Abstract
Nickel is widely applied in industrial settings and Ni (II) compounds have been classified as group one human carcinogens. The molecular basis of Ni (II) carcinogenicity has proved complex, for many stress response pathways are activated and yield unexpected Ni (II) specific toxicology profile. Ni (II) induced toxicogenomic change has been associated with altered activity of HIF, p53, c-MYC, NFκB and iron and 2-oxoglutarate dependent dioxygenases. Advancing high-throughput technology has indicated the toxicogenome of Ni (II) involves crosstalk between HIF, p53, c-MYC, NFκB and dioxygenases. This paper is intended to review the network of Ni (II) induced common transcription-factor-governed pathways by discussing transcriptome alteration, its governing transcription factors and the underlying mechanism. Finally, we propose a putative target network of Ni (II) as a human carcinogen.
Keywords: nickel, gene expression profile, microarray, next generation sequencing, p53, HIF, dioxygenase
Introduction
Nickel (Ni) is widely employed in industrial applications due to its corrosion resistance, physical strength, and special magnetic electronic properties. Ni (II) compounds have been classified as group one human carcinogens and metallic nickel has been listed as group 2b possible human carcinogens by the International Agency for Research on Cancer (IARC) (IARC 1990). The molecular basis of Ni (II) carcinogenicity has proved complex, for many stress response pathways are activated and yet yield unexpected Ni (II) specific toxicology profile.
Gene expression profiling has revealed a comprehensive image of differentially expressed genes (DEG) in cells after various stresses including Ni (II). The picture has become daunting with the advancing technology of microarray and next generation sequencing, turning the candidate gene pool from several thousands of genes to more than 30 thousands genes (Affimetrix). It is a useful strategy to focus on identification of modules (transcription factors, chromatin modifiers) that may regulate a set of genes involved in a biological process, and to work down the regulatory hierarchy to identify specific effectors. Toxins usually induce similar toxicogenomic change through common transcription-factor-governed pathways (Jennings et al. 2013). This paper is intended to review the crosstalk of Ni (II) induced common transcription-factor-governed pathways by discussing transcriptome alteration, its governing transcription factors and the underlying mechanism. Finally, we propose a putative target network of Ni (II) as a human carcinogen.
HIF governed DEG in Ni (II) exposed cells
An early study utilizing Affymetrix platform mouse U74 Genechip array to profile DEG in mouse embryonic fibroblast exposed to Ni (II) has found increased expression of genes involved in glucose metabolism including glycolytic enzymes and genes responsible for glucose transport (Salnikow et al. 2003). A comparison of DEG induced by Ni (II) and hypoxia revealed a significant role of hypoxia inducible factors (HIF) in altered transcription profile in both cases. Indeed, HIF has been found accumulated in various cell lines exposed to Ni (II) (Chen and Costa 2009; Yao et al. 2014). Glycolytic enzymes are among the genes that are inducible by HIF. A careful examination of DEG in Ni (II) exposed cells revealed that hexokinase I and glucose 6-phosphate dehydrogenase were two glycolytic enzymes that were not induced by nickel, and neither were they induced by hypoxia (Salnikow et al. 2003). In consistence with this, Ni (II) failed to induce glucose metabolism genes expression in HIF-1α deficient mouse fibroblast (Salnikow et al. 2002; Salnikow et al. 2003); indicating Ni (II) exerts its effect through up regulating HIF protein levels and their activities.
Ni (II) reportedly down regulates xenobiotic-response element containing genes, including CYP1B1, NQO1, UDP-glucuronyltransferase1A6, glutathione S-transferases, and aldehyde dehydrogenases (ALDH)(Davidson et al. 2003). These genes are regulated by aryl hydrocarbon receptor (AHR), a transcription factor that shares the same heterodimerization partner, ARNT/HIF-1β, with hypoxia inducible factors. It is conceivable that these two pathways might interact via competing ARNT/HIF-1β. However, evidence indicates this might not be true. Although the AHR protein amount is 4 to 10 fold higher than ARNT/HIF-1β in vivo and in vitro, the vast majority of liganded AHR is rapidly degraded, and only about 15% of the ARNT/HIF-1β pool is used when the AHR-signaling pathway is saturated (Pollenz 1996). Similarly, only about 12 to 15% of the ARNT/HIF-1β pool is sequestered in cells cultured under 1% oxygen tension for up to 16 hours (Pollenz et al. 1999). On the other hand, Ni (II) was capable of reducing AHR regulated genes’ expression to similar extent in the absence of HIF-1α (HIF-1α deficient mouse fibroblast); indicating HIF activity is dispensable to the suppression of AHR regulated genes induced by Ni (II) (Davidson et al. 2003).
