Hsp104 that promote thermotolerance but are not sufficient for prion propagation have been previously described.33 Interestingly, hsp104-G730D cells can propagate strong [PSI+] and m.d. high [RNQ+], but are unable to function in thermotolerance. Additionally, mutations in the C-terminal domain including K774E, L814S, L840Q, and 22 or 38 residue deletions of the C-terminus cause a loss of thermotolerance but these mutants are able to propagate [PSI+] to some extent.29,75 These previously characterized mutants are similar to Hsp104-G730D in that they exhibit reduced ATP hydrolysis and show defects in hexamerization.29 As yet, no function has been ascribed to the Hsp104 C-terminal domain, though some data suggest it is a site of substrate interaction.28
The hypothesis that Hsp104 is better able to remodel less stable aggregates can also be applied to the broad range of prion variants.57 Despite having the same sequence, prion variants are comprised of distinct structures that vary in stability and hence, may vary in their interaction with Hsp104. Hsp104 has previously been shown to play a role in prion variant selection.76 Continued expression of high levels of Hsp104 resulted in propagation of a [PSI+] variant dependent on overexpression of Hsp104.77 Additionally, recent data show that, like with disordered aggregates, less stable variants of [PSI+] are more efficiently remodeled by Hsp104 despite equal binding affinities for Hsp104.57 At low concentrations, Hsp104 catalyzed the generation of prion-competent seeds of strong variants but was unable to remodel weak variants. Therefore, the decreased activity of our Hsp104 mutants may specifically inhibit weak variants of [PSI+] in vivo due to the inability to interact with or remodel this specific amyloid structure or any structure that is more stable. Moreover, expressing the mutant concurrently with wild type, as seen in the diploids, caused mitotic loss of weak [PSI+] variants, thereby supporting previous results indicating that weak variants require a high level of cooperativity between hexameric subunits to propagate.66
In addition to causing unstable strong [PSI+] propagation, hsp104-G730D is also able to propagate m.d. high [RNQ+] to some extent. The m.d. high [RNQ+] variant has been shown to be less thermal-stable than the s.d. [RNQ+] variants.61,77 We have previously published another mutation in Hsp104, E190K, which also had a differential effect on the propagation of the [RNQ+] variants. Like hsp104-G730D, hsp104-E190K was unable to maintain the s.d. variants, but was able to propagate m.d. high [RNQ+].34 Additionally, both hsp104-G730D and hsp104-E190K showed defects in both [PSI+] propagation and thermotolerance. Our data from these 2 mutants suggest that variants of the [RNQ+] prion may be regulated by Hsp104 in a manner similar to the mechanisms elucidated for [PSI+]. Interestingly, the m.d. [RNQ+] variant that is the most resistant to alterations in Hsp104 activity also facilitates a high rate of [PSI+] induction. It’s interesting to theorize that changing environmental conditions may cause fluctuations in Hsp104 activity that in turn control the appearance and disappearance of prions, and as such, the environment may influence prion-dependent phenotypic variation, adaptability and survival in different conditions. Therefore, discerning Hsp104’s mechanism of disaggregation, as it applies to distinct prion variants, is critical to understanding how prions appear, disappear, and evolve.
As our understanding of self-propagating prions continues to develop, so too does the knowledge that the biological phenomenon of protein-only heritability is highly complex. In addition to mutations in the primary sequence affecting aggregation and propagation, we now understand that the same primary sequence can adopt multiple conformations, and these conformations differ in stability, heritability, and function. How a single protein sequence can adopt multiple conformations is a biological phenomenon that is still relatively unclear. Just as gene expression is regulated by an intricate network of transcription factors and promoter elements, prion variant propagation is regulated by a network of chaperones and in some cases, other prions. Understanding how changes in the chaperone network regulate prion variant formation and propagation will broaden our understanding of the mechanisms of prion variant generation, selection, and evolution.
Materials and Methods
Strain and plasmid construction
S. cerevisiae strains used in this study were derivatives of 74-D694 and were grown and manipulated using standard techniques. Yeast strains were either grown in rich media (1% yeast extract, 2% peptone, 2% glucose) or synthetic media lacking amino acids corresponding to plasmid selection (0.67% yeast nitrogen base, 2% glucose) or nonsense suppression analyses. Diploids were generated by mating haploids containing selectable plasmids and were verified by growth on minimal media and plating on haploid tester strains. Haploid progeny from diploids were isolated by micromanipulation and verified by phenotypic assays and mating type testing.
