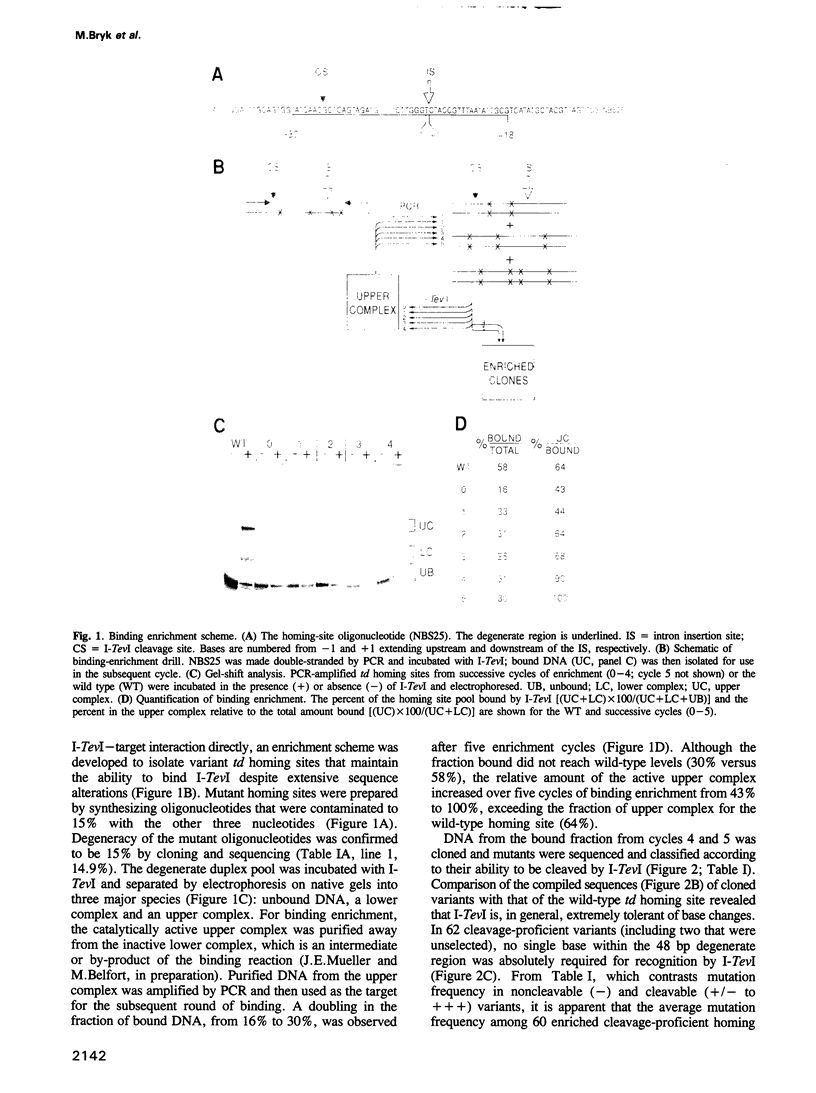
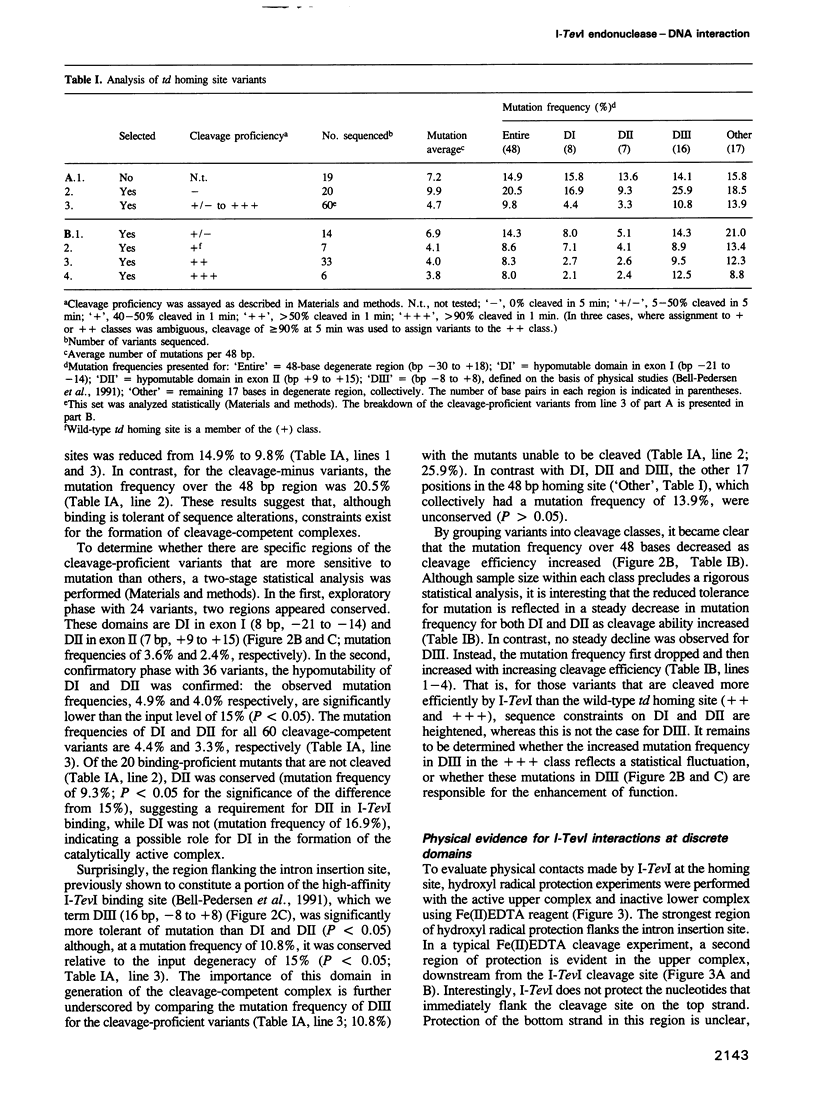
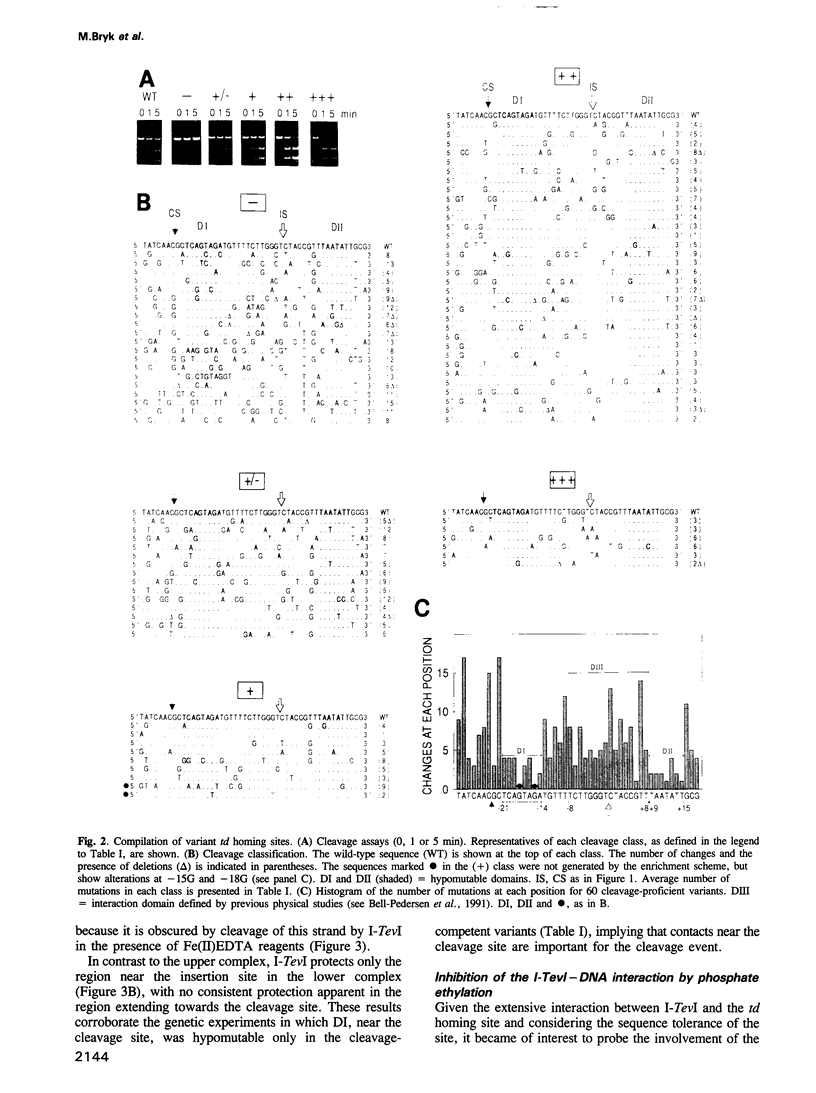
Abstract
I-TevI, a double-strand DNA endonuclease encoded by the mobile td intron of phage T4, has specificity for the intronless td allele. Genetic and physical studies indicate that the enzyme makes extensive contacts with its DNA substrate over at least three helical turns and around the circumference of the helix. Remarkably, no single nucleotide within a 48 bp region encompassing this interaction domain is essential for cleavage. Although two subdomains (DI and DII) contain preferred sequences, a third domain (DIII), a primary region of contact with the enzyme, displays much lower sequence preference. While DII and DIII suffice for recognition and binding of I-TevI, all three domains are important for formation of a cleavage-competent complex. Mutational, footprinting and interference studies indicate predominant interactions of I-TevI across the minor groove and phosphate backbone of the DNA. Contacts appear not to be at the single nucleotide level; rather, redundant interactions and/or structural recognition are implied. These unusual properties provide a basis for understanding how I-TevI recognizes T-even phage DNA, which is heavily modified in the major groove. These recognition characteristics may increase the range of natural substrates available to the endonuclease, thereby extending the invasive potential of the mobile intron.