Iron homeostasis and dioxygenase activity
The association of Ni (II) and HIF has been linked to iron homeostasis by further studies (Davidson et al. 2005). Iron homeostasis has to be meticulously balanced in cells to provide sufficient iron to achieve cellular functions, and to eliminate excessive accumulation of iron to avoid toxicity. Cells sequester extraneous iron by storing it in ferritin molecules (Nadadur et al. 2008). Ni (II) was found to block transferrin-dependent and -independent iron uptake, suggesting Ni (II) exerts its effects via perturbing iron homeostasis (Davidson et al. 2005). The prolyl hydroxylase PHD family hydroxylates the oxygen dependent disruption (ODD) domain of HIF. The hydroxylated proline residues in the ODD domain of HIF proteins can be recognized by E3 ligase Von Hippel-Lindau protein (VHL) and targeted for degradation. The PHD family is one of numerous members of iron and 2-oxoglutarate dependent dioxygenase family that also require oxygen and ascorbic acid for enzymatic activity. When the prolyl hydroxylases are not functional, no hydroxylation of the proline residues in the ODD domain occurs and VHL will not bind to it. This phenomenon was found inducible by Ni (II) in A549 cells (Davidson et al. 2005); indicating Ni (II) stabilizes HIF proteins via perturbing hydroxylation at ODD domain.
Iron and 2-oxoglutarate dependent dioxygenase is also a major component of histone demethylases. These jumonji C (JmjC)-domain containing enzymes can catalyze the oxidization of methyl groups on histones, and the resultant hydroxymethyl group is spontaneously lost in the form of formaldehyde, to remove one methyl group from the modified lysine(Labbe et al. 2013).
Ni (II) has been found to inhibit demethylase activity of KDM3A (JMJD1A) in vitro, and in turn led to an accumulation of dimethylation at histone H3 lysine 9 (H3K9me2) (Chen et al. 2010a). The same study also showed that the activity of DNA repair enzyme ABH2 was inhibited by Ni (II). Extended X-ray absorption fine structure showed that the best fit for iron binding ABH2 consists of five oxygen/nitrogen donor ligands of which two are histidine ligands; and when nickel binds to ABH2, the binding site is indistinguishable from the iron-binding site (Chen et al. 2010a). Isothermal titration calorimetry analysis suggested that Ni (II) binds to ABH2 with higher affinity than Fe (II). It is likely that dioxygenases such as PHD family, JmjC-domain containing histone demethylases and ABH DNA repair enzymes use the same iron coordinating motif (His-Asp-His) to bind ferrous iron at their active sites, and Ni(II) competes with Fe(II) and replaces it at the iron-binding site (Chen et al. 2010a; Gorres et al. 2009).
Histone mark governed DEG in Ni (II) exposed cells
The inhibition of JmjC-domain containing histone demethylases activity by Ni (II) was further evidenced by accumulation of other methylation modifications on histones in Ni (II) exposed cells, both in vivo and in vitro. H3K9me2 (substrate of JmjC-domain containing KDM3A, KDM4A, etc.), a mark of transcriptional repression; and H3K4me3 (substrate of JmjC-domain containing KDM2B, KDM5A, etc.), a mark of transcriptional activation, were found increased in gene promoter regions in cells exposed to Ni (II) (Chen et al. 2006; Chen et al. 2010b; Tchou-Wong et al. 2011). Human peripheral blood mononuclear cells (PBMC) from healthy subjects treated with Ni (II) ex vivo showed an increase in global levels of H3K4me3 and H3K9me2 (Arita et al. 2012b). On the other hand, in vivo study of PBMC of subjects with occupational exposure to high levels of nickel at a nickel refinery in China has found elevated global level of H3K4me3 (p = 0.0004) when compared to referent subjects(Arita et al. 2012a). In an independent in vivo study of workers in a steel plant, H3K4me2 (substrate of JmjC-domain containing KDM5A, KDM5B, etc.) were found increased in association with the years of steel plant employment of the study subjects. The increased H3K4me2 level was found associated with nickel exposure but not aluminum, manganese, zinc, lead exposure (Cantone et al. 2011). It is noticeable that H3K4me2 is also a substrate of a second group of histone demethylase, amine oxidase domain-containing flavin dependent enzymes KDM1A and KDM1B (Wojcieszynska et al. 2012). Whether Ni (II) exerts its effects through this group of enzyme needs to be investigated.