Hsp104 mutants from the original mutagenized strains were amplified by PCR and cloned into the pRS306 integrating vector. The mutants were integrated into a clean 74-D694 background by the pop-in/pop-out method and verified by DNA sequencing. The pProEx-Htb-Hsp104 purification plasmid has previously been described.29 Hsp104 mutations were cloned into pProEx-Htb-Hsp104 by restriction digest with BamHI and Bsu36I followed by ligation and sequencing to verify the mutation.
The “strong” and “weak” [PSI+] variants were characterized and kindly provided by the Chernoff and Liebman labs.17 The Sc37 [PSI+] variant was made by transforming Sup35NM fibers generated at 37 °C into [psi−] cells. These strains were made and kindly provided by the Weissman lab.49 The [RNQ+] variant strains were characterized as specific [PIN+] strains initially, but later confirmed to be [RNQ+], and were kindly provided by the Liebman lab.59,60 To analyze the Hsp104 mutants in both [RNQ+] and [PSI+] variants, each variant was mated to both hsp104-G254D [psi−][rnq−] and hsp104-G730D [psi−][rnq−] cells and diploids were selected. The diploids were sporulated and dissected by micromanipulation.
EMS mutagenesis screen
A strong [PSI+] yeast strain was subjected to EMS mutagenesis as previously described.34 Two cultures with viabilities of about 17% were plated to determine changes in color. Candidates were selected based on color phenotype and were initially identified as mutations in HSP104 by backcrossing to an hsp104Δ strain and analyzing the progeny for segregation of the phenotype. Genomic DNA was PCR amplified and sequenced to identify the point mutations in HSP104.
SDD-AGE
SDD-AGE analysis was performed as previously described. Cells were lysed by beadbeating in PEB buffer (25 mM Tris-HCl pH 7.5, 50 mM KCl, 10 mM MgCl2, 1 mM EDTA, 10% Glycerol plus mini EDTA-free protease inhibitors [Roche], Aprotinen [Sigma] and PMSF [Sigma]). Samples were incubated in sample buffer at room temperature for seven minutes then separated on a 1.5% agarose gel. The protein distribution was analyzed by western blot with anti-Sup35 antibodies or anti-Rnq1 antibodies.
Hsp104 purification
Recombinant Hsp104 was expressed in E. coli cells and purified as previously described with one further separation step added.23 Briefly, Hsp104, tagged with a 6xHis tag on the N-terminus, was first isolated on a Nickel-sepharose column, the 6 × His tag was cleaved off using the TEV protease, and Hsp104 was re-applied to the Nickel-sepharose column to separate the untagged Hsp104. Untagged Hsp104 was then applied to an anion exchange Q-sepharose column followed by an S-300 size exclusion column to isolate monomeric Hsp104 from any aggregated Hsp104 species. Untagged, monomeric Hsp104 was stored at −80 °C in a storage buffer (20 mM Tris pH 8.0, 100 mM NaCl, 10 mM MgCl2, 2 mM EDTA, 10% glycerol).
ATP hydrolysis assays
ATP hydrolysis was measured by the Malachite green assay as previously described.64 Briefly, 2 µg of purified protein was incubated in buffer (40 mM TrisHCl pH7.5, 175 mM NaCl, 5 mM MgCl2, 0.02% Triton X-100) with 5 mM ATP at 37 °C. At various times, Malachite green dye was added to the sample for one minute and the reaction was stopped by the addition of 34% citric acid. The dye absorbance was determined at 650 nm and the amount of free phosphate calculated based on a standard of KH2PO4.
Glycerol gradients
Purified Hsp104 was applied to a 4 mL linear (10–35%) glycerol gradient and centrifuged at 34k rpm for 18 h in a SLA-600 rotor. Gradients were fractionated and equal volumes of each fraction were analyzed by SDS-PAGE and western blot using an anti-Hsp104 antibody. Individual bands from each fraction were quantified using Image J and reported as a percent of total Hsp104.
