As modern humans migrated out of Africa, they encountered many different environmental conditions including temperature extremes, new pathogens, and high altitude. These diverse environments have likely acted as agents of natural selection and led to local adaptations. One of the most illustrious examples in humans is the adaptation of Tibetans to the hypoxic environment of the high-altitude Tibetan plateau1–3. A hypoxia pathway gene, EPAS1, was previously identified as having the most extreme signature of positive selection in Tibetans4–10, and was shown to be associated with differences in hemoglobin concentration at high altitude. Re-sequencing the region around EPAS1 in 40 Tibetan and 40 Han individuals, we find that this gene has a highly unusual haplotype structure that can only be convincingly explained by introgression of DNA from Denisovans or Denisovan-related individuals into humans. Scanning a larger set of worldwide populations, we find that the selected haplotype is only found in Denisovans and in Tibetans, and at very low frequency among Han Chinese. Furthermore, the length of the haplotype, and the fact that it is not found in any other populations, makes it unlikely that the Tibetan/Denisovan haplotype sharing was caused by incomplete ancestral lineage sorting rather than introgression. Our findings illustrate that admixture with other hominin species has provided genetic variation that helped humans adapt to new environments.
The Tibetan plateau (at greater than 4000m) is inhospitable to human settlement because of low atmospheric oxygen pressure (~ 40% lower than at sea level), cold climate and limited resources (e.g., sparse vegetation). Despite these extreme conditions, Tibetans have successfully settled in the plateau in part due to adaptations that confer lower infant mortality and higher fertility than acclimated women of low-altitude origin. The latter tend to have difficulty bearing children at high altitude, and their offspring typically have low birth weights compared to offspring from women of high altitude ancestry1,2. One well-documented pregnancy-related complication due to high altitude is the higher incidence of preeclampsia2,11 (hypertension during pregnancy). In addition, the physiological response to low oxygen differs between Tibetans and individuals of low-altitude origin. For most individuals, acclimatization to low oxygen involves an increase in blood hemoglobin levels. However, in Tibetans, the increase in hemoglobin levels is limited3, presumably because high hemoglobin concentrations are associated with increased blood viscosity and increased risk of cardiac events, thus resulting in a net reduction in fitness12,13.
Recently, the genetic basis underlying adaptation to high altitude in Tibetans was elucidated4–10 using exome and SNP array data. Several genes seem to be involved in the response but most studies identified EPAS1, a transcription factor induced under hypoxic conditions, as the gene with the strongest signal of Tibetan specific selection4–10. Furthermore, SNP variants in EPAS1 showed significant associations with hemoglobin levels in the expected direction in several of these studies; individuals carrying the derived allele have lower hemoglobin levels than individuals homozygous for the ancestral allele. Here, we re-sequence the complete EPAS1 gene in 40 Tibetan and 40 Han individuals at more than 200X coverage to further characterize this impressive example of human adaptation. Remarkably, we find the source of adaptation was likely due to the introduction of genetic variants from archaic Denisovan-like individuals (individuals closely related to the Denisovan individual from the Altai Mountains14) into the ancestral Tibetan gene pool.
Results
Exceptionally high genetic differentiation in EPAS1
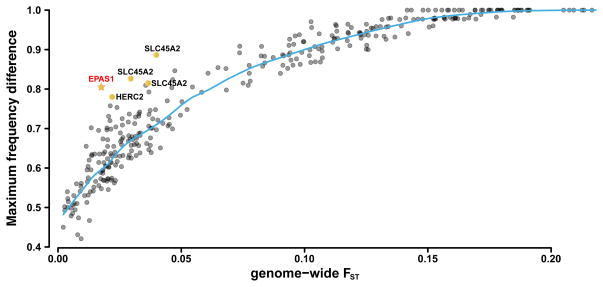
After applying standard next generation sequencing filters (see Methods), we call a total of 477 SNPs in a region of approximately 129kb in the combined Han and Tibetan samples (Tables S1, S2). We compute FST between Han and Tibetans, and confirm that it is highly elevated in the EPAS1 region as expected under strong local selection (Extended Data Fig. 1). Indeed, by comparison to 26 populations from the Human Genome Diversity Panel15,16, Figure 1, it is clear that the variants in this region are far more differentiated than one would expect given the average genome-wide differentiation between Han and Tibetans (FST ~0.02, Yi et al. 20104). The only other genes with comparably large frequency differences between any closely related populations are the previously identified loci associated with lighter skin pigmentation in Europeans, SLCA45A2 and HERC217–20, although in these examples the populations compared (e.g. Hazara and French, Brahui and Russians) are more genetically differentiated than Han and Tibetans. In populations as closely related as Han and Tibetans, we find no examples of SNPs with as much differentiation as seen in EPAS1, illustrating the uniqueness of its selection signal.
Figure 1. Genome-wide Fst vs maximal allele frequency.
The relationship between genome-wide FST (x-axis) computed for each pair of the 26 populations and maximal allele frequency (y-axis), first explored in Coop et al. (19). Maximal allele frequency is defined as the largest frequency difference observed for any SNP between a population pair. The 26 populations are from the Human Genome Diversity Panel (HGDP). The labels highlight genes that harbor SNPs previously identified as having strong local adaptation.