A high throughput screening of KDM3A (JMJD1A) targeted genes in human bronchial epithelial BEAS-2B cells exposed to Ni (II) using ChIP-on-Chip Affymetrix GeneChip® Human Promoter 1.0R Array revealed 620 potential genes that are in close association with KDM3A (supplemental table 1 in Chen et al. 2010b), 67 of which were repressed more than two-fold when siRNA against KDM3A was used to knockdown the histone demethylase expression in BEAS-2B cells (supplemental table 2 in Chen et al. 2010b), indicating these 67 genes are most likely to be affected by Ni (II) through inhibiting KDM3A activity. We cross-referenced the list of genes that were down regulated in nickel refinery workers in vivo when compared to referent subjects (supplemental table 1 in Arita et al. 2012a), and found 10 genes were repressed in both cases (Table 1). Given these two independent studies were conducted in different systems (in vitro and in vivo) and different cell types (bronchial epithelial cells and PBMC), we argue that this observation suggests in vitro research of Ni (II) provides valuable information to help understand the consequences of human exposure to Ni (II) and their underlying mechanism.
Table 1.
KDM3A target genes that were repressed in nickel refinery workers.
| Expression |
|||||
|---|---|---|---|---|---|
| Gene Symbol |
Control RNAi |
KDM3A RNAi |
Referents | Nickel refinery workers |
Description |
| IL6R | 1 | 0.37 | 1 | 0.61 | Interleukin 6 (IL6) receptor |
| TAF1 | 1 | 0.2 | 1 | 0.81 | TATA box binding protein (TBP)- associated factor |
| ITGB2 | 1 | 0.28 | 1 | 0.77 | Integrin, beta 2 |
| FAM48A | 1 | 0.28 | 1 | 0.68 | Family with sequence similarity 48, member A |
| AMACR | 1 | 0.33 | 1 | 0.83 | Alpha-methylacyl-CoA racemase |
| EXOSC2 | 1 | 0.34 | 1 | 0.78 | Exosome component 2 |
| EDEM3 | 1 | 0.41 | 1 | 0.65 | ER degradation enhancer, mannosidase alpha-like 3 |
| SMC2 | 1 | 0.45 | 1 | 0.67 | Structural maintenance of chromosomes 2 |
| MTAP | 1 | 0.47 | 1 | 0.84 | Methylthioadenosine phosphorylase |
| C10orf57 | 1 | 0.49 | 1 | 0.66 | Also known as TMEM254, transmembrane protein 254 |
Ni (II) exposure has been shown to induce accumulation of histone marks of both repressive and active transcription. While decreased gene transcription levels probably result from accumulation of H3K9me2 at gene promoter regions, the increased transcription levels might be caused by an increase of H3K4me3 at gene transcription starting sites (TSS)(Barski et al. 2007). Despite the dramatic increase of global H3K4me3 in Ni (II) exposed human lung adenocarcinoma A549 cell line (detected by Western Blot) (Zhou et al. 2009), the global H3K4me3 profile at TSS wasn’t affected by Ni (II) in A549 cells (detected by ChIP-Seq) (Tchou-Wong et al. 2011). It was rather the post-TSS peak of nickel-treated cells remained higher than that of control cells over a broader region spanning over 4,000 bp downstream of TSS of the genes that were up-regulated in nickel-treated cells (Tchou-Wong et al. 2011).