Thermotolerance
An equal number of cells from cultures of HSP104, hsp104-G254D, hsp104-G730D, and hsp104Δ grown to mid-log phase were pre-treated at 37 °C to induce HSP104 expression then heat-shocked at 50 °C. At the indicated time intervals, samples were taken and spotted on rich media in a 5-fold serial dilution.
Disclosure of Potential Conflicts of Interest
The authors declare no competing financial interests.
Acknowledgments
We thank Lisa Underwood for performing the EMS mutagenesis screen. We also thank JR Glover, B Bukau, J Weissman, Y Chernoff, and S Liebman for generously providing yeast strains, plasmids, and protocols. Additionally we thank members of the True lab and A Cashikar for comments on the manuscript. This work was supported by funding from the National Institutes of Health and the National Science Foundation.
Author Contributions
Dulle JE and True HL designed the experiments. Dulle JE performed all the experiments. Dulle JE and True HL analyzed the data. Dulle JE and True HL wrote the manuscript.
References
- 1.Collinge J. Prion diseases of humans and animals: their causes and molecular basis. Annu Rev Neurosci. 2001;24:519–50. doi: 10.1146/annurev.neuro.24.1.519. [DOI] [PubMed] [Google Scholar]
- 2.Prusiner SB, Scott MR, DeArmond SJ, Cohen FE. Prion protein biology. Cell. 1998;93:337–48. doi: 10.1016/S0092-8674(00)81163-0. [DOI] [PubMed] [Google Scholar]
- 3.Paushkin SV, Kushnirov VV, Smirnov VN, Ter-Avanesyan MD. In vitro propagation of the prion-like state of yeast Sup35 protein. Science. 1997;277:381–3. doi: 10.1126/science.277.5324.381. [DOI] [PubMed] [Google Scholar]
- 4.Tuite MF, Cox BS. Propagation of yeast prions. Nat Rev Mol Cell Biol. 2003;4:878–90. doi: 10.1038/nrm1247. [DOI] [PubMed] [Google Scholar]
- 5.Bousset L, Melki R. Similar and divergent features in mammalian and yeast prions. Microbes Infect. 2002;4:461–9. doi: 10.1016/S1286-4579(02)01561-7. [DOI] [PubMed] [Google Scholar]
- 6.Patino MM, Liu JJ, Glover JR, Lindquist S. Support for the prion hypothesis for inheritance of a phenotypic trait in yeast. Science. 1996;273:622–6. doi: 10.1126/science.273.5275.622. [DOI] [PubMed] [Google Scholar]
- 7.Paushkin SV, Kushnirov VV, Smirnov VN, Ter-Avanesyan MD. Propagation of the yeast prion-like [psi+] determinant is mediated by oligomerization of the SUP35-encoded polypeptide chain release factor. EMBO J. 1996;15:3127–34. [PMC free article] [PubMed] [Google Scholar]
- 8.Liebman SW, Derkatch IL. The yeast [PSI+] prion: making sense of nonsense. J Biol Chem. 1999;274:1181–4. doi: 10.1074/jbc.274.3.1181. [DOI] [PubMed] [Google Scholar]
- 9.True HL, Berlin I, Lindquist SL. Epigenetic regulation of translation reveals hidden genetic variation to produce complex traits. Nature. 2004;431:184–7. doi: 10.1038/nature02885. [DOI] [PubMed] [Google Scholar]
- 10.True HL, Lindquist SL. A yeast prion provides a mechanism for genetic variation and phenotypic diversity. Nature. 2000;407:477–83. doi: 10.1038/35035005. [DOI] [PubMed] [Google Scholar]
- 11.Derkatch IL, Bradley ME, Zhou P, Chernoff YO, Liebman SW. Genetic and environmental factors affecting the de novo appearance of the [PSI+] prion in Saccharomyces cerevisiae. Genetics. 1997;147:507–19. doi: 10.1093/genetics/147.2.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sondheimer N, Lindquist S. Rnq1: an epigenetic modifier of protein function in yeast. Mol Cell. 2000;5:163–72. doi: 10.1016/S1097-2765(00)80412-8. [DOI] [PubMed] [Google Scholar]
