Abstract
The laminin-binding integrin α3β1 is highly expressed in epidermal keratinocytes where it regulates both cell-autonomous and paracrine functions that promote wound healing and skin tumorigenesis. However, roles for α3β1 in regulating gene expression programs that control the behaviors of immortalized or transformed keratinocytes remain underexplored. In the current study, we used a microarray approach to identify genes that are regulated by α3β1 in immortalized keratinocytes. α3β1-responsive genes included several that are involved in extracellular matrix proteolysis or remodeling, including fibulin-2 and SPARC. However, α3β1-dependent induction of specific target genes was influenced by the genetic lesion that triggered immortalization, as α3β1-dependent fibulin-2 expression occurred in cells immortalized by either SV40 large T antigen or p53-null mutation, while α3β1-dependent SPARC expression occurred only in the former cells. Interestingly, qPCR arrays did not reveal strong patterns of α3β1-dependent gene expression in freshly isolated primary keratinocytes, suggesting that this regulation is acquired during immortalization. p53-null keratinocytes transformed with oncogenic RasV12 retained α3β1-dependent fibulin-2 expression, and RNAi-mediated knockdown of fibulin-2 in these cells reduced invasion, although not their tumorigenic potential. These findings demonstrate a prominent role for α3β1 in immortalized/transformed keratinocytes in regulating fibulin-2 and other genes that promote matrix remodeling and invasion.
Keywords: α3β1 integrin, keratinocyte, immortalization, epidermis, fibulin-2
INTRODUCTION
Integrins are the major cell surface receptors for adhesion to extracellular matrix (ECM), and they activate intracellular signaling pathways that control a wide variety of cell functions, including cell polarity, proliferation, survival, and motility (Hynes, 2002). Numerous studies have demonstrated roles for keratinocyte integrins in mediating epidermal function during both normal and pathological skin remodeling processes, including wound healing, tumorigenesis, tumor invasion and various skin diseases (Grose et al., 2002; Margadant et al., 2010; Missan and DiPersio, 2012; Watt, 2002). However, roles for integrins in regulating genes that control these processes remain unclear. Integrin α3β1 is highly expressed in the epidermis where it mediates keratinocyte adhesion to laminin-332 (LN-332) in the basement membrane (Carter et al., 1991; Delwel et al., 1994; Kreidberg, 2000). α3β1 is also expressed highly in wound epidermis (Margadant et al., 2010), and in squamous cell carcinoma (SCC) where it contributes to tumor growth, invasion and metastasis (Janes and Watt, 2006; Sachs et al., 2012). Indeed, numerous studies using primary or immortalized keratinocytes have identified roles for α3β1 in a range of cell functions important for wound healing and tumor progression, including cell polarization and migration, ECM organization, invasion, survival, and paracrine stimulation of endothelial cells (Choma et al., 2007; Choma et al., 2004; deHart et al., 2003; Iyer et al., 2005; Kreidberg, 2000; Lamar et al., 2008a; Margadant et al., 2009; Mitchell et al., 2009). Furthermore, in vivo studies using global or epidermis-specific α3 knockout mice have identified roles for α3β1 in basement membrane organization and epidermal-dermal adhesion (DiPersio et al., 1997; Longmate et al., 2014), hair follicle morphogenesis (Conti et al., 2003), wound reepithelialization (Margadant et al., 2009), induction of wound angiogenesis (Mitchell et al., 2009), and tumor growth (Abramoff et al., 2013; Lamar et al., 2008b).
Many α3β1-mediated keratinocyte functions can be attributed to its roles in mediating cell spreading, migration, and cytoskeletal reorganization (Kreidberg, 2000). However, α3β1 also regulates expression of secreted proteases or growth factors in immortalized keratinocytes that can alter ECM or stimulate other cellular compartments, thereby allowing epidermal cells to modify the wound or tumor microenvironment (Iyer et al., 2005; Mitchell et al., 2009). In the current study, we sought to identify α3β1-dependent changes in global gene expression that may contribute to ECM remodeling or invasive properties of immortalized keratinocytes. Microarray analysis and quantitative PCR (qPCR) validation revealed substantial changes in gene expression in immortalized mouse keratinocytes (MK cells) that lack α3β1 due to null mutation of the Itga3 gene, which encodes the α3 subunit. Many of these α3β1-responsive genes are involved in normal or pathological skin remodeling, including wound healing and epidermal carcinogenesis, and several encode proteins with known roles in modulating the skin microenvironment through changes in ECM organization, ECM proteolysis, or paracrine stimulation of other cells. One such protein, fibulin-2, is a secreted matricellular protein that can bind several ECM proteins including perlecan, fibrillin-1, aggrecan, fibronectin, and γ2 chain-containing laminins (Timpl et al., 2003; Pan et al., 1993). Another example is SPARC (secreted protein acidic and rich in cysteine), an ECM-associated protein with roles in tumor angiogenesis, invasion and metastasis (Arnold and Brekken, 2009). Interestingly, custom qPCR arrays did not reveal statistically significant differences in most genes upon deletion of α3β1 from non-immortalized, primary keratinocytes, suggesting that α3β1-dependent gene regulation was acquired by immortalized keratinocytes, as we described previously for MMP-9 (DiPersio et al., 2000; Lamar et al., 2008b). However, while α3β1-dependent regulation of fibulin-2 was observed in cells immortalized by either SV40 large T antigen or p53-null mutation, α3β1-dependence of SPARC and other genes was influenced by the immortalizing genetic lesion. siRNA-mediated suppression of fibulin-2 reduced invasion of p53-null MK cells, consistent with pro-invasive/pro-metastatic roles for fibulin-2 in some cancers (Baird et al., 2013; Senapati et al., 2012). However, stable knockdown of fibulin-2 in RasV12-transformed keratinocytes did not inhibit α3β1-dependent tumor growth in a subcutaneous allograft model. Taken together, these findings support a role for integrin α3β1 in the regulation of fibulin-2 and other genes that contribute to the invasive properties of transformed keratinocytes.
RESULTS
Microarrays of immortalized keratinocytes reveal α3β1-dependent changes in genes that modulate the tumor microenvironment
We used Affymetrix whole genome arrays (Mouse Exon ST 1.0) to compare global gene expression between MK cells that were derived from either wild-type mice (MK+/+ cells) or mice that lack α3β1 (MK−/− cells), and were immortalized with SV40 Large T antigen (LTAg) (DiPersio et al., 2000). MK cells were cultured in low calcium (0.05 mM CaCl2) on collagen-coated plates, since under these conditions they deposit abundant LN-332 regardless of α3β1 expression (DiPersio et al., 2000). Initially, we identified 739 genes that increased or decreased by at least 1.5-fold in MK−/− cells compared to MK+/+ cells. However, some differences could be due to clonal variations that arose during expansion of these cell lines (DiPersio et al., 2000). Therefore, we narrowed our list of candidate genes to those that changed in MK−/− cells but were rescued to original expression levels after restoring α3β1 in this line (i.e., MK−/−:hα3 cells) by stable transfection with human α3 (Iyer et al., 2005). This analysis identified a subset of 252 genes that were rescued by α3β1, most of which were restored to within 1.5-fold of their expression in MK+/+ cells. Select genes that fulfilled these initial criteria are shown in Figure S1. From genes that fit this pattern, 86 were validated as α3β1-dependent using customized qPCR arrays to compare expression in MK+/+ cells versus either MK−/− cells or MK−/−:hα3 cells. A selection of validated genes that fulfilled these final criteria are shown in Figures 1a and 1b, and listed in Figure 1c.