A highly diverged EPAS1 haplotype
FST is particularly elevated in a 32.7 kb region containing the 32 most differentiated SNPs (green box in Extended Data Fig. 1; Table S3), which is the best candidate region for the advantageous mutation(s). We therefore focus the subsequent analyses primarily on this region. Phasing the data (see Methods) to identify Han and Tibetan haplotypes in this region (Figure 2) we find that Tibetans carry a high frequency haplotype pattern that is strikingly different from both their minority haplotypes and the common haplotype observed in the Han. For example, the region harbors a highly differentiated 5-SNP haplotype motif (AGGAA) within a 2.5kb window that is only seen in Tibetan samples and in none of the Han (the first 5 SNPs in Table S3, and blue arrows in Figure 2). The pattern of genetic variation within Tibetans appears even more unusual because none of the variants in the five-SNP motif is present in any of the minority haplotypes of Tibetans. Even when subject to a selective sweep, we would not generally expect a single haplotype to contain so many unique mutations not found on other haplotypes.
Figure 2. Haplotype pattern in a region defined by SNPs that are at high frequency in Tibet and at low frequency in the Han (see Table S3).
Each column is a polymorphic genomic location (95 in total), each row is a phased haplotype (80 Han and 80 Tibetan haplotypes), and the colored column on the left denotes the population identity of the individuals (Han in orange, Tibetans in pink). The top two rows (in dark green) are the haplotypes of the Denisovan individual. The dark cells represent the presence of the derived allele and the grey space represents the presence of the ancestral allele (see Methods). The first column corresponds to the first positions in Table S3 and the last column corresponds to the last position in Table S3. The red and blue arrows at the top indicate the 32 sites in Table S3. The blue arrows represent a five-SNP haplotype block defined by the first five SNPs in the 32.7kb region. The stars beneath the arrows point to sites where Tibetans share a derived allele with the Denisovan individual.
We investigate whether a model of selection on either a de novo mutation (SDN) or selection on standing variation (SSV) could possibly lead to so many fixed differences between haplotype classes in such a short region within a single population. To do so, we simulate a 32.7kb region under these models assuming different strengths of selection and conditioning on the current allele frequency in the sample (see Methods). We find that the observed number of fixed differences between the haplotype classes is significantly higher than what is expected by simulations under any of the models explored (Extended Data Fig. 2). Thus the degree of differentiation between haplotypes is significantly larger than expected from mutation, genetic drift and directional selection alone. In other words, it is unlikely (p<0.02 under either a SSV scenario or under a SDN scenario) that the high degree of haplotype differentiation could be caused by a single beneficial mutation landing by chance on a background of rare SNPs, which are then brought to high frequency by selection. The remaining explanations are the presence of strong epistasis between many mutations, or that a divergent population introduced the haplotype into Tibetans by gene flow or through ancestral lineage sorting.
Gene flow from other populations
We search for potential donor populations in two different data sets: the 1000 Genomes project21 and whole genome data from Meyer et al. 201214. We originally defined the EPAS1 32.7kb region boundaries by the level of observed differentiation between the Tibetans and Han only (Table S3, Extended Data Fig. 1 and Figure 2) as described in the previous section. In that region, the most common haplotype in Tibetans is tagged by the distinctive 5-SNP motif (AGGAA; the first 5 SNPs in Figure 2), not found in any of our 40 Han samples. We first focus on this 5-SNP motif and determine whether it is unique to Tibetans or if it is found in other populations.
Intriguingly, when we examine the 1000 Genomes Project data set, we discover that the Tibetan 5-SNP motif (AGGAA) is not present in any of these populations, except for a single CHS (Southern Han Chinese) and a single CHB (Beijing Han Chinese) individual. Extended Data Fig. 3 contains the frequencies of all the haplotypes present in the fourteen 1000 genomes populations21 at these five SNP positions. Furthermore, when we examine the data set from Meyer et al. 201214 containing both modern (Papuan, San, Yoruba, Mandeka, Mbuti, French, Sardinian, Han Dai, Dinka, Karitiana, CEU) and archaic (high coverage Denisovan and low coverage Croatian Neanderthal) human genomes14, we discover that the 5-SNP motif is completely absent in all of their modern human population samples (Table S4). Therefore, apart from one CHS and one CHB individual, none of the other extant human populations sampled to date carry this 5-SNP haplotype. Strikingly, the Denisovan haplotype at these 5 sites (AGGAA) exactly matches the 5-SNP Tibetan motif (Table S4 and Extended Data Fig. 3).
We observe the same pattern when focusing on the entire 32.7 kb region and not just the 5-SNP motif. Twenty SNPs in this region have unusually high frequency differences of at least 0.65 between Tibetans and all the other populations from the 1000 Genomes project (Extended Data Fig. 4). However, in Tibetans, 15 out of these 20 SNPs are identical to the Denisovan haplotype generating an overall pattern of high haplotype similarity between the selected Tibetan haplotype and the Denisovan haplotype (Tables S5–S7). Interestingly, 5 of these SNPs in the region are private SNPs shared between Tibetans and the Denisovan, but not shared with any other population worldwide, except for two SNPs at low frequency in Han Chinese (Extended Data Fig. 4 and Table S7).