Full text
PDF
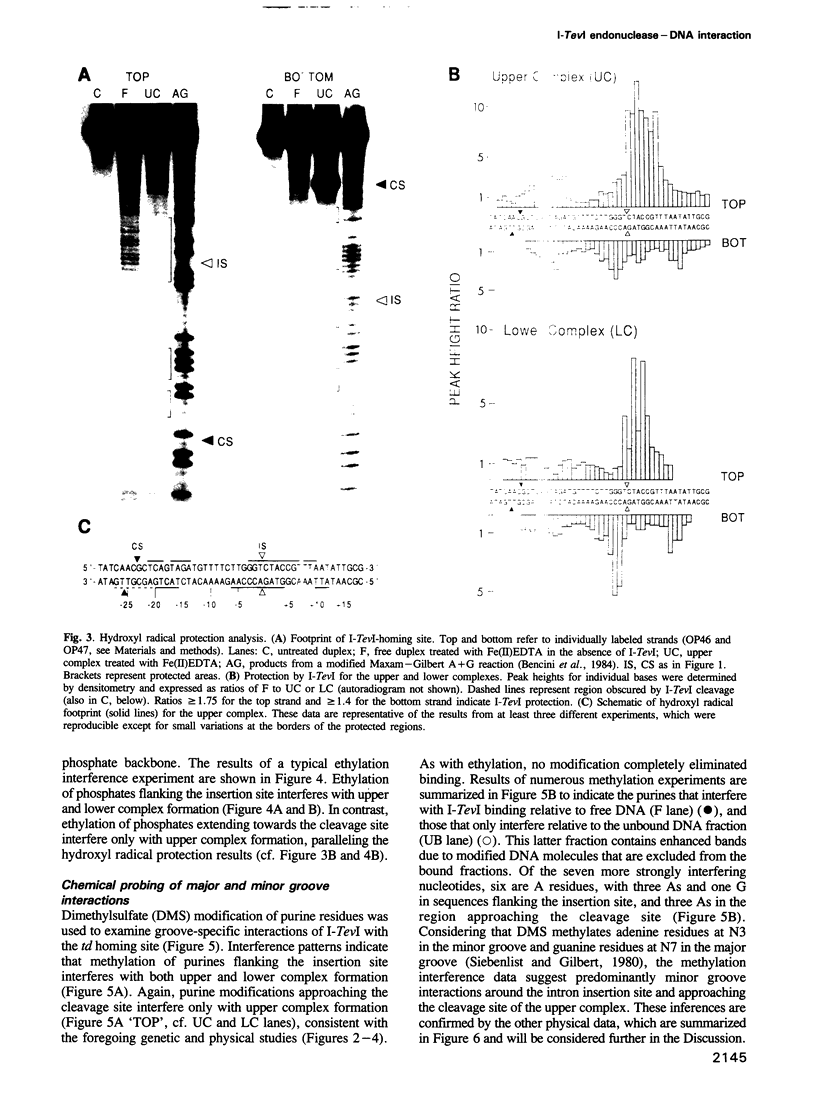

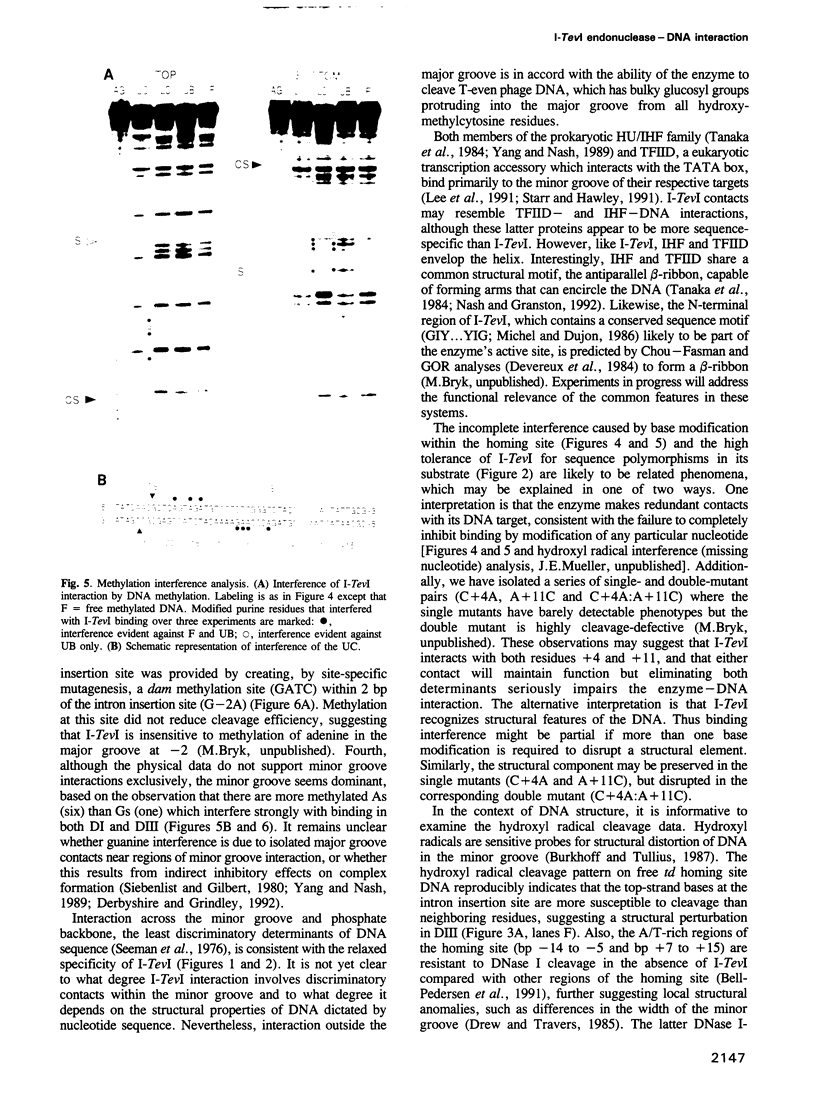
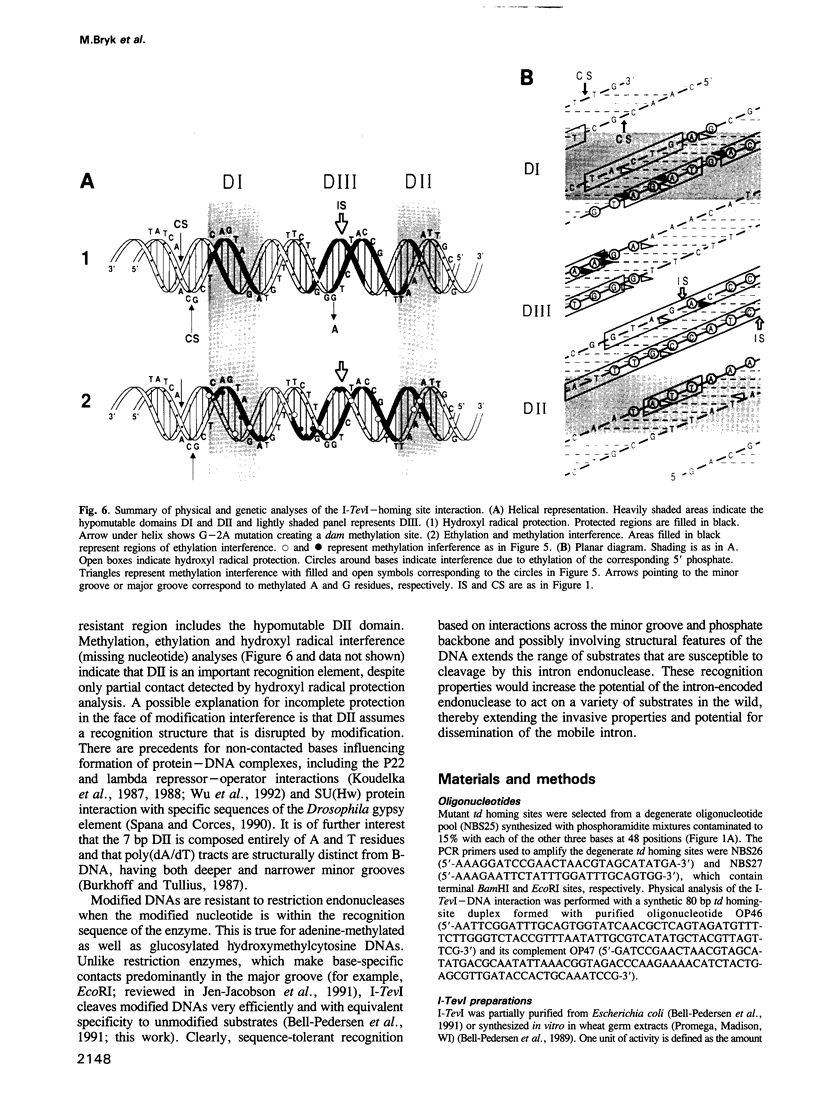

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Belfort M. Phage T4 introns: self-splicing and mobility. Annu Rev Genet. 1990;24:363–385. doi: 10.1146/annurev.ge.24.120190.002051. [DOI] [PubMed] [Google Scholar]
- Bell-Pedersen D., Quirk S. M., Bryk M., Belfort M. I-TevI, the endonuclease encoded by the mobile td intron, recognizes binding and cleavage domains on its DNA target. Proc Natl Acad Sci U S A. 1991 Sep 1;88(17):7719–7723. doi: 10.1073/pnas.88.17.7719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell-Pedersen D., Quirk S., Clyman J., Belfort M. Intron mobility in phage T4 is dependent upon a distinctive class of endonucleases and independent of DNA sequences encoding the intron core: mechanistic and evolutionary implications. Nucleic Acids Res. 1990 Jul 11;18(13):3763–3770. doi: 10.1093/nar/18.13.3763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkhoff A. M., Tullius T. D. The unusual conformation adopted by the adenine tracts in kinetoplast DNA. Cell. 1987 Mar 27;48(6):935–943. doi: 10.1016/0092-8674(87)90702-1. [DOI] [PubMed] [Google Scholar]