P53 and MYC governed DEG in Ni (II) exposed cells attenuated by HIF
Increased gene expression of p53 (Salnikow et al. 2002), and its downstream CDK inhibitor p21 (CDKN1A) (Arita et al. 2012a; Green et al. 2013; Salnikow et al. 2002; Wong et al. 2013), GADD45 (Arita et al. 2012a; Arita et al. 2012b; Salnikow et al. 2002; Wong et al. 2013) has been reported in Ni (II) exposed cells, which includes in vitro studies on human lung cell lines BEAS-2B, H460, and in vivo study on PBMC from nickel exposed human subjects, indicating the activation of p53 is a highly reproducible phenomenon from nickel exposure. However, p53 transcription factor binding site was not over-represented in the genes that had elevated expression in nickel refinery workers (Yao et al. unpublished data), this is consistent with an early report that nickel (both soluble and insoluble form) did not induce p53 driven reporter gene (Huang et al. 2001). Moreover, no change has been found in expression of BCL2, BCL-XL, and MCL1 (transactivated by p53) in Ni (II) exposed cells (Arita et al. 2012a; Green et al. 2013).
The activity of p53 is regulated at multiple layers, including the phosphorylation at Ser15 and protein levels, which have been found up-regulated and coordinated with an increase of p21 expression in cells exposed to Ni (II) (Ding et al. 2009; Green et al. 2013; Wong et al. 2013). The induction of p21 has been found to be p53-dependent and HIF-independent in Ni (II) exposed cells (Salnikow et al. 2002; Wong et al. 2013). On the other hand, the induction of GADD45 has been found to be independent of p53 and HIF (Salnikow et al. 2002; Wong et al. 2013).
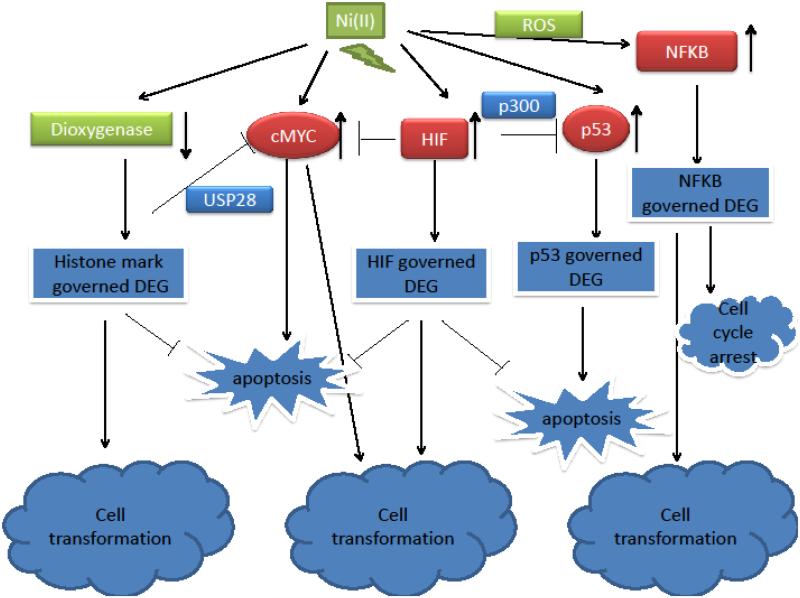
Interestingly, p53 is activated under hypoxia but its activity is usually attenuated at the same time (Achison and Hupp 2003; Crowder et al. 2013; Koumenis et al. 2001). Hypoxia environment induced p53 accumulation and caused an enhanced interaction of p53 with mSin3A transcription co-repressor complex but not p300 transcription activator complex (Koumenis et al. 2001). It was later proven that HIF and p53 indeed compete for p300 for their transactivation activity, ectopic expression of p300 could relieve the competition (Schmid et al. 2004). We propose that the seemingly contradictory phenomena of p53 activation (accumulated p53 protein level versus transactivation of limited p53 binding genes) observed in Ni (II) exposed cells was rather due to limited amount of p300 occupied by dramatically elevated HIF protein, which led to limited transactivation of p53 binding genes (Figure 1).
Figure 1.
Putative target network of nickel carcinogenicity.