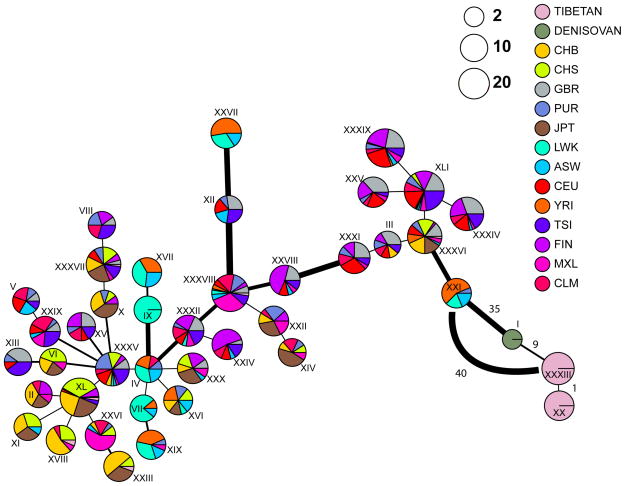
If we consider all SNPs (not just the most differentiated) in the 32.7kb region annotated in humans, to build a haplotype network22 using the 40 most common haplotypes, we observe a clear pattern in which the Tibetan haplotype is much closer to the Denisovan haplotype than any modern human haplotype (Figure 3 and Extended Figures 5a; see Extended Data Figures 6a–b for haplotype networks constructed using other criteria). Furthermore, we find that the Tibetan haplotype is slightly more divergent from other modern human populations than the Denisovan haplotype is, a pattern expected under introgression (see Methods and Extended Data Fig. 5b). Raw sequence divergence for all sites and all haplotypes shows a similar pattern (Extended Data Fig. 7). Moreover, the divergence between the common Tibetan haplotype and Han haplotypes is larger than expected for comparisons among modern humans, but well within the distribution expected from human-Denisovan comparisons (Extended Data Fig. 8). Notably, sequence divergence between the Tibetans’ most common haplotype and Denisovan is significantly lower (p=0.0028) than what we expect from human-Denisovan comparisons (Extended Data Fig. 8). We also find that the number of pairwise differences between the common Tibetan haplotype and the Denisovan haplotype (n=12) is compatible with the levels one would expect from mutation accumulation since the introgression event (see Methods for Extended Data Fig. 8). Finally, if we compute D14 and S23,24, two statistics that have been designed to detect archaic introgression into modern humans, we obtain significant values (D-statistic p-values < 0.001, and S* p-values <=0.035) for the 32.7 kb region using multiple null models of no gene-flow (see Methods, Tables S8–S10, and Extended Data Figures 9 and 10a).
Figure 3. A haplotype network based on the number of pairwise differences between the 40 most common haplotypes.
The haplotypes were defined from all the SNPs present in the combined 1000 Genomes and Tibetan samples: 515 SNPs in total within the 32.7kb EPAS1 region. The Denisovan haplotypes were added to the set of the common haplotypes. The R software package pegas23 was used to generate the figure, using pairwise differences as distances. Each pie chart represents one unique haplotype, labeled with Roman numerals, and the radius of the pie chart is proportional to the log2(number of chromosomes with that haplotype) plus a minimum size so that it is easier to see the Denisovan haplotype. The sections in the pie provide the breakdown of the haplotype representation amongst populations. The width of the edges is proportional to the number of pairwise differences between the joined haplotypes; the thinnest edge represents a difference of 1 mutation. The legend shows all the possible haplotypes among these populations (see Methods for definition of population acronyms). The numbers next to an edge in the bottom right are the number of pairwise differences between the corresponding haplotypes. We added an edge afterwards between the Tibetan haplotype XXXIII and its closest non-Denisovan haplotype (XXI) to indicate its divergence from the other modern human groups. Extended Data Fig. 5a contains all the pairwise differences between the haplotypes presented in this figure.
Thus, we conclude that the haplotype associated with altitude adaptation in Tibetans is likely a product of introgression from Denisovans or Denisovan-related populations. The only other possible explanation is ancestral lineage sorting. However, this explanation is exceedingly unlikely as it cannot explain the significant D and S values and because it would require a long haplotype to be maintained without recombination since the time of divergence between Denisovans and humans (estimated to be at least 200,000 years14). The chance of maintaining a 32.7 kb fragment in both lineages throughout 200,000 years is conservatively estimated at p=0.0012 assuming a constant recombination of 2.3e-8 per bp per generation (see Methods). Furthermore, the haplotype would have to have been independently lost in all African and non-African populations, except for Tibetans and Han Chinese.
Discussion
We have re-sequenced the EPAS1 region and found that Tibetans harbor a highly differentiated haplotype that is only found at very low frequency in the Han population among all the 1000 Genomes populations, and is otherwise only observed in a previously sequenced Denisovan individual14. As the haplotype is observed in a single individual in both CHS and CHB samples, it suggests that it was introduced into humans prior to the separation of Han and Tibetan populations, but subject to selection in Tibetans after the Tibetan plateau was colonized. Alternatively, recent admixture from Tibetans to Hans may have introduced the haplotype to nearby Han populations outside Tibet. The CHS and CHB individuals carrying the 5-SNP Tibetan/Denisovan haplotype (Extended Data Fig. 3) show no evidence of being recent migrants from Tibet (see Methods and Extended Data Fig. 10b), suggesting that if the haplotype was carried from Tibet to China by migrants, this migration did not occur within the last few generations.