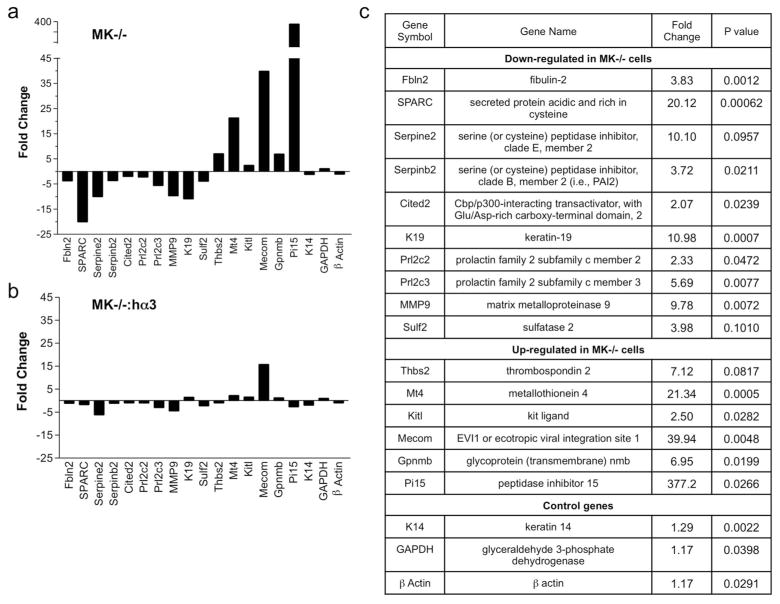
Figure 1.
qPCR validation of α3β1-dependent genes in immortalized MK cells. A subset of α3β1-regulated genes identified by microarray analysis (Fig. S1) was analyzed by qPCR. Analysis was focused on a selection of genes confirmed as α3β1-dependent through restoration of original expression levels in MK−/− cells following stable expression of human α3. (a, b) Graphs depict gene expression changes as fold decrease (negative value) or increase (positive value) in (a) MK−/− cells or (b) MK−/−:hα3 cells, each relative to MK+/+ cells. (c) Summary of changes in genes that are down-regulated or up-regulated in MK−/− cells compared to MK+/+ cells. Data are mean of three experiments; P values calculated based on Student’s t-test of replicate 2^(−Delta Ct) values for each gene in control group and treatment groups.
Among validated α3β1-dependent genes were many that contribute to ECM remodeling in tumors or wounds (Fig. 1), including ECM-associated proteins (e.g., fibulin-2, SPARC, thrombospondin-2) and ECM proteases/sulfatases or their inhibitors (e.g., sulf-2, serpine2, serpinB2/PAI-2, Pi15). α3β1-dependent regulation of fibulin-2 is particularly intriguing since this protein can bind the γ2 chain of laminin-332, the major ECM ligand of α3β1 (Utani et al., 1997), and we recently implicated fibulin-2 in α3β1-dependent epidermal functions during development and wound healing (Longmate et al., 2014). Although SPARC is overexpressed in esophageal SCC (Che et al., 2006; Porte et al., 1998), the literature regarding SPARC function in cancer is controversial, and both pro-tumorigenic and tumor suppressor functions have been described (Arnold and Brekken, 2009; Briggs et al., 2002; Gerson et al., 2012; Koblinski et al., 2005; Ledda et al., 1997). Expression of the protease inhibitor Pi15 decreased most substantially in α3β1-deficient cells on both gene microarrays and qPCR arrays. While the functional impact of this change is not known, it is noteworthy that altered Pi15 expression has been associated with wound healing (Roupe et al., 2010). α3β1 deletion also altered expression of transcriptional regulators, such as mecom and cited2. Of note, cited2 is a transcriptional coactivator that induces MMP-9 gene expression in response to TGF-β (Chou et al., 2006), which could conceivably contribute to cooperative induction of MMP-9 mRNA by α3β1 and TGFβ that we previously described in MK cells (Lamar et al., 2008a).
α3β1 also regulated genes that encode factors for paracrine stimulation of other cell types (e.g., kitl, Prl2c2/Prl2c3). Indeed, members of the mitogen-regulated protein (MRP)/proliferin family were among the α3β1-dependent genes detected by microarray (Fig. S1) and validated by qPCR (Fig. 1). This finding validates our approach to identify α3β1-dependent genes, as it is consistent with our previous report that α3β1-dependent MRP-3/Prl2c3 gene expression promotes MK cell crosstalk to endothelial cells (Mitchell et al., 2009). Nevertheless, not all α3β1-dependent genes were revealed by the initial microarray, since α3β1-dependent MMP-9 expression described previously (Iyer et al., 2005) was confirmed by qPCR (Fig. 1) but not detected by gene microarray.
α3β1-dependent gene regulation is associated with keratinocyte immortalization
Our earlier work showed that α3β1-dependent MMP-9 gene expression is acquired during keratinocyte immortalization (DiPersio et al., 2000; Lamar et al., 2008b). To determine whether α3β1-dependent regulation of other genes is similarly acquired by immortalized cells, we utilized our custom qPCR array to compare gene expression in freshly isolated primary keratinocytes from neonatal mice that either express α3β1 (control mice) or lack α3β1 in epidermis (α3eKO mice) due to deletion of a floxed Itga3 allele by Cre recombinase under control of the keratin-14 promoter (Mitchell et al., 2009). α3 protein was readily detected by immunoblot of primary cultures from control mice (albeit at variable levels) but was uniformly undetectable in cultures from α3eKO mice (Fig. S2a). Interestingly, microarrays of cells isolated from three individual mice of each genotype revealed no statistically significant differences between control and α3eKO cells for genes that had been identified as α3β1-responsive in immortalized MK cells (Fig. S2b). Notably, we observed a trend towards decreased expression of fibulin-2 and thrombospondin-2 in α3eKO primary cells, although the magnitude was variable and did not reach statistical significance. These findings indicate that α3β1-dependent regulation of most genes was acquired by immortalized keratinocytes.
α3β1-dependent gene regulation is influenced by the genetic lesion that drives keratinocyte immortalization
To determine how α3β1-mediated gene regulation observed in LTAg-immortalized MK cells is influenced by other genetic lesions that drive immortalization/transformation, we utilized an independently derived set of mouse keratinocyte lines (Fig. 2a). IMK cells are immortalized by p53 knockout and either express α3β1 (IMKα3+/+) or lack α3β1 (IMKα3−/−), as described (Lamar et al., 2008b). TMK cells are transformed, tumorigenic derivatives of IMK cells that were stably transduced with oncogenic H-RasV12 (Lamar et al., 2008b). Since p53 loss and oncogenic activation of H-Ras are common genetic lesions in cutaneous SCC (Azzoli et al., 1998; Yuspa, 1998), these IMK and TMK lines provide a useful model for assessing integrin-dependent gene expression in SCC progression. qPCR showed dramatically reduced fibulin-2 mRNA in IMKα3−/− cells compared with IMKα3+/+ cells (Fig. 2b), indicating similar α3β1-dependent regulation to that in LTAg-immortalized MK cells (Fig. 1). Other genes that were α3β1-dependent in both p53-null and LTAg-immortalized keratinocytes included MMP-9, Serpine2, Sulf2, and Mt4 (data not shown) (DiPersio et al., 2000; Lamar et al., 2008b). In contrast, SPARC and certain other genes that were α3β1-dependent in LTAg-immortalized cells were not α3β1-dependent in p53-null IMK cells (Fig. 2c, and data not shown), indicating an influence of the genetic lesion that initiates immortalization. RasV12-transformed TMK cells retained expression patterns for fibulin-2 and SPARC that were observed in the parental IMK cells (Fig. 2d, e).