Similar activation-attenuation phenomena have been observed in c-MYC pathway in Ni (II) exposed cells, but through different mechanism. Ni (II) induced c-MYC protein as well as mRNA transcription in BEAS-2B cells while HIF (both HIF-1α and HIF-2α) attenuates c-MYC activity by promoting proteasomal degradation of c-MYC in lung carcinoma A549 cells (Li et al. 2009a; Li et al. 2009b). Ubiquitin specific peptidase 28 (USP28) is a deubiquitinating enzyme bound to c-MYC that prevent salvaging signal to be accumulated on the protein (Wasylishen et al. 2013). Ni (II) and hypoxia increased the levels of the gene silencing mark H3K9me2 (substrate of iron dependent dioxygenase) at USP28 promoter region, which suppressed USP28 gene expression (Li et al. 2009a). The decreased level of USP28 in Ni (II) exposed cells further led to enhanced proteasomal degradation of c-MYC (Figure 1). Although both HIF-1α and HIF-2α have been found essential for c-MYC degradation in A549 cells, no evidence support HIF regulates the expression of USP28 (Li et al. 2009a). Interestingly, MYC and p53 activation in Ni (II) exposed cells is known to lead to cell apoptosis, the attenuation of both pathways by HIF presumably would help cells to bypass apoptosis.
Reactive oxygen species (ROS) governed DEG and pathways
Nickel induces weak oxidative stress that depletes glutathione and activates nuclear factor kappa B (NF-κB) and other oxidatively sensitive transcription factors (Salnikow et al. 1994). However, induced ROS is a very common phenotype in cells that encounter various toxins, so we chose to discuss ROS governed DEG and pathway alterations induced by Ni (II). ROS have been implicated in a wide array of cellular processes by mediating signal transduction. ROS induced by Ni (II) was able to promote phosphorylation of AKT (protein kinase B). The activated AKT in turn activates apoptosis signal-regulating kinase 1 (ASK1) by promoting the phosphorylation at threonine 838 residue, this association is specific in Ni (II) exposed cells but not in unexposed cells (Pan et al. 2010). The signal is then relayed to downstream kinase p38, which activates AP-1 and NFAT, leading to a p53 independent G2-M growth arrest (Ding et al. 2009; Huang et al. 2001).
The activated NF-κB by Ni (II) serves as a transcription repressor, leading to decreased expression of key regulators of the interferon-dependent transcriptional cascade, IRF3, IRF7 and IKKε. The activated NF-κB also results in a down-modulation of N-terminal truncated (ΔN) p63, which lacks the transactivation domain (Zhang et al. 2011). A supplement of ΔNp63 suppressed p21 induction by Ni (II), indicating there is crosstalk between p53 and NF-κB pathways in Ni (II) exposed cells (Zhang et al. 2011). Ni (II) up-regulates p21 through p53 and NF-κB pathways. A direct activation of p53 as well as an inhibition of p21 negative regulator ΔNp63 via NF-κB lead to decreased proliferation in cells exposed to Ni (II). It was proposed that the inhibitory effect of ΔNp63 on p21 promoter may be either through p53 binding site or other sites beyond p53 (Zhang et al. 2011). The cell cycle arrest and apoptosis induced by activation of p53, NF-κB and c-MYC might be critical and serve as a selection force during Ni (II) carcinogenicity, because only the cells have compromised p53, NF-κB and c-MYC will be selected for during long term Ni (II) exposure. This hypothesis has been supported by a selection for Ni (II) resistant cells in mouse fibroblast, only those with lower NF-κB, AP-1 DNA binding capacity were selected and an altered oxidative stress response was found in these cells, which provided a possible mechanism of cell transformation (Salnikow et al. 1994). Nevertheless, it is critical for cancer cells to arrest their growth and survive in secondary organs as micrometastases until suitable environmental conditions are present to reactivate tumor cell outgrowth (Tran et al. 2011).