Previous studies examining the genetic contributions of Denisovans to modern humans14,25 suggest that Melanesians have a much larger Denisovan component than either Han or Mongolians, even though the latter populations are geographically much closer to the Altai mountains14,25. Interestingly, the putatively beneficial Denisovan EPAS1 haplotype is not observed in modern day Melanesians or in the high coverage Altai Neanderthal26 (Table S4). Skoglund and Jakobsson27 found evidence for Denisovan admixture throughout Southeast Asia (as well as in Melanesians) based on a global analysis of SNP array data from 1600 individuals from a diverse set of populations27, and this finding has been recently confirmed by Prufer et al. 201426. Therefore, it appears that sufficient archaic admixture into populations near the Tibetan region occurred to explain the presence of this Denisovan haplotype outside Melanesia. Furthermore, the haplotype may have been maintained in some human populations, including Tibetans and their ancestors, through the action of natural selection.
Recently a few studies have supported the notion of adaptive introgression from archaic humans to modern humans as playing a role in the evolution of immunity-related genes (HLA28 and STAT229) and in the evolution of skin pigmentation genes (BNC223, 30). Our findings imply that one of the most clear-cut examples of human adaptation is likely due to a similar mechanism of gene-flow from archaic hominins into modern humans. With our increased understanding that human evolution has involved a substantial amount of gene-flow from various archaic species, we are now also starting to understand that adaptation to local environments may have been facilitated by gene-flow from other hominins that may already have been adapted to those environments.
Methods
DNA samples
DNA samples included in this work were extracted from peripheral blood of 41 unrelated Tibetan individuals living at > 4300-meter above sea level within the Himalayan Plateau. Tibetan identity was based on self-reported family ancestry. The individuals were from two villages of Dingri (20 samples prefixed DR; 4300m altitude) and Naqu (21 samples prefixed NQ; 4600m altitude). All participants gave a self-report of at least three generations living at the sampling site, and provided informed consent for this study. These individuals are a subset of the 50 individuals exome-sequenced from Yi et al. 20104. Samples of Han Chinese (CHB) are from 1000 Genomes Project21 (40 samples prefixed NA).
A combined strategy of long-range PCR and next-generation sequencing was used to decipher the whole EPAS1 gene and it’s +/− 30Kb flanking region (147Kb in total). We designed 38 pairs of long-range PCR primers to amplify the region in 41 Tibetan and 40 Han individuals. PCR products from all individuals were fragmented and indexed, then sequenced to higher than 260-fold depth for each individual with the Illumina Hiseq2000 sequencer. The reads were aligned to the UCSC human reference genome (hg18) using the SOAPaligner31. Genotypes of each individual at every genomic location of the EPAS1 gene and the flanking region were called by SOAPsnp32. To make comparisons easier with the Han samples, we only used 40 Tibetan samples for this study.
Data filtering
The coverage for each individual is roughly 200X. Genotypes of each individual at every site in the EPAS1 gene and the flanking region were called by SOAPsnp32 which resulted in 700 SNPs in the combined Tibetan-Han sample. However, we filtered out some sites post-genotype calling by performing standard filters that are applied in the analyses of next generation sequencing data. Specifically, of the 700 SNPs called, we removed SNPs that 1) were not in Hardy Weinberg equilibrium, 2) were located in hard to map regions (the SNP is located at a position with mappability=0, using the Duke Unique 35 track), 3) had heterozygote individuals where the distribution of counts for the two alleles were different (the counts of the two alleles deviate from the expectation of 50% assuming a binomial distribution), 4) had different quality score distributions for the two alleles, 5) fell on or near a deletion or insertion and 6) were tri-allelic. A total of 477 SNPs in the combined sample remained after applying all the filters. Within Tibetans, 354 sites (out of the 477 sites) were SNPs, and within the Han, 364 sites (out of the 477) were SNPs. After filtering, we used Beagle to phase the Tibetan and Han individuals together33.
HGDP Data, Figure 1
We downloaded the Human Genome Diversity Panel (HGDP) SNP data from the University of Chicago website (http://hgdp.uchicago.edu/data/plink_data/) and followed the filtering criteria in Coop et al. 200934. We used the formula of Reynolds et al. 198335 to compute FST between pairs of populations. We used the intersection of SNPs between the 50 Tibetan exomes from Yi et al. 20107 and the HGDP SNPs, resulting in 8362 SNPs. Note, the number 354 quoted in the previous section refers to Tibetan SNPs from the full re-sequencing of the EPAS1 gene in this study.
We calculated FST for each pair of populations and also scored the frequencies of the SNP with the largest frequency difference between pairs of populations. Using the genotypes from the 26 populations we have re-created Figure 2A in Coop et al. 200934 using the SNPs overlapping in two data sets: the 50 Tibetan exomes data set and the HGDP15,16 data set. The figure displays the maximum SNP frequency difference against the mean FST across all SNPs for each pair of the HGDP populations. We added one data point to this figure consisting of the mean Fst between Tibetans and Han (Fst ≈ .018) and the SNP with the largest frequency difference between Han and Tibetans (~ .8), which is a SNP in the EPAS1 gene.