- Chu F. K., Maley F., Wang A. M., Pedersen-Lane J., Maley G. Purification and substrate specificity of a T4 phage intron-encoded endonuclease. Nucleic Acids Res. 1991 Dec 25;19(24):6863–6869. doi: 10.1093/nar/19.24.6863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu F. K., Maley G., Pedersen-Lane J., Wang A. M., Maley F. Characterization of the restriction site of a prokaryotic intron-encoded endonuclease. Proc Natl Acad Sci U S A. 1990 May;87(9):3574–3578. doi: 10.1073/pnas.87.9.3574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colleaux L., D'Auriol L., Galibert F., Dujon B. Recognition and cleavage site of the intron-encoded omega transposase. Proc Natl Acad Sci U S A. 1988 Aug;85(16):6022–6026. doi: 10.1073/pnas.85.16.6022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derbyshire K. M., Grindley N. D. Binding of the IS903 transposase to its inverted repeat in vitro. EMBO J. 1992 Sep;11(9):3449–3455. doi: 10.1002/j.1460-2075.1992.tb05424.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dervan P. B. Design of sequence-specific DNA-binding molecules. Science. 1986 Apr 25;232(4749):464–471. doi: 10.1126/science.2421408. [DOI] [PubMed] [Google Scholar]
- Devereux J., Haeberli P., Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drew H. R., Travers A. A. Structural junctions in DNA: the influence of flanking sequence on nuclease digestion specificities. Nucleic Acids Res. 1985 Jun 25;13(12):4445–4467. doi: 10.1093/nar/13.12.4445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dujon B., Belfort M., Butow R. A., Jacq C., Lemieux C., Perlman P. S., Vogt V. M. Mobile introns: definition of terms and recommended nomenclature. Gene. 1989 Oct 15;82(1):115–118. doi: 10.1016/0378-1119(89)90035-8. [DOI] [PubMed] [Google Scholar]
- Dujon B. Group I introns as mobile genetic elements: facts and mechanistic speculations--a review. Gene. 1989 Oct 15;82(1):91–114. doi: 10.1016/0378-1119(89)90034-6. [DOI] [PubMed] [Google Scholar]
- Eddy S. R., Gold L. The phage T4 nrdB intron: a deletion mutant of a version found in the wild. Genes Dev. 1991 Jun;5(6):1032–1041. doi: 10.1101/gad.5.6.1032. [DOI] [PubMed] [Google Scholar]
- Hendrickson W., Schleif R. A dimer of AraC protein contacts three adjacent major groove regions of the araI DNA site. Proc Natl Acad Sci U S A. 1985 May;82(10):3129–3133. doi: 10.1073/pnas.82.10.3129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koudelka G. B., Harbury P., Harrison S. C., Ptashne M. DNA twisting and the affinity of bacteriophage 434 operator for bacteriophage 434 repressor. Proc Natl Acad