Conclusion remark
Ni (II) modulates an intricate network containing several modules including transcription factors and histone modifiers through perturbing iron homeostasis, oxidative stress; ultimately resulting in early transformation, as elaborated in Figure 1. Ni (II) promotes proliferation and cell survival by increasing the accumulation of HIF, and hence HIF governed gene expression. HIF directly inhibits transactivation of p53 by competing p300 transcription activator, further down-regulating the expression of p53 governed gene expression and apoptosis. Nickel treatment leads to an accumulation of c-MYC, however, the accumulation is attenuated by Ni (II) induced H3K9me2 driven transcription repression of USP28. Oxidative stress generated by Ni (II) activates NFκB, p53 and c-MYC pathway, promoting cell cycle arrest and apoptosis, which might serve as a selection force to shift the population to one with impaired tumor suppressor genes as well as keeping the cells as dormant micrometastases until suitable microenvironment presents.
Acknowledgement
We thank colleagues in the lab for their valuable discussion. This work is supported by NIEHS ES000260 and NIEHS ES023174.
Reference
- Achison M, Hupp TR. Hypoxia attenuates the p53 response to cellular damage. Oncogene. 2003;22(22):3431–40. doi: 10.1038/sj.onc.1206434. doi:10.1038/sj.onc.1206434. [DOI] [PubMed] [Google Scholar]
- Affimetrix Technical Note:Design and Performance of the GeneChip® Human Genome U133 Plus 2.0 and Human Genome U133A 2.0 Arrays.
- Arita A, Niu J, Qu Q, et al. Global levels of histone modifications in peripheral blood mononuclear cells of subjects with exposure to nickel. Environmental health perspectives. 2012a;120(2):198–203. doi: 10.1289/ehp.1104140. doi:10.1289/ehp.1104140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arita A, Shamy MY, Chervona Y, et al. The effect of exposure to carcinogenic metals on histone tail modifications and gene expression in human subjects. Journal of trace elements in medicine and biology : organ of the Society for Minerals and Trace Elements. 2012b;26(2-3):174–8. doi: 10.1016/j.jtemb.2012.03.012. doi:10.1016/j.jtemb.2012.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barski A, Cuddapah S, Cui K, et al. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129(4):823–37. doi: 10.1016/j.cell.2007.05.009. doi:10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- Cantone L, Nordio F, Hou L, et al. Inhalable metal-rich air particles and histone H3K4 dimethylation and H3K9 acetylation in a cross-sectional study of steel workers. Environmental health perspectives. 2011;119(7):964–9. doi: 10.1289/ehp.1002955. doi:10.1289/ehp.1002955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Costa M. Iron- and 2-oxoglutarate-dependent dioxygenases: an emerging group of molecular targets for nickel toxicity and carcinogenicity. Biometals : an international journal on the role of metal ions in biology, biochemistry, and medicine. 2009;22(1):191–6. doi: 10.1007/s10534-008-9190-3. doi:10.1007/s10534-008-9190-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Giri NC, Zhang R, et al. Nickel ions inhibit histone demethylase JMJD1A and DNA repair enzyme ABH2 by replacing the ferrous iron in the catalytic centers. The Journal of biological chemistry. 2010a;285(10):7374–83. doi: 10.1074/jbc.M109.058503. doi:10.1074/jbc.M109.058503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Ke Q, Kluz T, Yan Y, Costa M. Nickel ions increase histone H3 lysine 9 dimethylation and induce transgene silencing. Molecular and cellular biology. 2006;26(10):3728–37. doi: 10.1128/MCB.26.10.3728-3737.2006. doi:10.1128/MCB.26.10.3728-3737.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Kluz T, Zhang R, Costa M. Hypoxia and nickel inhibit histone demethylase JMJD1A and repress Spry2 expression in human bronchial epithelial BEAS-2B cells. Carcinogenesis. 2010b;31(12):2136–44. doi: 10.1093/carcin/bgq197. doi:10.1093/carcin/bgq197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowder SW, Horton LW, Lee SH, et al. Passage-dependent cancerous transformation of human mesenchymal stem cells under carcinogenic hypoxia. FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 2013;27(7):2788–98. doi: 10.1096/fj.13-228288. doi:10.1096/fj.13-228288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson T, Chen H, Garrick MD, D'Angelo G, Costa M. Soluble nickel interferes with cellular iron homeostasis. Molecular and cellular biochemistry. 2005;279(1-2):157–62. doi: 10.1007/s11010-005-8288-y. doi:10.1007/s11010-005-8288-y. [DOI] [PubMed] [Google Scholar]