Tibetan and Han haplotypes at the 32.7kb highly differentiated region, Figure 2
The 32.7kb region was identified as the region of highest genetic differentiation between Tibetans and Han (green box in Extended Data Fig. 1), providing the strongest candidate region for the location of the selective sweep. To examine the haplotypes in this region, we first filtered out SNPs that were <= 5% or >=95% frequency in both the Tibetan and Han samples, i.e. SNPs that were very common or very rare in both populations simultaneously. We computed the number of pairwise differences between every pair of haplotypes. Then we ordered the haplotypes based on their number of pairwise distances from the most common haplotype in each population separately. Figure 2 is generated using the heatmap.2 function from the gplots package of the statistical computing platform R36, with derived and ancestral alleles colored black and light grey, respectively. We used the chimp sequence to define the ancestral state. However, the chimp allele was missing at one of the 32 most differentiated sites (see arrows in Figure 2), so in that case we used the orangutan and rhesus macaque alleles to define the ancestral allele.
Simulations, Selection on a de novo mutation (SDN) and on standing variation (SSV), Extended Data Fig. 2
We used msms37 to simulate data for a population of constant size with mutation, recombination, and positive selection affecting a single site. We conditioned on a current allele frequency in the population of 69/80, the observed value in the real EPAS1 data. We estimated a Tibetan effective population size of N=7000 (see supplementary section titled “Estimate of population size”). In addition, we assumed three different selection coefficients: 2Ns =200, 500, 1000, and a recombination rate per base pair per generation of 2.3e-8 (this is the average recombination rate in the EPAS1 gene using the fine-scale estimates from the map of Myers et al. 200538. This recombination estimate is very similar to the estimate from the African American map39 for the EPAS1 gene which is 2.4e-8. We set the mutation rate to 2.0e-8 per base pair per generation because this is what we estimated using the patterns of genetic diversity in the EPAS1 gene in Tibetans under an Approximate Bayesian Computation (ABC) framework (see supplementary Information titled “Estimate of the mutation rate” for details). This mutation rate estimate is similar to the phylogenetic estimates reviewed in Scally et al. 201140. We note that the human-chimp differences in other intronic regions in the genome of the same size has a mean (417) and median (410) close to that found for the EPAS1 32.7kb region (see supplementary section titled “Distribution of human-chimp differences in 32.7kb regions” for details), suggesting that this region does not have an unusual mutation rate. In the simulations, we examined a region of 32.7kb around the selected site and grouped the haplotypes into two groups: those that carried the beneficial allele and those that did not. If k is the number of chromosomes carrying the beneficial mutation, we counted how many mutations within the 32.7kb region had frequency bigger or equal to (k/80 – 0.05) in the group that harbored the beneficial allele (i.e. fixed or almost fixed in that group), and simultaneously had frequency 0 in the other group.
To simulate data for a sweep from standing variation, we used mbs41 and the same parameters as in the previous set of simulations, but assuming an initial allele frequency of the advantageous allele of 1% when selection starts. To be able to compare the number of almost fixed sites from these simulations to the observed data, we needed to make a call in the Han-Tibetan dataset of what could plausibly be the selected site. The most straightforward choice is the site that has the highest Han-Tibetan differentiation; see the circled SNP in Extended Data Fig. 1 (this site also has the largest frequency difference (~0.85) between Tibetans and any of the 1000 Genomes populations). Tibetan individuals with the derived mutation at this site were defined as carrying the selected haplotype, and the remaining individuals were defined as not carrying the selected haplotype. Then we performed the same counting of “almost fixed” sites between these two groups as was done for the simulations. The simulated distribution of almost fixed differences and the real data are shown in Extended Data Fig. 2 (histograms of almost fixed differences).
For the SDN model under a selection coefficient of 2Ns =200, 500, 1000, the p-values are 0.004, 0.006 and 0.006 respectively. Under SSV with a selection parameter of 2Ns =200, 500, 1000, the p-values are 0.002, 0.012 and 0.015 respectively. We note that increasing the initial frequency of the selected allele (to 5%) also leads to a smaller number of fixed differences than what we observe in the real data, thereby making the simulated SSV scenario similarly unlikely (p-values are 0.007, 0.01 and 0.006 for 2Ns =200, 500, 1000 respectively). We also note that simulating data with a smaller mutation rate will not result in an increase in the number of fixed differences.
5 SNP motif
We identified the contiguous 5-SNP haplotype motif that is most common in the 40 Tibetan samples, but entirely absent in the 40 Han individuals (see the 5-SNP haplotype defined by the first five blue arrows in Figure 2). The 5 SNPs comprising this motif (positioned at 46421420, 46422184, 46422521, 46423274 and 46423846), span a 2.5kb region (46423846 – 46421420 ~ 2.5 kb) containing no other SNPs (even when including low and high frequency SNPs). The genomic positions of this 5-SNP motif were then examined in the phased 1000 Genomes21 dataset to compute the frequency of this 5-SNP haplotype in the populations sequenced in the 1000 Genomes project (see Extended Data Fig. 3, and Methods section titled “Haplotype frequencies at the 5-SNP motif in 1000 Genomes data, Extended Data Fig. 3”). In the following, we will refer to this 5 SNP motif as the ‘core’ Tibetan haplotype.