- 13.Derkatch IL, Bradley ME, Masse SV, Zadorsky SP, Polozkov GV, Inge-Vechtomov SG, Liebman SW. Dependence and independence of [PSI(+)] and [PIN(+)]: a two-prion system in yeast? EMBO J. 2000;19:1942–52. doi: 10.1093/emboj/19.9.1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Osherovich LZ, Weissman JS. Multiple Gln/Asn-rich prion domains confer susceptibility to induction of the yeast [PSI(+)] prion. Cell. 2001;106:183–94. doi: 10.1016/S0092-8674(01)00440-8. [DOI] [PubMed] [Google Scholar]
- 15.Derkatch IL, Bradley ME, Hong JY, Liebman SW. Prions affect the appearance of other prions: the story of [PIN(+)] Cell. 2001;106:171–82. doi: 10.1016/S0092-8674(01)00427-5. [DOI] [PubMed] [Google Scholar]
- 16.Stein KC, True HL. The [RNQ+] prion: a model of both functional and pathological amyloid. Prion. 2011;5:291–8. doi: 10.4161/pri.5.4.18213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chernoff YO, Lindquist SL, Ono B, Inge-Vechtomov SG, Liebman SW. Role of the chaperone protein Hsp104 in propagation of the yeast prion-like factor [psi+] Science. 1995;268:880–4. doi: 10.1126/science.7754373. [psi+] [DOI] [PubMed] [Google Scholar]
- 18.Alberti S, Halfmann R, King O, Kapila A, Lindquist S. A systematic survey identifies prions and illuminates sequence features of prionogenic proteins. Cell. 2009;137:146–58. doi: 10.1016/j.cell.2009.02.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tuite MF, Marchante R, Kushnirov V. Fungal prions: structure, function and propagation. Top Curr Chem. 2011;305:257–98. doi: 10.1007/128_2011_172. [DOI] [PubMed] [Google Scholar]
- 20.Glover JR, Lindquist S. Hsp104, Hsp70, and Hsp40: a novel chaperone system that rescues previously aggregated proteins. Cell. 1998;94:73–82. doi: 10.1016/S0092-8674(00)81223-4. [DOI] [PubMed] [Google Scholar]
- 21.Sanchez Y, Lindquist SL. HSP104 required for induced thermotolerance. Science. 1990;248:1112–5. doi: 10.1126/science.2188365. [DOI] [PubMed] [Google Scholar]
- 22.Lum R, Niggemann M, Glover JR. Peptide and protein binding in the axial channel of Hsp104. Insights into the mechanism of protein unfolding. J Biol Chem. 2008;283:30139–50. doi: 10.1074/jbc.M804849200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lum R, Tkach JM, Vierling E, Glover JR. Evidence for an unfolding/threading mechanism for protein disaggregation by Saccharomyces cerevisiae Hsp104. J Biol Chem. 2004;279:29139–46. doi: 10.1074/jbc.M403777200. [DOI] [PubMed] [Google Scholar]
- 24.Tessarz P, Mogk A, Bukau B. Substrate threading through the central pore of the Hsp104 chaperone as a common mechanism for protein disaggregation and prion propagation. Mol Microbiol. 2008;68:87–97. doi: 10.1111/j.1365-2958.2008.06135.x. [DOI] [PubMed] [Google Scholar]
- 25.Hung GC, Masison DC. N-terminal domain of yeast Hsp104 chaperone is dispensable for thermotolerance and prion propagation but necessary for curing prions by Hsp104 overexpression. Genetics. 2006;173:611–20. doi: 10.1534/genetics.106.056820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grimminger-Marquardt V, Lashuel HA. Structure and function of the molecular chaperone Hsp104 from yeast. Biopolymers. 2010;93:252–76. doi: 10.1002/bip.21301. [DOI] [PubMed] [Google Scholar]
- 27.Winkler J, Tyedmers J, Bukau B, Mogk A. Chaperone networks in protein disaggregation and prion propagation. J Struct Biol. 2012;179:152–60. doi: 10.1016/j.jsb.2012.05.002. [DOI] [PubMed] [Google Scholar]
- 28.Cashikar AG, Schirmer EC, Hattendorf DA, Glover JR, Ramakrishnan MS, Ware DM, Lindquist SL. Defining a pathway of communication from the C-terminal peptide binding domain to the N-terminal ATPase domain in a AAA protein. Mol Cell. 2002;9:751–60. doi: 10.1016/S1097-2765(02)00499-9. [DOI] [PubMed] [Google Scholar]
- 29.Tkach JM, Glover JR. Amino acid substitutions in the C-terminal AAA+ module of Hsp104 prevent substrate recognition by disrupting oligomerization and cause high temperature inactivation. J Biol Chem. 2004;279:35692–701. doi: 10.1074/jbc.M400782200. [DOI] [PubMed] [Google Scholar]