- Davidson T, Salnikow K, Costa M. Hypoxia inducible factor-1 alpha-independent suppression of aryl hydrocarbon receptor-regulated genes by nickel. Molecular pharmacology. 2003;64(6):1485–93. doi: 10.1124/mol.64.6.1485. doi:10.1124/mol.64.6.1485. [DOI] [PubMed] [Google Scholar]
- Ding J, He G, Gong W, et al. Effects of nickel on cyclin expression, cell cycle progression and cell proliferation in human pulmonary cells. Cancer epidemiology, biomarkers & prevention : a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology. 2009;18(6):1720–9. doi: 10.1158/1055-9965.EPI-09-0115. doi:10.1158/1055-9965.EPI-09-0115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorres KL, Pua KH, Raines RT. Stringency of the 2-His-1-Asp active-site motif in prolyl 4-hydroxylase. PloS one. 2009;4(11):e7635. doi: 10.1371/journal.pone.0007635. doi:10.1371/journal.pone.0007635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green SE, Luczak MW, Morse JL, DeLoughery Z, Zhitkovich A. Uptake, p53 pathway activation, and cytotoxic responses for Co(II) and Ni(II) in human lung cells: implications for carcinogenicity. Toxicological sciences : an official journal of the Society of Toxicology. 2013;136(2):467–77. doi: 10.1093/toxsci/kft214. doi:10.1093/toxsci/kft214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C, Li J, Costa M, et al. Hydrogen peroxide mediates activation of nuclear factor of activated T cells (NFAT) by nickel subsulfide. Cancer research. 2001;61(22):8051–7. [PubMed] [Google Scholar]
- IARC . IARC Monographs on the Evaluation of Carcinogenic Risks to Humans. Vol. 49. World Health Organization; Lyon, France: 1990. Chromium, Nickel and Welding. [PMC free article] [PubMed] [Google Scholar]
- Jennings P, Limonciel A, Felice L, Leonard MO. An overview of transcriptional regulation in response to toxicological insult. Archives of toxicology. 2013;87(1):49–72. doi: 10.1007/s00204-012-0919-y. doi:10.1007/s00204-012-0919-y. [DOI] [PubMed] [Google Scholar]
- Koumenis C, Alarcon R, Hammond E, et al. Regulation of p53 by hypoxia: dissociation of transcriptional repression and apoptosis from p53-dependent transactivation. Molecular and cellular biology. 2001;21(4):1297–310. doi: 10.1128/MCB.21.4.1297-1310.2001. doi:10.1128/MCB.21.4.1297-1310.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labbe RM, Holowatyj A, Yang ZQ. Histone lysine demethylase (KDM) subfamily 4: structures, functions and therapeutic potential. American journal of translational research. 2013;6(1):1–15. [PMC free article] [PubMed] [Google Scholar]
- Li Q, Kluz T, Sun H, Costa M. Mechanisms of c-myc degradation by nickel compounds and hypoxia. PloS one. 2009a;4(12):e8531. doi: 10.1371/journal.pone.0008531. doi:10.1371/journal.pone.0008531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Suen TC, Sun H, Arita A, Costa M. Nickel compounds induce apoptosis in human bronchial epithelial Beas-2B cells by activation of c-Myc through ERK pathway. Toxicology and applied pharmacology. 2009b;235(2):191–8. doi: 10.1016/j.taap.2008.12.005. doi:10.1016/j.taap.2008.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadadur SS, Srirama K, Mudipalli A. Iron transport & homeostasis mechanisms: their role in health & disease. The Indian journal of medical research. 2008;128(4):533–44. [PubMed] [Google Scholar]