Haplotype frequencies at the 5-SNP motif in 1000 Genomes data, Extended Data Figure 3
For all samples/populations in the 1000 genomes project21, we interrogated the 5 sites in the “core” Tibetan haplotype identified in EPAS1, and counted the frequencies of each of the unique haplotypes within each population group of the 1000 genomes. The barplot in Extended Data Fig. 3 is a summary of these frequencies within each population, colored by the unique haplotype sequences present.
Haplotype network, Figure 3
We constructed a haplotype network including Tibetans, Denisovans and the 1000Genomes samples (YRI, Yoruban; LWK, Luhya; ASW, African American from the South West; TSI, Toscani; CEU, Utah Residents with Northern and Western European ancestry; GBR, British; FIN, Finnish; JPT, Japanese; CHS, Southern Han Chinese; CHB, Han Chinese from Beijing; MXL, Mexican; PUR, Puerto Rican; CLM, Colombian) for the 32.7kb EPAS1 region in the combined 1000Genomes samples. To limit the number of haplotypes to display, we identified the 40 most common haplotypes. There are a total of 515 SNPs in the 32.7kb EPAS1 region that pass all quality filters. We used the R36 software package “pegas”22 to build a tree that connects haplotypes based on pairwise differences (see Figure 3). The Denisovan individual is homozygous at all the 515 sites. Note, Figure 3 does not include the Iberians (IBS) for clarity (to reduce the number of colors needed for the plot), and because the small sample of Iberians (18) only contain haplotypes observed in other European samples. We find that the Denisovan haplotype is closest to the Tibetan haplotypes. Extended Data Fig. 5a contains all the pairwise differences between the 41 (40 from modern humans and 1 from Denisovan) haplotypes in Figure 3.
The observed haplotype structure is compatible with the introgression hypothesis. As expected under the introgression hypothesis, the Tibetan haplotype is more distant to the non-Tibetan haplotypes than the Denisovan haplotype because, after the admixture event, the introgressed haplotype accumulated extra mutations. In contrast, the Denisovan haplotype, being the product of DNA extracted from an ancient specimen, did not have time to accumulate a similar number of mutations. This effect is illustrated in Extended Data Fig. S5b. For example, the divergence between the introgressed haplotype (i.e the Tibetan (Tib) haplotype) and the Yoruban haplotype would be larger than between the observed Denisovan haplotype and the Yoruban (YRI) haplotype (see Extended Data Fig. S5b and Supplementary Information titled “Extended Data Figure 5b”).
Haplotype network, Extended Data Fig. 6a–b
Figure 3 plots the network of the 40 most common haplotypes. Alternatively, we also used the 20 sites such that the frequency difference between Tibetans and each of the 14 populations from the 1000 Genomes project21 is at least 0.65 (see Extended Data Fig. 4) to construct a haplotype network (Extended Data Fig. 6a). The resulting region spanned by these SNPs is the same 32.7kb region as previously identified by considering sites that are the most differentiated between Tibetans and Han (Table S3). For more details about Extended Data Figures 6a and 6b, see Supplementary Information titled “Haplotype networks constructed using other criteria”).
Denisovan-Human number of pairwise differences at the EPAS1 32.7kb region, Extended Data Fig. 7
We computed the number of pairwise differences as described in Supplementary Information titled “Number of pairwise differences.” We removed all the Denisovan sites that had a genotype quality smaller than 40 and a mapping quality smaller than 30, using the same thresholds as in the Denisovan paper14. This filtering resulted in the removal of 782 sites (out of 32746). We also removed another 27 sites within the 32.7kb region that did not pass our quality filters in Tibetans (see Data Filtering section). The total number of SNPs in the combined Tibetan, 1000Genomes and the Denisovan samples is 520. For the 32.7kb region in EPAS1, we computed the number of pairwise differences between the Denisovan haplotypes and each of Tibetan haplotypes (red histograms, Extended Data Fig. 7). We also computed the number of pairwise differences between the Denisovan haplotypes and each of the haplotypes in the 1000 Genomes Project’s populations (CHS, CHB, CEU, JPT, ASW, FIN, PUR, GBR, LWK, MXL, CLM, IBS and YRI, see blue histograms in Extended Data Fig. 7). Notice that for this comparison, we compared every site that passes the quality filters even if the site is not polymorphic in modern humans. This is in contrast to Figure 3 where we only considered the sites that are polymorphic in modern humans. Furthermore, if a site is not polymorphic in our sample, we assumed that all of our samples carry the human reference allele. We plot two histograms in each panel of Extended Data Fig. 7: the distribution of Tibetan-Denisovan comparison (red histogram) and the distribution of pairwise differences between the Denisovan haplotype and each population (blue histogram) from the 1000 Genomes Project (Extended Data Fig. 7).
Denisovan/modern human divergence and modern human/modern human divergence, Extended Data Fig. 8
To compute the genomic distribution of modern human/Denisovan pairwise differences we examined all windows of intronic sequence of size 32.7kb (using a table from Ensembl with the exon boundaries for all genes) from chromosomes 1 to 6. Within each 32.7kb region, we removed all the Denisovan sites that had a genotype quality smaller than 40 and a mapping quality smaller than 30. We computed divergence by computing all the pairwise differences between a human haplotype and the Denisovan haplotypes (see supplementary section titled “Number of pairwise differences”) and dividing by the effective sequence length (i.e. all the sites in the 32.7kb region that passed all the filters - a mapping quality higher or equal to 30 and a genotype quality higher or equal to 40). We only kept the 32.7kb regions where at least 20,000 sites passed these quality criteria. The modern humans used in these comparisons were the first 80 CEU chromosomes, the first 80 CHS chromosomes and the first 80 CHB chromosomes from the 1000 Genomes data. If a site was not polymorphic in modern humans, we assumed that they carried the reference allele.