- 30.Kryndushkin DS, Alexandrov IM, Ter-Avanesyan MD, Kushnirov VV. Yeast [PSI+] prion aggregates are formed by small Sup35 polymers fragmented by Hsp104. J Biol Chem. 2003;278:49636–43. doi: 10.1074/jbc.M307996200. [DOI] [PubMed] [Google Scholar]
- 31.Wegrzyn RD, Bapat K, Newnam GP, Zink AD, Chernoff YO. Mechanism of prion loss after Hsp104 inactivation in yeast. Mol Cell Biol. 2001;21:4656–69. doi: 10.1128/MCB.21.14.4656-4669.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Satpute-Krishnan P, Langseth SX, Serio TR. Hsp104-dependent remodeling of prion complexes mediates protein-only inheritance. PLoS Biol. 2007;5:e24. doi: 10.1371/journal.pbio.0050024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Takahashi A, Hara H, Kurahashi H, Nakamura Y. A systematic evaluation of the function of the protein-remodeling factor Hsp104 in [PSI+] prion propagation in S. cerevisiae by comprehensive chromosomal mutations. Prion. 2007;1:69–77. doi: 10.4161/pri.1.1.4060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bardill JP, Dulle JE, Fisher JR, True HL. Requirements of Hsp104p activity and Sis1p binding for propagation of the [RNQ(+)] prion. Prion. 2009;3:151–60. doi: 10.4161/pri.3.3.9662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hattendorf DA, Lindquist SL. Cooperative kinetics of both Hsp104 ATPase domains and interdomain communication revealed by AAA sensor-1 mutants. EMBO J. 2002;21:12–21. doi: 10.1093/emboj/21.1.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hattendorf DA, Lindquist SL. Analysis of the AAA sensor-2 motif in the C-terminal ATPase domain of Hsp104 with a site-specific fluorescent probe of nucleotide binding. Proc Natl Acad Sci U S A. 2002;99:2732–7. doi: 10.1073/pnas.261693199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Helsen CW, Glover JR. Insight into molecular basis of curing of [PSI+] prion by overexpression of 104-kDa heat shock protein (Hsp104) J Biol Chem. 2012;287:542–56. doi: 10.1074/jbc.M111.302869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.True HL. The battle of the fold: chaperones take on prions. Trends Genet. 2006;22:110–7. doi: 10.1016/j.tig.2005.12.004. [DOI] [PubMed] [Google Scholar]
- 39.Inoue Y, Taguchi H, Kishimoto A, Yoshida M. Hsp104 binds to yeast Sup35 prion fiber but needs other factor(s) to sever it. J Biol Chem. 2004;279:52319–23. doi: 10.1074/jbc.M408159200. [DOI] [PubMed] [Google Scholar]
- 40.Shorter J, Lindquist S. Hsp104 catalyzes formation and elimination of self-replicating Sup35 prion conformers. Science. 2004;304:1793–7. doi: 10.1126/science.1098007. [DOI] [PubMed] [Google Scholar]
- 41.Sweeny EA, Shorter J. Prion proteostasis: Hsp104 meets its supporting cast. Prion. 2008;2:135–40. doi: 10.4161/pri.2.4.7952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Newnam GP, Wegrzyn RD, Lindquist SL, Chernoff YO. Antagonistic interactions between yeast chaperones Hsp104 and Hsp70 in prion curing. Mol Cell Biol. 1999;19:1325–33. doi: 10.1128/mcb.19.2.1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sharma D, Masison DC. Functionally redundant isoforms of a yeast Hsp70 chaperone subfamily have different antiprion effects. Genetics. 2008;179:1301–11. doi: 10.1534/genetics.108.089458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Aron R, Higurashi T, Sahi C, Craig EA. J-protein co-chaperone Sis1 required for generation of [RNQ+] seeds necessary for prion propagation. EMBO J. 2007;26:3794–803. doi: 10.1038/sj.emboj.7601811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Higurashi T, Hines JK, Sahi C, Aron R, Craig EA. Specificity of the J-protein Sis1 in the propagation of 3 yeast prions. Proc Natl Acad Sci U S A. 2008;105:16596–601. doi: 10.1073/pnas.0808934105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jones GW, Masison DC. Saccharomyces cerevisiae Hsp70 mutations affect [PSI+] prion propagation and cell growth differently and implicate Hsp40 and tetratricopeptide repeat cochaperones in impairment of [PSI+] Genetics. 