Figure 2.
α3β1 regulates gene expression of fibulin-2, but not SPARC, in p53-null immortalized IMK cells and RasV12-transformed TMK cells. (a) Chart indicates the genetic lesion(s) that were used to effect the immortalization or transformation of different mouse keratinocyte-derived cell lines (see text for details). (b–e) Graphs show qPCR analysis of (b, d) fibulin-2 (Fbln-2) or (c, e) SPARC gene expression in (b, c) IMK cells that either express α3β1 (IMKα3+/+) or lack α3β1 (IMKα3−/−), or (d, e) derivatives of the latter cells that are transformed with RasV12 (TMKα3+/+ and TMKα3−/−). Relative qPCR signals are shown after normalization to those for β-actin mRNA. Data are shown as mean ± s.e.m. for three separate experiments; *P<0.05, unpaired t-test.
Suppression of fibulin-2 reduces invasion of RasV12-transformed keratinocytes
Fibulin-2 can be expressed as a monomer or a disulfide-linked homodimer. The monomer can range in size from 160 kDa to 195 kDa due to variations in mRNA splicing, post-translational modification, or proteolysis (Pan et al., 1993; Sasaki et al., 1997; Yi et al., 2007; Sicot et al., 2008). Immunoblots of non-reduced lysates from LTAg-immortalized MK cells with anti-fibulin-2 revealed a ~160 kDa band corresponding to the monomeric form, as well as a high molecular weight band corresponding to the ~375 kDa dimeric form (Fig. 3a, non-reducing). As expected, only the monomeric form was detected on a reducing blot (Fig. 3a, reducing). Consistent with our qPCR results, absence of α3β1 was associated with reduced expression of total fibulin-2 protein, defined as the sum of dimeric and monomeric forms (Fig. 3a, b). Similarly, total fibulin-2 protein was reduced in both p53-null immortalized IMK cells and RasV12-transformed TMK cells that lack α3β1 (Fig. 3c). Notably, IMK cells and TMK cells each showed considerable variation in the relative abundance of monomeric and dimeric fibulin-2 over several experiments. While the reason for this variation is unclear, it could be due to sensitivity of disulfide-linkage to subtle differences in culture conditions.
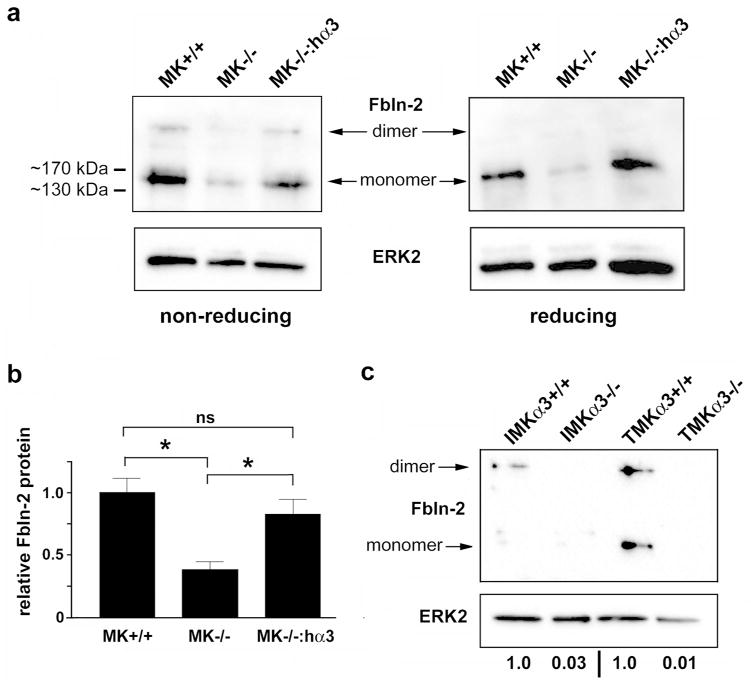
Figure 3.
Absence of α3β1 is associated with reduced fibulin-2 protein. (a) Total lysates from LTAg-immortalized MK+/+ cells, MK−/− cells, or MK−/−:hα3 cells were resolved on non-reducing or reducing gels and immunoblotted with anti-fibulin-2 (Fbln-2, top), or anti-ERK2 (bottom) as loading control. Arrows indicate positions of disulfide-linked homodimeric and monomeric fibulin-2. Molecular weight markers shown at left. (b) Graph shows quantification in MK cells of total fibulin-2 (dimeric plus monomeric), normalized to ERK-2. Data are mean ± s.e.m.; n=4; one-way ANOVA, *P<0.05; Tukey’s multiple comparison; ns, not significant. (c) Cell lysates (non-reduced) from p53-null immortalized MK cells (IMKα3+/+ or IMKα3−/−) or RasV12-transformed cells (TMKα3+/+ or TMKα3−/−) were immunoblotted as above; representative of three experiments. Relative signal intensity for total fibulin-2, normalized to ERK-2, is listed below lanes.
Mutations in p53 and Ras are associated with increased invasion (Campbell et al., 1993; Papathoma et al., 2001), and both IMKα3−/− cells and TMKα3−/− cells showed reduced invasion through Matrigel compared with their α3-expressing counterparts (Fig. S3) (Lamar et al., 2008b). Since fibulin-2 may promote invasion in some cancers (Baird et al., 2013; Senapati et al., 2012), we reasoned that α3β1-mediated invasion might require fibulin-2. We showed that treatment of α3β1-expressing IMK cells or TMK cells with either of two siRNAs to knockdown fibulin-2 (Fig. 4a, b) significantly reduced invasion compared to treatment with control siRNA (Fig. 4c, d). Thus, α3β1-dependent fibulin-2 expression enhances invasion of keratinocytes that harbor a loss-of-function p53 mutation and oncogenic RasV12.
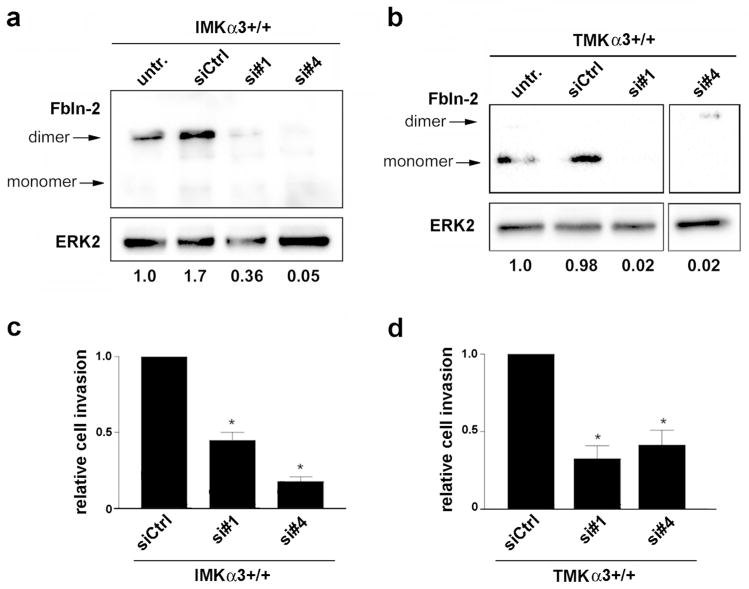
Figure 4.