We also computed modern human/modern human divergence at the same intronic regions. In this case, we compare modern human populations (CHB vs CHS, CHB vs CEU, CHS vs CEU) by comparing all 80 haplotypes in one group to all 80 haplotypes in the other group for a total of 3×80×80 comparisons. The distributions of modern human/Denisovan and modern human/modern human pairwise differences are both plotted in Extended Data Fig. 8. We also display the distribution of Tibetan-Han pairwise differences in the 32.7kb region of the EPAS1 locus (80 Tibetan and 80 Han for a total of 6400 comparisons). Finally, we include the pairwise differences between the Denisovan and the Tibetans computed as in Extended Data Fig. 7, standardized by the number of sites that passed all quality filters. This number (12/31937) leads to a sequence divergence of 0.000375 for the most common Tibetan haplotypes, and this is indeed significantly lower (p-value = 0.0028) than what is expected under the distribution of human/Denisovan divergence (see Extended Data Fig. 8). Table S11 contains the details regarding the 12 differences between the Tibetan and the Denisovan haplotypes.
To further address the issue as to whether a difference of 12 differences between the Denisovans and Tibetans is expected under the introgression hypothesis, we computed the number of mutations theoretically expected for an introgressed region of this size, given published estimates of the age of the sample, and the coalescence time within Denisovans. We assumed that mutations occur as a Poisson process and used the estimates of split times from Prufer et. al. 201426 between the called introgressed Denisovan haplotypes and the Denisovan haplotypes (subsection titled “The introgressing Denisovan and Siberian Denisovan split < 394 kya assuming mu=0.5×10−9/bp/year” on page 114 of the Supplementary Information S13.2 of Prufer et al. 201426). Using these estimates, the number of expected mutations between the Denisovan haplotype and our introgressed haplotype (the Tibetans’ most common haplotype) is simply: [2*tMRCA-age]*L*mu = 11.25, where tMRCA is the time to the most recent common ancestor estimated at 394 kya, mu=0.5×10−9/bp/year, L=32.7kb, and age is the age of the Denisovan sample which we conservatively set to 100,000 years. Clearly, the observed value of 12 mutations is remarkably close to the expected number (11.25). In fact, we would need to observe 17 or more mutations to be able to reject the introgression hypothesis at the 5% significance level. If we use our estimate of the mutation rate in the EPAS1 gene, mu=1.0×10−9/bp/year (2.0×10−8/bp/generation), then the expected number of differences is 22.5. Therefore we conclude the number of differences we observe are compatible with the previous estimates of introgressed Denisovan versus sampled Denisovan sequence divergence.
Probability of 32.7kb haplotype block from shared ancestral lineage
We calculate the probability of a haplotype, of length at least 32.7kb, shared by modern Tibetans and the archaic Denisovan due to incomplete ancestral lineage sorting. Let r be the recombination rate per generation per base pair (bp). Let t be the length of the human and Denisovan branches since divergence. The expected length of a shared ancestral sequence is 1/(r×t). Let this expected length = L. Assuming an exponential distribution of admixture tracts, the probability of seeing a shared fragment of length ≥ m is exp(−m/L). However, conditional on observing the Denisovan nucleotide at position j, the expected length is the sum of two exponential random variables with expected lengths L, therefore it follows a Gamma distribution with shape parameter 2, and rate parameter lambda=1/L. Inserting numbers for human branch length after divergence at a conservative lower estimate of 200kyr, and the Denisovan branch of 100kya (divergence minus the estimated age of the Denisovan sample which can be as old as 100kya14,26), and assuming a generation time of 25 yrs, we get L = 1/(2.3e-8*(300e3/25)) = 3623.18bp, and the probability of a length of at least m = 32,700 bp is 1-GammaCDF(32700, shape=2, rate=1/L) = 0.0012. Here the recombination of 2.3e-8 is the average recombination rate in EPAS1 calculated from the estimates in Myers et al. 201214. We should mention, both this divergence estimate for the Denisovan/human split and the age of the Denisovan sample are highly conservative14,25,26, so the actual probability may be considerably less. Also, the haplotype would have to have been independently lost in all African and non-African populations, except for Tibetans and Han Chinese.
Null Distribution of D statistics under models of no gene flow, Extended Data Figure 9
As another approach to assess the probability of an ancestral lineage having given rise to the 32.7kb haplotype we observe in Tibetans in the absence of gene flow, we compared D-statistics between human populations under simulations42 of several demographic models described in Sankararaman et al. 201243. D-statistics were calculated according to equation 2 in Durand et al. 201144. The two modern human populations used in computing D-statistics are Tibetans and either CHB, CEU or YRI. See Supplementary Information titled “D statistics under Models of no gene flow” for more details. All simulations results result in a p-value < 0.001 for all comparisons (see Extended Data Fig. 9, Supplementary Tables S8–S10 and Supplementary Material “D statistics under models of no gene flow.”