2003;163:495–506. doi: 10.1093/genetics/163.2.495. [PSI+] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mathur V, Hong JY, Liebman SW. Ssa1 overexpression and [PIN(+)] variants cure [PSI(+)] by dilution of aggregates. J Mol Biol. 2009;390:155–67. doi: 10.1016/j.jmb.2009.04.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Krishnan R, Lindquist SL. Structural insights into a yeast prion illuminate nucleation and strain diversity. Nature. 2005;435:765–72. doi: 10.1038/nature03679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tanaka M, Chien P, Naber N, Cooke R, Weissman JS. Conformational variations in an infectious protein determine prion strain differences. Nature. 2004;428:323–8. doi: 10.1038/nature02392. [DOI] [PubMed] [Google Scholar]
- 50.Toyama BH, Kelly MJ, Gross JD, Weissman JS. The structural basis of yeast prion strain variants. Nature. 2007;449:233–7. doi: 10.1038/nature06108. [DOI] [PubMed] [Google Scholar]
- 51.Collinge J, Clarke AR. A general model of prion strains and their pathogenicity. Science. 2007;318:930–6. doi: 10.1126/science.1138718. [DOI] [PubMed] [Google Scholar]
- 52.Tanaka M, Chien P, Yonekura K, Weissman JS. Mechanism of cross-species prion transmission: an infectious conformation compatible with two highly divergent yeast prion proteins. Cell. 2005;121:49–62. doi: 10.1016/j.cell.2005.03.008. [DOI] [PubMed] [Google Scholar]
- 53.Derkatch IL, Chernoff YO, Kushnirov VV, Inge-Vechtomov SG, Liebman SW. Genesis and variability of [PSI] prion factors in Saccharomyces cerevisiae. Genetics. 1996;144:1375–86. doi: 10.1093/genetics/144.4.1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhou P, Derkatch IL, Uptain SM, Patino MM, Lindquist S, Liebman SW. The yeast non-Mendelian factor [ETA+] is a variant of [PSI+], a prion-like form of release factor eRF3. EMBO J. 1999;18:1182–91. doi: 10.1093/emboj/18.5.1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tanaka M, Collins SR, Toyama BH, Weissman JS. The physical basis of how prion conformations determine strain phenotypes. Nature. 2006;442:585–9. doi: 10.1038/nature04922. [DOI] [PubMed] [Google Scholar]
- 56.Tessier PM, Lindquist S. Prion recognition elements govern nucleation, strain specificity and species barriers. Nature. 2007;447:556–61. doi: 10.1038/nature05848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.DeSantis ME, Shorter J. Hsp104 drives “protein-only” positive selection of Sup35 prion strains encoding strong [PSI(+)] Chem Biol. 2012;19:1400–10. doi: 10.1016/j.chembiol.2012.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tipton KA, Verges KJ, Weissman JS. In vivo monitoring of the prion replication cycle reveals a critical role for Sis1 in delivering substrates to Hsp104. Mol Cell. 2008;32:584–91. doi: 10.1016/j.molcel.2008.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bradley ME, Edskes HK, Hong JY, Wickner RB, Liebman SW. Interactions among prions and prion “strains” in yeast. Proc Natl Acad Sci U S A. 2002;99(Suppl 4):16392–9. doi: 10.1073/pnas.152330699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bradley ME, Liebman SW. Destabilizing interactions among [PSI(+)] and [PIN(+)] yeast prion variants. Genetics. 2003;165:1675–85. doi: 10.1093/genetics/165.4.1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bardill JP, True HL. Heterologous prion interactions are altered by mutations in the prion protein Rnq1p. J Mol Biol. 2009;388:583–96. doi: 10.1016/j.jmb.2009.