Knockdown of fibulin-2 leads to reduced invasion of IMKα3+/+ and TMKα3+/+ cells. (a, b) Representative immunoblots show suppression of fibulin-2 (Fbln-2) protein in (a) IMKα3+/+ cells or (b) TMKα3+/+ cells using two distinct siRNAs that target fibulin-2 (si#1 and si#4), compared with control siRNA (siCtrl) or untreated cells (untr.); blot for ERK2 served as a loading control. Relative signal intensity for total fibulin-2 (dimeric plus monomeric), normalized to ERK-2, is listed under each lane. Fourth lane in panel (b) is from the same blot as first three lanes. (c, d) Graphs show relative invasion of (c) IMKα3+/+ cells or (d) TMKα3+/+ cells treated with fibulin-2-targeting siRNAs, normalized to cells treated with control siRNA; data are mean ± s.e.m; n=3; *P<0.05, two-way ANOVA.
Stable knockdown of fibulin-2 in tumorigenic TMKα3+/+ cells does not inhibit in vivo tumor growth
We previously showed that TMKα3−/− cells display reduced subcutaneous tumor growth compared to TMKα3+/+ cells (Lamar et al., 2008b), consistent with a recently described pro-tumorigenic role for α3β1 in SCC development (Sachs et al., 2012). To determine whether fibulin-2 facilitates α3β1-dependent tumor growth, we stably transduced TMKα3+/+ cells with lentiviral vectors expressing two independent fibulin-2 shRNAs, each of which efficiently suppressed fibulin-2 as determined by qPCR (data not shown) and immunoblot (Fig. 5a). Following subcutaneous injection into nude mice, TMKα3+/+ cells transduced with control shRNA showed significantly faster tumor growth than did TMKα3−/− cells transduced with the same shRNA (Fig. 5b), confirming α3β1-dependent tumor growth in this allograft model (Lamar et al., 2008b). However, stable transduction of TMKα3+/+ cells with either fibulin-2-targeting shRNA did not reduce tumor growth compared with control shRNA (Fig. 5b), indicating that suppression of fibulin-2 does not inhibit α3β1-mediated tumor growth of TMK cells. Although we observed that TMKα3+/+ cells transduced with one fibulin-2-targeting shRNA formed larger tumors than control TMKα3+/+ cells, we did not observe this effect with the other fibulin-2-targeting shRNA.
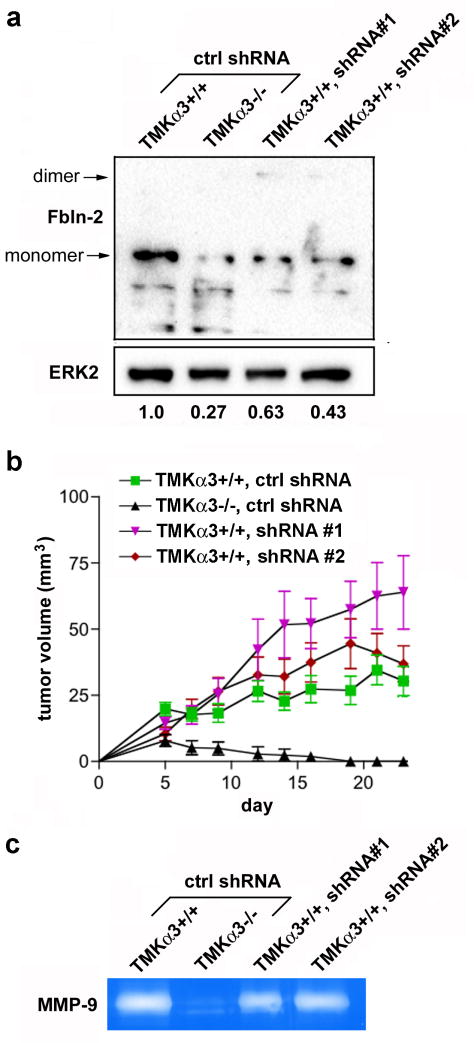
Figure 5.
Fibulin-2 suppression in RasV12-transformed TMK cells does not inhibit tumor growth. (a) Immunoblot shows fibulin-2 (Fbln-2) knockdown in TMKα3+/+ cells transduced with two shRNAs, compared with control shRNA (ctrl). Relative signal intensity for total fibulin-2, normalized to ERK-2, is listed under each lane. (b) TMKα3−/− or TMKα3+/+ cells transduced with indicated shRNA were injected subcutaneously into nude mice. Graph shows average tumor volume ± s.e.m. over 23 days for each variant. TMKα3+/+ with ctrl shRNA, n=12; others, n=10. Each TMKα3+/+ variant showed a significant increase in tumor size (averaged over last three time-points) compared with TMKα3−/− cells; *P<0.05; one-way ANOVA, Dunnett’s post-test. (c) Gelatin zymography of culture medium shows reduced MMP-9 secretion in TMKα3−/− cells compared with TMKα3+/+ cells, but no effect of fibulin-2-targeting shRNAs in the latter cells.
Since we previously showed that α3β1-dependent MMP-9 expression is important for TMKα3+/+ cell invasion (Lamar et al., 2008b), and some fibulins (including fibulin-2) have been associated with MMP expression (Creighton and Hanash, 2003; Obaya et al., 2012), we performed gelatin zymography to assess MMP-9 secretion in the TMK variants. TMKα3−/− cells secreted substantially less MMP-9 compared with TMKα3+/+ cells (Fig. 5c) (Lamar et al., 2008b). However, MMP-9 secretion was not altered by shRNA-mediated suppression of fibulin-2 (Fig. 5c), suggesting that MMP-9 and fibulin-2 each have necessary but independent roles in α3β1-mediated TMK cell invasion.
DISCUSSION
It is well established that integrins can regulate the ability of tumor cells to modulate the ECM of the tumor microenvironment through regulation of proteases or expression of ECM proteins (Missan and DiPersio, 2012; Yue et al., 2012). In particular, integrin α3β1 has long been known to regulate expression or activities of extracellular proteases involved in tumor progression or wound healing, including MMPs and the urokinase plasminogen activator (uPA) system (Ghosh et al., 2006; Iyer et al., 2005; Lamar et al., 2008b; Morini et al., 2000; Sugiura and Berditchevski, 1999). Here, we have identified a role for α3β1 in regulating keratinocyte genes that encode ECM proteins, extracellular proteases or their inhibitors, growth factors, or transcriptional regulators, many of which are known to contribute directly or indirectly to microenvironmental modifications of cutaneous tumors or wounds. Interestingly, some genes (e.g., fibulin-2 and MMP-9) showed similar α3β1-dependent expression in cells immortalized by either LTAg expression or p53 knockout, while other genes (e.g., SPARC) show dissimilar regulation in these two cases. Taken together, our findings suggest that α3β1-mediated regulation of some target genes is influenced by the genetic lesion that triggers immortalization.