Sci U S A. 1988 Jul;85(13):4633–4637. doi: 10.1073/pnas.85.13.4633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koudelka G. B., Harrison S. C., Ptashne M. Effect of non-contacted bases on the affinity of 434 operator for 434 repressor and Cro. 1987 Apr 30-May 6Nature. 326(6116):886–888. doi: 10.1038/326886a0. [DOI] [PubMed] [Google Scholar]
- Lambowitz A. M., Belfort M. Introns as mobile genetic elements. Annu Rev Biochem. 1993;62:587–622. doi: 10.1146/annurev.bi.62.070193.003103. [DOI] [PubMed] [Google Scholar]
- Lambowitz A. M. Infectious introns. Cell. 1989 Feb 10;56(3):323–326. doi: 10.1016/0092-8674(89)90232-8. [DOI] [PubMed] [Google Scholar]
- Lee D. K., Horikoshi M., Roeder R. G. Interaction of TFIID in the minor groove of the TATA element. Cell. 1991 Dec 20;67(6):1241–1250. doi: 10.1016/0092-8674(91)90300-n. [DOI] [PubMed] [Google Scholar]
- Michel F., Dujon B. Genetic exchanges between bacteriophage T4 and filamentous fungi? Cell. 1986 Aug 1;46(3):323–323. doi: 10.1016/0092-8674(86)90651-3. [DOI] [PubMed] [Google Scholar]
- Nash H. A., Granston A. E. Similarity between the DNA-binding domains of IHF protein and TFIID protein. Cell. 1991 Dec 20;67(6):1037–1038. doi: 10.1016/0092-8674(91)90280-c. [DOI] [PubMed] [Google Scholar]