- Pan J, Chang Q, Wang X, et al. Reactive oxygen species-activated Akt/ASK1/p38 signaling pathway in nickel compound-induced apoptosis in BEAS 2B cells. Chemical research in toxicology. 2010;23(3):568–77. doi: 10.1021/tx9003193. doi:10.1021/tx9003193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollenz RS. The aryl-hydrocarbon receptor, but not the aryl-hydrocarbon receptor nuclear translocator protein, is rapidly depleted in hepatic and nonhepatic culture cells exposed to 2,3,7,8-tetrachlorodibenzo-p-dioxin. Molecular pharmacology. 1996;49(3):391–8. [PubMed] [Google Scholar]
- Pollenz RS, Davarinos NA, Shearer TP. Analysis of aryl hydrocarbon receptor-mediated signaling during physiological hypoxia reveals lack of competition for the aryl hydrocarbon nuclear translocator transcription factor. Molecular pharmacology. 1999;56(6):1127–37. doi: 10.1124/mol.56.6.1127. [DOI] [PubMed] [Google Scholar]
- Salnikow K, Davidson T, Costa M. The role of hypoxia-inducible signaling pathway in nickel carcinogenesis. Environmental health perspectives. 2002;110(Suppl 5):831–4. doi: 10.1289/ehp.02110s5831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salnikow K, Davidson T, Kluz T, Chen H, Zhou D, Costa M. GeneChip analysis of signaling pathways effected by nickel. Journal of environmental monitoring : JEM. 2003;5(2):206–9. doi: 10.1039/b210262p. [DOI] [PubMed] [Google Scholar]
- Salnikow K, Gao M, Voitkun V, Huang X, Costa M. Altered oxidative stress responses in nickel-resistant mammalian cells. Cancer research. 1994;54(24):6407–12. [PubMed] [Google Scholar]
- Schmid T, Zhou J, Kohl R, Brune B. p300 relieves p53-evoked transcriptional repression of hypoxia-inducible factor-1 (HIF-1) The Biochemical journal. 2004;380:289–95. doi: 10.1042/BJ20031299. Pt 1. doi:10.1042/BJ20031299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tchou-Wong KM, Kiok K, Tang Z, et al. Effects of nickel treatment on H3K4 trimethylation and gene expression. PloS one. 2011;6(3):e17728. doi: 10.1371/journal.pone.0017728. doi:10.1371/journal.pone.0017728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran DD, Corsa CA, Biswas H, Aft RL, Longmore GD. Temporal and spatial cooperation of Snail1 and Twist1 during epithelial-mesenchymal transition predicts for human breast cancer recurrence. Molecular cancer research : MCR. 2011;9(12):1644–57. doi: 10.1158/1541-7786.MCR-11-0371. doi:10.1158/1541-7786.MCR-11-0371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasylishen AR, Chan-Seng-Yue M, Bros C, et al. MYC phosphorylation at novel regulatory regions suppresses transforming activity. Cancer research. 2013;73(21):6504–15. doi: 10.1158/0008-5472.CAN-12-4063. doi:10.1158/0008-5472.CAN-12-4063. [DOI] [PubMed] [Google Scholar]
- Wojcieszynska D, Hupert-Kocurek K, Guzik U. Flavin-dependent enzymes in cancer prevention. International journal of molecular sciences. 2012;13(12):16751–68. doi: 10.3390/ijms131216751. doi:10.3390/ijms131216751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong VC, Morse JL, Zhitkovich A. p53 activation by Ni(II) is a HIF-1alpha independent response causing caspases 9/3-mediated apoptosis in human lung cells. Toxicology and applied pharmacology. 2013;269(3):233–9. doi: 10.1016/j.taap.2013.03.023. doi:10.1016/j.taap.2013.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Y, Lu Y, Chen WC, et al. Cobalt and nickel stabilize stem cell transcription factor OCT4 through modulating its sumoylation and ubiquitination. PloS one. 2014;9(1):e86620. doi: 10.1371/journal.pone.0086620. doi:10.1371/journal.pone.0086620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Li W, Cheng S, et al. Nickel-induced down-regulation of DeltaNp63 and its role in the proliferation of keratinocytes. Toxicology and applied pharmacology. 2011;253(3):235–43. doi: 10.1016/j.taap.2011.03.024. doi:10.1016/j.taap.2011.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Li Q, Arita A, Sun H, Costa M. Effects of nickel, chromate, and arsenite on histone 3 lysine methylation. Toxicology and applied pharmacology. 2009;236(1):78–84. doi: 10.1016/j.taap.2009.01.009. doi:10.1016/j.taap.2009.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]