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kalastavadi T, True HL. Analysis of the [RNQ+] prion reveals stability of amyloid fibers as the key determinant of yeast prion variant propagation. J Biol Chem. 2010;285:20748–55. doi: 10.1074/jbc.M110.115303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bagriantsev SN, Kushnirov VV, Liebman SW. Analysis of amyloid aggregates using agarose gel electrophoresis. Methods Enzymol. 2006;412:33–48. doi: 10.1016/S0076-6879(06)12003-0. [DOI] [PubMed] [Google Scholar]
- 64.Schirmer EC, Queitsch C, Kowal AS, Parsell DA, Lindquist S. The ATPase activity of Hsp104, effects of environmental conditions and mutations. J Biol Chem. 1998;273:15546–52. doi: 10.1074/jbc.273.25.15546. [DOI] [PubMed] [Google Scholar]
- 65.Parsell DA, Kowal AS, Lindquist S. Saccharomyces cerevisiae Hsp104 protein. Purification and characterization of ATP-induced structural changes. J Biol Chem. 1994;269:4480–7. [PubMed] [Google Scholar]
- 66.DeSantis ME, Leung EH, Sweeny EA, Jackrel ME, Cushman-Nick M, Neuhaus-Follini A, Vashist S, Sochor MA, Knight MN, Shorter J. Operational plasticity enables hsp104 to disaggregate diverse amyloid and nonamyloid clients. Cell. 2012;151:778–93. doi: 10.1016/j.cell.2012.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Krzewska J, Melki R. Molecular chaperones and the assembly of the prion Sup35p, an in vitro study. EMBO J. 2006;25:822–33. doi: 10.1038/sj.emboj.7600985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Romanova NV, Chernoff YO. Hsp104 and prion propagation. Protein Pept Lett. 2009;16:598–605. doi: 10.2174/092986609788490078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Glover JR, Lum R. Remodeling of protein aggregates by Hsp104. Protein Pept Lett. 2009;16:587–97. doi: 10.2174/092986609788490087. [DOI] [PubMed] [Google Scholar]
- 70.Du Z, Park KW, Yu H, Fan Q, Li L. Newly identified prion linked to the chromatin-remodeling factor Swi1 in Saccharomyces cerevisiae. Nat Genet. 2008;40:460–5. doi: 10.1038/ng.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Patel BK, Gavin-Smyth J, Liebman SW. The yeast global transcriptional co-repressor protein Cyc8 can propagate as a prion. Nat Cell Biol. 2009;11:344–9. doi: 10.1038/ncb1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wickner RB, Taylor KL, Edskes HK, Maddelein ML, Moriyama H, Roberts BT. Prions of yeast as heritable amyloidoses. J Struct Biol. 2000;130:310–22. doi: 10.1006/jsbi.2000.4250. [DOI] [PubMed] [Google Scholar]
- 73.Doyle SM, Shorter J, Zolkiewski M, Hoskins JR, Lindquist S, Wickner S. Asymmetric deceleration of ClpB or Hsp104 ATPase activity unleashes protein-remodeling activity. Nat Struct Mol Biol. 2007;14:114–22. doi: 10.1038/nsmb1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Schaupp A, Marcinowski M, Grimminger V, Bösl B, Walter S. Processing of proteins by the molecular chaperone Hsp104. J Mol Biol. 2007;370:674–86. doi: 10.1016/j.jmb.2007.04.070. [DOI] [PubMed] [Google Scholar]
- 75.Mackay RG, Helsen CW, Tkach JM, Glover JR. The C-terminal extension of Saccharomyces cerevisiae Hsp104 plays a role in oligomer assembly. Biochemistry. 2008;47:1918–27. doi: 10.1021/bi701714s. [DOI] [PubMed] [Google Scholar]
- 76.Kryndushkin DS, Engel A, Edskes H, Wickner RB. Molecular chaperone Hsp104 can promote yeast prion generation. Genetics. 2011;188:339–48. doi: 10.1534/genetics.111.127779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Borchsenius AS, Müller S, Newnam GP, Inge-Vechtomov SG, Chernoff YO. Prion variant maintained only at high levels of the Hsp104 disaggregase. Curr Genet. 2006;49:21–9. doi: 10.1007/s00294-005-0035-0. [DOI] [PubMed] [Google Scholar]