Interestingly, most genes that showed α3β1-dependent regulation in immortalized keratinocytes were not altered in primary, non-immortalized keratinocytes by deletion of α3β1, suggesting that this regulation was acquired as part of the immortalized phenotype. The acquisition of α3β1-dependent regulation of fibulin-2 and other genes in immortalized MK cells resembles that which we reported previously for the MMP-9 gene (DiPersio et al., 2000), which we subsequently linked to invasive potential of oncogenically transformed MK cells (Lamar et al., 2008b). Our current findings suggest an important role for α3β1 in mediating adaptive changes in gene expression that facilitate carcinogenesis, where the exact profile of acquired gene changes may depend on the oncogenic or tumor suppressor mutations involved.
There is a wide range of potential mechanisms whereby integrins may regulate tumor cell behavior (Missan and DiPersio, 2012). Our results suggest that α3β1-mediated induction of fibulin-2 in transformed keratinocytes contributes to invasive behavior. Roles for fibulins in cancer are complex, and family members have been described to have either tumor suppressive or oncogenic roles, and in some cases both (Gallagher et al., 2005; Schiemann et al., 2002). For example, fibulin-1 has both tumor suppressive and tumor promoting functions, and fibulin-4 may be oncogenic (Gallagher et al., 2005; Schiemann et al., 2002). Fibulin-5 may also have cell type-specific effects on cell growth and angiogenesis (Albig and Schiemann, 2004; Schiemann et al., 2002). Although little is known about roles of fibulin-2 in cancer progression, it has been described both as a promoter of malignancy and a tumor suppressor (Obaya et al., 2012). Indeed, fibulin-2 has been associated with metastasis of adenocarcinoma (Ramaswamy et al., 2003) and malignant progression of lung adenocarcinoma (Baird et al., 2013). On the other hand, loss of fibulin-2 has been associated with breast cancer progression (Yi et al., 2007) and Kaposis sarcoma (Alcendor et al., 2011), and one study described anti-angiogenic and tumor suppressive roles for fibulin-2 in nasopharyngeal carcinoma (Law et al., 2012). Stromal cells also deposit fibulin-2 into the ECM, which may influence tumor progression. Importantly, our MK cell model allowed us to focus on the functional importance of fibulin-2 that is expressed by immortalized/transformed keratinocytes. Interestingly, suppression of fibulin-2 in transformed keratinocytes reduced invasion, but not in vivo tumor growth, suggesting that keratinocyte-derived fibulin-2 is dispensable at early stages of tumorigenesis but may become important at later stages of invasion.
In summary, the ability of α3β1 in immortalized/transformed keratinocytes to regulate expression of fibulin-2, as well as other genes associated with ECM remodeling, supports an important role for this integrin in SCC development and invasion. This regulation may be related to a role for α3β1 that we recently described in epidermis, where its ability to regulate fibulin-2 appears important for proper skin development and wound healing (Longmate et al., 2014). Given the similarities in tissue remodeling that occurs during skin carcinogenesis and wound healing (Arwert et al., 2012; Dvorak, 1986; Schäfer and Werner, 2008), it will be interesting to determine whether other α3β1-dependent genes in immortalized keratinocytes are similarly regulated in wound keratinocytes.
MATERIALS AND METHODS
Cell culture
LTAg-immortalized MK cell lines were derived from wild-type (MK+/+) or α3-null (MK−/−) neonatal mice harboring an SV40 large T antigen transgene, as described (DiPersio et al., 2000). MK−/−:hα3 cells were derived from MK−/− cells stably transfected with human α3 (Iyer et al., 2005). p53-deficient IMKα3+/+ and IMKα3−/− cells were similarly derived from wild-type or α3-null neonatal mice, respectively, harboring a p53-null mutation (Lamar et al., 2008b). To generate transformed TMK cells, IMKα3+/+ or IMKα3−/− cells were transduced with a retrovirus encoding oncogenic RasV12 (Lamar et al., 2008b). Epidermis-specific α3 knockout mice (α3eKO mice) are homozygous for a floxed α3 allele (α3flx/flx) and express a Cre recombinase transgene under control of the keratin 14 promoter (K14-Cre), as described (Mitchell et al., 2009). Primary keratinocytes were isolated from α3eKO mice (genotype K14-Cre:α3flx/flx) or control littermates lacking K14-Cre (genotype α3flx/flx) using established protocols (DiPersio et al., 2000). Culture conditions for primary keratinocytes and MK cell lines were described elsewhere (DiPersio et al., 2000; Lamar et al., 2008b).
RNA isolation, gene microarrays and qPCR
Total RNA was isolated using RNeasy Plus isolation kit (Qiagen, Valencia, CA), and quality was confirmed on an Agilent Bioanalyzer. Affymetrix whole genome arrays (Mouse Exon ST 1.0) include probe coverage over the full length of mouse coding sequences. Each of the 28,853 genes is represented on the array by replicate (27 each) probes spanning each gene. Microarray data acquisition and processing was performed using Affymetrix Command Console Operating Software, and the quality of array data assessed using metrics derived with Affymetrix Expression Console (Intensity Distribution, Mean Signal, BAC Spike, polyA Spike, Pos Vs Neg AUC, Mad Residual Signal, RLE Mean, and Hierarchical Clustering of Samples). Data was imported into GeneSpring v11 for advanced analysis. First, raw data was quantile-normalized using the PLIER algorithm, subjected to a Principal component analysis to again remove outliers and filtered on expression values to exclude probe sets in the bottom 20th percentile of expression across all conditions. Normalized data were subjected to ANOVA (P<0.05) incorporating a multiple correction factor (Benjamini-Hochberg) to remove false positives. Lists were compiled for genes expressed differentially in pair-wise comparisons of MK variants or primary cultures of different genotype. The complete gene list was submitted to Gene Expression Omnibus (series GSE42041).
α3β1-dependent genes identified by microarray were validated using custom qPCR arrays (SABiosciences, Valencia, CA), designed to include 87 genes that were differentially expressed on the microarrays, 2 house keeping controls (ACTB and GAPDH), and controls for evaluating mouse genomic DNA contamination, reverse transcription, and a positive control for qPCR. Total RNA (2 μg each from three independent experiments) was reverse transcribed using reagents kits from SABiosciences, and resulting cDNAs (20 ng) were used for qPCR. 384-well plates were run on an ABI 7900HT instrument using standard protocol and data analyzed using templates provided by SABiosciences. Fold changes were determined using the 2^−(delta Ct) method.
Individual qPCR for fibulin-2 and SPARC was performed using iQ SYBR green Supermix (Bio-rad, Hercules, CA). cDNA was generated using iScript Reverse Transcription Supermix (Bio-Rad). Conditions for fibulin-2: forward primer, 5′-TGTTGTTGGGGACACAGCTA-3′; reverse primer, 5′-CGTCTGTGCATTCACCATCT-3′; 95°C, 10 min, 1 cycle; followed by 94°C, 30 sec; 53°C, 30 sec; 72°C, 30 sec; 40 cycles. Conditions for SPARC: forward primer, 5′-TTCGACTCTTCCTGCCACTT-3′; reverse primer, 5′CCAGTGGACAGGGAAGATGT-3′; 95°C, 10 min, 1 cycle; followed by 94°C, 1 min; 53°C, 1 min; 72°C, 1 min; 40 cycles. Conditions for β-actin: forward primer, 5′-AGGGAAATCGTGCGTGACAT-3′; reverse primer, 5′-CATCTGCTGGAAGGTGGACA-3′; 95°C, 10 min, 1 cycle; followed by 94°C, 1 min; 58°C, 90 sec; 72°C, 90 sec; 35 cycles. Relative mRNA levels were calculated using the formula [2^−(Ct target gene − Ct β-actin gene)] × 100.