- Perlman P. S., Butow R. A. Mobile introns and intron-encoded proteins. Science. 1989 Dec 1;246(4934):1106–1109. doi: 10.1126/science.2479980. [DOI] [PubMed] [Google Scholar]
- Quirk S. M., Bell-Pedersen D., Belfort M. Intron mobility in the T-even phages: high frequency inheritance of group I introns promoted by intron open reading frames. Cell. 1989 Feb 10;56(3):455–465. doi: 10.1016/0092-8674(89)90248-1. [DOI] [PubMed] [Google Scholar]
- Sargueil B., Hatat D., Delahodde A., Jacq C. In vivo and in vitro analyses of an intron-encoded DNA endonuclease from yeast mitochondria. Recognition site by site-directed mutagenesis. Nucleic Acids Res. 1990 Oct 11;18(19):5659–5665. doi: 10.1093/nar/18.19.5659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeman N. C., Rosenberg J. M., Rich A. Sequence-specific recognition of double helical nucleic acids by proteins. Proc Natl Acad Sci U S A. 1976 Mar;73(3):804–808. doi: 10.1073/pnas.73.3.804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siebenlist U., Gilbert W. Contacts between Escherichia coli RNA polymerase and an early promoter of phage T7. Proc Natl Acad Sci U S A. 1980 Jan;77(1):122–126. doi: 10.1073/pnas.77.1.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spana C., Corces V. G. DNA bending is a determinant of binding specificity for a Drosophila zinc finger protein. Genes Dev. 1990 Sep;4(9):1505–1515. doi: 10.1101/gad.4.9.1505. [DOI] [PubMed] [Google Scholar]