Immunoblots
Cells were lysed in non-reducing lysis buffer (Cell Signaling Technology, Beverly, MA) and protein concentrations determined by BCA Protein Assay (Pierce, Rockford, IL). Reducing lysis buffer contained 200 mM dithiothreitol (DTT). Equal protein was assayed by immunoblot with antibodies at the indicated dilutions: rabbit polyclonal anti-α3 integrin, 1:1000 (DiPersio et al., 1995); rabbit polyclonal anti-fibulin-2, 1:1000 (gift from Dr. Mon-Li Chu, Thomas Jefferson University); rabbit polyclonal anti-ERK2, 1:1000 (Santa Cruz, Santa Cruz, CA). Horseradish peroxidase-conjugated secondary antibody was goat anti-rabbit IgG (Cell Signaling, Danvers, MA), 1:2000. Chemiluminescence was performed using SuperSignal chemiluminescent substrate (ThermoScientific, Rockford, IL). Blots were quantitated using Image J software (NIH) or Biorad Imagelab software.
siRNA transfection
Cells were plated in full MK medium on collagen-coated 6-well dishes 24 hours prior to transfection. GenMute siRNA transfection reagent for primary keratinocytes (SignaGen Laboratories, Rockville, MD) was used to transfect cells with 80nM siRNA (Dharmacon Inc, Lafayette, CO). Two days later, cells were lysed for immunoblot or seeded for invasion assays. Of four fibulin-2-targeting siRNAs, we chose two that showed efficient suppression (siRNA #1-GCGAAGGCUACCAGUACUA, siRNA #4-CCAAUAGCCUGCCGGGAGA). siRNA against luciferase was used as control.
Matrigel invasion assays
Cells (8 × 104) were seeded onto growth factor-reduced Matrigel invasion chambers (8 μm pore; BD Biosciences, San Jose, CA) in MK medium without interferon-γ. Cells were allowed to invade for 16 h, then fixed in 3.7% formaldehyde. Cells that invaded through the filter were permeabilized in 0.05% triton X-100, stained with 4′,6-diamidino-2-phenylindole (DAPI; 1 μg/ml), and quantified manually by imaging four fields/well at 20X magnification on a Nikon eclipse TE2000-U inverted microscope. Data are from 4 experiments, each performed in duplicate.
Lentiviral transduction with shRNA
pGIPZ lentiviral vectors containing fibulin-2-specific shRNA sequences or non-targeting control shRNA were purchased from Thermo Scientific Open Biosystems (Lafayette, CO). 293FT packaging cells were co-transfected with packaging plasmids pCMV-dR8.2 and pCMV-VSV-G (Addgene plasmids 8455 and 8454). TMK cells were infected with viral particles plus antennapedia peptide (Anaspec, Fremont, CA), and stable transductants were selected in 10 μg/ml puromycin.
Gelatin zymography
Cells (4 × 105) were plated onto collagen coated 6-well dishes in full MK medium overnight, then washed and cultured in 2 ml serum-free medium without antibiotics for 24 hours. Culture medium was collected and incubated with gelatin-agarose beads (Sigma) overnight, then assessed by gelatin zymography, as described (DiPersio et al., 2000).
Subcutaneous injections
TMK cells (5 × 106) were suspended in 200 μl complete MK medium and injected into right flanks of 10–12 female NCR nude mice per test group (Taconic, Hudson, NY). Tumor length (l) and width (w) were measured over 23 days using a Vernier caliper (Bel-Art Scienceware). Tumor volume was estimated using the formula (w2 × l)/2. Statistical analysis was performed by first averaging tumor size from the last three time points for each TMK variant, then using one-way ANOVA with Dunnett’s post-test to compare each TMKα3+/+ variant (i.e., transduced with control or fibulin-2-targeting shRNA) to the TMKα3−/− control. Animal studies were approved by the Institutional Animal Care and Use Committee of Albany Medical College.
Supplementary Material
Acknowledgments
We thank Dr. Mon-Li Chu (Thomas Jefferson University, PA) for graciously providing the antibody for fibulin-2. We also thank Drs. My Mahoney (Thomas Jefferson University, PA), Susan LaFlamme, Livingston Van De Water, Whitney Longmate and Sita Subbaram for critical reading of the manuscript, Dr. Peter Vincent for assistance with statistics, and Sean Hammond for excellent technical assistance. This research was supported by NIH grants to C.M.D. from the National Cancer Institute (R01CA129637) and the National Institute of Arthritis and Musculoskeletal and Skin Diseases (R21AR056390).
Abbreviations
- ECM
extracellular matrix
- Fbln-2
fibulin-2
- MRP
mitogen-regulated protein
- MMP
matrix metalloproteinase
- LN-332
laminin-332
- EMT
epithelial-to-mesenchymal transition
- MK
mouse keratinocyte
- SPARC
secreted protein acidic and rich in cysteine
- SCC
squamous cell carcinoma
- qPCR
quantitative real time polymerase chain reaction
- LTAg
SV40 large T antigen
- TGF-β
transforming growth factor β
- DAPI
4′,6-diamidino-2-phenylindole
- uPA
urokinase plasminogen activator
Footnotes
CONFLICT OF INTEREST
The authors declare no conflicts of interest.
References
- Abramoff MD, Folk JC, Han DP, et al. Automated analysis of retinal images for detection of referable diabetic retinopathy. JAMA ophthalmology. 2013;131:351–7. doi: 10.1001/jamaophthalmol.2013.1743. [DOI] [PubMed] [Google Scholar]
- Albig AR, Schiemann WP. Fibulin-5 antagonizes vascular endothelial growth factor (VEGF) signaling and angiogenic sprouting by endothelial cells. DNA Cell Biol. 2004;23:367–79. doi: 10.1089/104454904323145254. [DOI] [PubMed] [Google Scholar]
- Alcendor DJ, Knobel S, Desai P, et al. KSHV regulation of fibulin-2 in Kaposi’s sarcoma: implications for tumorigenesis. Am J Pathol. 2011;179:1443–54. doi: 10.1016/j.ajpath.2011.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold SA, Brekken RA. SPARC: a matricellular regulator of tumorigenesis. Journal of cell communication and signaling. 2009;3:255–73. doi: 10.1007/s12079-009-0072-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arwert EN, Hoste E, Watt FM. Epithelial stem cells, wound healing and cancer. Nat Rev Cancer. 2012;12:170–80. doi: 10.1038/nrc3217. [DOI] [PubMed] [Google Scholar]
- Azzoli CG, Sagar M, Wu A, et al. Cooperation of p53 loss of function and v-Ha-ras in transformation of mouse keratinocyte cell lines. Mol Carcinog. 1998;21:50–61. doi: 10.1002/(sici)1098-2744(199801)21:1<50::aid-mc7>3.0.co;2-t. [DOI] [PubMed] [Google Scholar]
- Baird BN, Schliekelman MJ, Ahn YH, et al. Fibulin-2 is a driver of malignant progression in lung adenocarcinoma. PLoS One. 2013;8:e67054. doi: 10.1371/journal.pone.0067054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briggs J, Chamboredon S, Castellazzi M, et al. Transcriptional upregulation of SPARC, in response to c-Jun overexpression, contributes to increased motility and invasion of MCF7 breast cancer cells. Oncogene. 2002;21:7077–91. doi: 10.1038/sj.onc.1205857. [DOI] [PubMed] [Google Scholar]
- Campbell C, Quinn AG, Ro YS, et al. p53 mutations are common and early events that precede tumor invasion in squamous cell neoplasia of the skin. J Invest Dermatol. 1993;100:746–8. doi: 10.1111/1523-1747.ep12475717. [DOI] [PubMed] [Google Scholar]
- Carter WG, Ryan MC, Gahr PJ. Epiligrin, a new cell adhesion ligand for integrin α3β1 in epithelial basement membranes. Cell. 1991;65:599–610. doi: 10.1016/0092-8674(91)90092-d. [DOI] [PubMed] [Google Scholar]
- Che Y, Luo A, Wang H, et al. The differential expression of SPARC in esophageal squamous cell carcinoma. International journal of molecular medicine. 2006;17:1027–33. [PubMed] [Google Scholar]
- Choma DP, Milano V, Pumiglia KM, et al. Integrin α3β1-dependent activation of FAK/Src regulates Rac1-mediated keratinocyte polarization on laminin-5. J Invest Dermatol. 2007;127:31–40. doi: 10.1038/sj.jid.5700505. [DOI] [PubMed] [Google Scholar]
- Choma DP, Pumiglia K, DiPersio CM. Integrin α3β1 directs the stabilization of a polarized lamellipodium in epithelial cells through activation of Rac1. J Cell Sci. 2004;117:3947–59. doi: 10.1242/jcs.01251. [DOI] [PubMed] [Google Scholar]
- Chou YT, Wang H, Chen Y, et al. Cited2 modulates TGF-beta-mediated upregulation of MMP9. Oncogene. 2006;25:5547–60. doi: 10.1038/sj.onc.1209552. [DOI] [PubMed] [Google Scholar]
- Conti FJ, Rudling RJ, Robson A, et al. alpha3beta1-integrin regulates hair follicle but not interfollicular morphogenesis in adult epidermis. J Cell Sci. 2003;116:2737–47. doi: 10.1242/jcs.00475. [DOI] [PubMed] [Google Scholar]
- Creighton C, Hanash S. Expression of matrix metalloproteinase 9 (MMP-9/gelatinase B) in adenocarcinomas strongly correlated with expression of immune response genes. In Silico Biol. 2003;3:301–11. [PubMed] [Google Scholar]
- deHart GW, Healy KE, Jones JC. The role of alpha3beta1 integrin in determining the supramolecular organization of laminin-5 in the extracellular matrix of keratinocytes. Exp Cell Res. 2003;283:67–79. doi: 10.1016/s0014-4827(02)00028-9. [DOI] [PubMed] [Google Scholar]
- Delwel GO, de Melker AA, Hogervorst F, et al. Distinct and overlapping ligand specificities of the α3Aβ1 and α6Aβ1 integrins: recognition of laminin isoforms. Mol Biol Cell. 1994;5:203–15. doi: 10.1091/mbc.5.2.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiPersio CM, Hodivala-Dilke KM, Jaenisch R, et al. α3β1 integrin is required for normal development of the epidermal basement membrane. J Cell Biol. 1997;137:729–42. doi: 10.1083/jcb.137.3.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiPersio CM, Shah S, Hynes RO. α3Aβ1 integrin localizes to focal contacts in response to diverse extracellular matrix proteins. J Cell Sci. 1995;108:2321–36. doi: 10.1242/jcs.108.6.2321. [DOI] [PubMed] [Google Scholar]
- DiPersio CM, Shao M, Di Costanzo L, et al. Mouse keratinocytes immortalized with large T antigen acquire α3β1 integrin-dependent secretion of MMP-9/gelatinase B. J Cell Sci. 2000;113:2909–21. doi: 10.1242/jcs.113.16.2909. [DOI] [PubMed] [Google Scholar]
- Dvorak HF. Tumors: wounds that do not heal. Similarities between tumor stroma generation and wound healing. N Engl J Med. 1986;315:1650–9. doi: 10.1056/NEJM198612253152606. [DOI] [PubMed] [Google Scholar]
- Gallagher WM, Currid CA, Whelan LC. Fibulins and cancer: friend or foe? Trends Mol Med. 2005;11:336–40. doi: 10.1016/j.molmed.2005.06.001. [DOI] [PubMed] [Google Scholar]
- Gerson KD, Shearstone JR, Maddula VS, et al. Integrin beta4 regulates SPARC protein to promote invasion. J Biol Chem. 2012;287:9835–44. doi: 10.1074/jbc.M111.317727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh S, Johnson JJ, Sen R, et al. Functional relevance of urinary-type plasminogen activator receptor-alpha3beta1 integrin association in proteinase regulatory pathways. J Biol Chem. 2006;281:13021–9. doi: 10.1074/jbc.M508526200. [DOI] [PubMed] [Google Scholar]
- Grose R, Hutter C, Bloch W, et al. A crucial role of beta 1 integrins for keratinocyte migration in vitro and during cutaneous wound repair. Development. 2002;129:2303–15. doi: 10.1242/dev.129.9.2303. [DOI] [PubMed] [Google Scholar]
- Hynes RO. Integrins: bidirectional, allosteric signaling machines. Cell. 2002;110:673–87. doi: 10.1016/s0092-8674(02)00971-6. [DOI] [PubMed] [Google Scholar]
- Iyer V, Pumiglia K, DiPersio CM. α3β1 integrin regulates MMP-9 mRNA stability in immortalized keratinocytes: a novel mechanism of integrin-mediated MMP gene expression. J Cell Sci. 2005;118:1185–95. doi: 10.1242/jcs.01708. [DOI] [PubMed] [Google Scholar]
- Janes SM, Watt FM. New roles for integrins in squamous-cell carcinoma. Nat Rev Cancer. 2006;6:175–83. doi: 10.1038/nrc1817. [DOI] [PubMed] [Google Scholar]
- Koblinski JE, Kaplan-Singer BR, VanOsdol SJ, et al. Endogenous osteonectin/SPARC/BM-40 expression inhibits MDA-MB-231 breast cancer cell metastasis. Cancer Res. 2005;65:7370–7. doi: 10.1158/0008-5472.CAN-05-0807. [DOI] [PubMed] [Google Scholar]