- Starr D. B., Hawley D. K. TFIID binds in the minor groove of the TATA box. Cell. 1991 Dec 20;67(6):1231–1240. doi: 10.1016/0092-8674(91)90299-e. [DOI] [PubMed] [Google Scholar]
- Tanaka I., Appelt K., Dijk J., White S. W., Wilson K. S. 3-A resolution structure of a protein with histone-like properties in prokaryotes. Nature. 1984 Aug 2;310(5976):376–381. doi: 10.1038/310376a0. [DOI] [PubMed] [Google Scholar]
- Tullius T. D., Dombroski B. A. Iron(II) EDTA used to measure the helical twist along any DNA molecule. Science. 1985 Nov 8;230(4726):679–681. doi: 10.1126/science.2996145. [DOI] [PubMed] [Google Scholar]
- Wernette C., Saldanha R., Smith D., Ming D., Perlman P. S., Butow R. A. Complex recognition site for the group I intron-encoded endonuclease I-SceII. Mol Cell Biol. 1992 Feb;12(2):716–723. doi: 10.1128/mcb.12.2.716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L., Vertino A., Koudelka G. B. Non-contacted bases affect the affinity of synthetic P22 operators for P22 repressor. J Biol Chem. 1992 May 5;267(13):9134–9139. [PubMed] [Google Scholar]
- Yang C. C., Nash H. A. The interaction of E. coli IHF protein with its specific binding sites. Cell. 1989 Jun 2;57(5):869–880. doi: 10.1016/0092-8674(89)90801-5. [DOI] [PubMed] [Google Scholar]