- Kreidberg JA. Functions of α3β1 integrin. Curr Opin Cell Biol. 2000;12:548–53. doi: 10.1016/s0955-0674(00)00130-7. [DOI] [PubMed] [Google Scholar]
- Lamar JM, Iyer V, DiPersio CM. Integrin α3β1 potentiates TGFβ-mediated induction of MMP-9 in immortalized keratinocytes. J Invest Dermatol. 2008a;128:575–86. doi: 10.1038/sj.jid.5701042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamar JM, Pumiglia KM, DiPersio CM. An immortalization-dependent switch in integrin function up-regulates MMP-9 to enhance tumor cell invasion. Cancer Res. 2008b;68:7371–9. doi: 10.1158/0008-5472.CAN-08-1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law EW, Cheung AK, Kashuba VI, et al. Anti-angiogenic and tumor-suppressive roles of candidate tumor-suppressor gene, Fibulin-2, in nasopharyngeal carcinoma. Oncogene. 2012;31:728–38. doi: 10.1038/onc.2011.272. [DOI] [PubMed] [Google Scholar]
- Ledda MF, Adris S, Bravo AI, et al. Suppression of SPARC expression by antisense RNA abrogates the tumorigenicity of human melanoma cells. Nat Med. 1997;3:171–6. doi: 10.1038/nm0297-171. [DOI] [PubMed] [Google Scholar]
- Longmate W, Monichan R, Chu ML, et al. Reduced fibulin-2 contributes to loss of basement membrane integrity and skin blistering in mice lacking integrin α3β1 in the epidermis. J Invest Dermatol. 2014 doi: 10.1038/jid.2014.10. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margadant C, Charafeddine RA, Sonnenberg A. Unique and redundant functions of integrins in the epidermis. FASEB J. 2010;24:4133–52. doi: 10.1096/fj.09-151449. [DOI] [PubMed] [Google Scholar]
- Margadant C, Raymond K, Kreft M, et al. Integrin α3β1 inhibits directional migration and wound re-epithelialization in the skin. J Cell Sci. 2009;122:278–88. doi: 10.1242/jcs.029108. [DOI] [PubMed] [Google Scholar]
- Missan DS, DiPersio M. Integrin control of tumor invasion. Crit Rev Eukaryot Gene Expr. 2012;22:309–24. doi: 10.1615/critreveukargeneexpr.v22.i4.50. [DOI] [PubMed] [Google Scholar]
- Mitchell K, Szekeres C, Milano V, et al. α3β1 integrin in epidermis promotes wound angiogenesis and keratinocyte-to-endothelial-cell crosstalk through the induction of MRP3. J Cell Sci. 2009;122:1778–87. doi: 10.1242/jcs.040956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morini M, Mottolese M, Ferrari N, et al. The α3β1 integrin is associated with mammary carcinoma cell metastasis, invasion, and gelatinase B (MMP-9) activity. Int J Cancer. 2000;87:336–42. [PubMed] [Google Scholar]
- Obaya AJ, Rua S, Moncada-Pazos A, et al. The dual role of fibulins in tumorigenesis. Cancer Lett. 2012;325:132–8. doi: 10.1016/j.canlet.2012.06.019. [DOI] [PubMed] [Google Scholar]
- Pan T-C, Sasaki T, Zhang R-Z, et al. Structure and expression of fibulin-2, a novel extracellular matrix protein with multiple EGF-like repeats and consensus motifs for calcium binding. J Cell Biol. 1993;123:1269–77. doi: 10.1083/jcb.123.5.1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papathoma AS, Zoumpourlis V, Balmain A, et al. Role of matrix metalloproteinase-9 in progression of mouse skin carcinogenesis. Mol Carcinog. 2001;31:74–82. doi: 10.1002/mc.1042. [DOI] [PubMed] [Google Scholar]
- Porte H, Triboulet JP, Kotelevets L, et al. Overexpression of stromelysin-3, BM-40/SPARC, and MET genes in human esophageal carcinoma: implications for prognosis. Clinical Cancer Research. 1998;4:1375–82. [PubMed] [Google Scholar]
- Ramaswamy S, Ross KN, Lander ES, et al. A molecular signature of metastasis in primary solid tumors. Nat Genet. 2003;33:49–54. doi: 10.1038/ng1060. [DOI] [PubMed] [Google Scholar]
- Roupe KM, Alberius P, Schmidtchen A, et al. Gene expression demonstrates increased resilience toward harmful inflammatory stimuli in the proliferating epidermis of human skin wounds. Exp Dermatol. 2010;19:e329–32. doi: 10.1111/j.1600-0625.2009.01038.x. [DOI] [PubMed] [Google Scholar]
- Sachs N, Secades P, van Hulst L, et al. Loss of integrin alpha3 prevents skin tumor formation by promoting epidermal turnover and depletion of slow-cycling cells. Proc Natl Acad Sci U S A. 2012;109:21468–73. doi: 10.1073/pnas.1204614110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki T, Mann K, Wiedemann H, et al. Dimer model for the microfibrillar protein fibulin-2 and identification of the connecting disulfide bridge. EMBO J. 1997;16:3035–43. doi: 10.1093/emboj/16.11.3035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schäfer M, Werner S. Cancer as an overhealing wound:an old hypothesis revisited. Nat Rev Cell Biol. 2008;9:628–38. doi: 10.1038/nrm2455. [DOI] [PubMed] [Google Scholar]
- Schiemann WP, Blobe GC, Kalume DE, et al. Context-specific effects of fibulin-5 (DANCE/EVEC) on cell proliferation, motility, and invasion. Fibulin-5 is induced by transforming growth factor-beta and affects protein kinase cascades. J Biol Chem. 2002;277:27367–77. doi: 10.1074/jbc.M200148200. [DOI] [PubMed] [Google Scholar]
- Senapati S, Gnanapragassam VS, Moniaux N, et al. Role of MUC4-NIDO domain in the MUC4-mediated metastasis of pancreatic cancer cells. Oncogene. 2012;31:3346–56. doi: 10.1038/onc.2011.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sicot F-X, Tsuda T, Markova D, et al. Fibulin-2 is dispensable for mouse development and elastic fiber formation. Mol Cell Biol. 2008;28:1061–7. doi: 10.1128/MCB.01876-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiura T, Berditchevski F. Function of α3β1-tetraspan protein complexes in tumor cell invasion. Evidence for the role of the complexes in production of matrix metalloproteinase 2 (MMP-2) J Cell Biol. 1999;146:1375–89. doi: 10.1083/jcb.146.6.1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timpl R, Sasaki T, Kostka G, et al. Fibulins: a versatile family of extracellular matrix proteins. Nat Rev Mol Cell Biol. 2003;4:479–89. doi: 10.1038/nrm1130. [DOI] [PubMed] [Google Scholar]
- Utani A, Nomizu M, Yamada Y. Fibulin-2 Binds to the Short Arms of Laminin-5 and Laminin-1 via Conserved Amino Acid Sequences. J Biol Chem. 1997;272:2814–20. doi: 10.1074/jbc.272.5.2814. [DOI] [PubMed] [Google Scholar]
- Watt FM. Role of integrins in regulating epidermal adhesion, growth and differentiation. EMBO J. 2002;21:3919–26. doi: 10.1093/emboj/cdf399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi CH, Smith DJ, West WW, et al. Loss of fibulin-2 expression is associated with breast cancer progression. Am J Pathol. 2007;170:1535–45. doi: 10.2353/ajpath.2007.060478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue J, Zhang K, Chen J. Role of integrins in regulating proteases to mediate extracellular matrix remodeling. Cancer Microenviron. 2012;5:275–83. doi: 10.1007/s12307-012-0101-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuspa SH. The pathogenesis of squamous cell cancer: lessons learned from studies of skin carcinogenesis. J Dermatol Sci. 1998;17:1–7. doi: 10.1016/s0923-1811(97)00071-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.