Genome-wide value of D statistics
D-statistics have been employed to assess genome-wide levels of archaic introgression in previous studies14, 25. To assess whether Tibetans carry more Denisovan admixture than other populations (CEU or CHB), we used the SNP genotype data from Simonson et al. 201045 and computed D-statistics as in Durand et al. 201144: D(chimp, Denisovan, Tibetan and CHB) and D(chimp, Denisovan, Tibetan and CEU). At the genome-wide level, using the D-statistic, we found no evidence that there is more Denisovan admixture in Tibetans than in the Han (D = 0.000504688). We also did not find evidence that there is more Denisovan admixture in Tibetans than in the Europeans (D = 0.001898642).
Empirical distribution of D-statistics for 32.7kb intronic regions
The EPAS1 32.7kb region was chosen due to its positive selection signal, and not based on a genome-wide analysis of Denisovan introgression. Therefore, we only performed one test when testing for introgression and did not have to correct p-values for multiple testing. We do not have Tibetan whole genome sequence data, but as shown in the previous section, genotype array data suggests that the level of Denisovan introgression between Han and Tibetans is similar. Moreover, Tibetans and Han are closely related populations. Therefore, using Han data as a proxy, we can determine whether the observed D-values at the EPAS1 region (D(TIB, YRI, DEN, Chimp) = −0.8818433) is an outlier compared to the distribution of D-values at other 32.7kb intronic regions. Using the empirical distribution of D-values across chromosomes 1 to 22, substituting the 80 Han chromosomes for our 80 Tibetan chromosomes and computing D(HAN, YRI, DEN, Chimp) for each 32.7kb intronic region, we obtain a p-value < 0.008. However, as the variance in D depends on the number of informative sites, this is probably an overestimate of the true p-value. In fact, there are no other regions in the region with as many informative sites and as extreme a D-value as that observed for EPAS1. This region is clearly a strong outlier.
Null distribution of S* statistics under models of no gene flow, Extended Data Fig. 10a
As a final approach for eliminating the hypothesis of ancestral lineage sorting, we follow the methods of Vernot et al. 201423 to compute S* (originally derived by Plagnol et al. 200624). S* was designed to identify regions of archaic introgression. As in the previous section, we used all the 4 models of Sankararaman et al. 201243 that do not include gene flow and simulated data to compute the null distributions of S*. Distributions are generated from 1000 simulations, and within each simulation we have representation of the 80 Tibetan chromosomes, and 20 Yoruban chromosomes as the outgroup. For each simulated data set we follow Vernot et al. 201423 and compute S* on a per chromosome basis, after sampling at random 20 chromosomes from the Tibetan group and removing SNPs that are observed in the Yoruban chromosomes, and then the maximum S* is recorded. The above process is carried out for 10 random samplings of 20 Tibetan chromosomes and the maximum of the 10 is the final recorded S*. The exact same procedure is applied to the simulated data and the real data of 80 Tibetan chromosomes. Extended Data Fig. 10a shows that under all four models, S* is significantly different from the null distribution with all the empirical p-values lying below 0.035. The grey vertical line is the S* value computed for the real data. The p-values are 0.035, 0.028, 0.019 and 0.017 respectively for each model (top to bottom).
Principal Component Analyses using 1000 Genomes Chinese samples and Tibetans from Simonson et al. 2010, Extended Data Fig. 10b
Since one single CHB individual carries a haplotype that is very similar to the Denisovan haplotype in EPAS1 (Extended Data Fig. 7), we wanted to assess whether this similarity might be due to recent gene-flow from Tibetans to CHB. If that were true, then we would expect to observe similarities at other loci. Therefore we compute the first and second principal components using all of chromosome 2. For simplicity, we only used chromosome 2 because it contains the EPAS1 gene and has a sufficiently high number of SNPs to carry out the PCA analysis. We do not have genome-wide genotype calls for the 40 Tibetan samples considered in this study. Therefore, as a proxy, we used the Tibetan genotype data from Simonson et al. 201045 and compared their Tibetan samples to the CHB and CHS individuals from 1000 Genomes. Extended Data Fig. 10b shows that all the CHB and the CHS individuals cluster together and principal component 1 clearly separates Tibetans from CHB and CHS individuals. Furthermore, the CHB individual with the Denisovan EPAS1 haplotype (Extended Data Figures 6a and 6b) clearly clusters with other CHB and CHS individuals and do not show any closer genetic affinity with Tibetans. This suggests that the CHB individual with a Denisovan-like haplotype in EPAS1 is not a descendant of a recent immigrant from Tibet.
Supplementary Material
Extended Data Figure 1. FST calculated for each SNP between Tibetan and Han populations. Each dot represents the FST value for each SNP in EPAS1. The x-axis is the physical position in the gene. Positions are based on the hg18 build of the human genome. The green box defines a 32.7kb region where we observe the largest genetic differentiation between Han and Tibetans. The first and last positions of this 32.7kb region correspond to the first and last position of the SNPs listed in Table S3. For comparison, from Yi et al. 20104, the genome-wide FST between Han and Tibetans is 0.02. The site with the largest frequency difference (and therefore largest FST) is circled.
Extended Data Figure 2 Distribution of fixed differences. The left panel is the distribution of fixed differences between two haplotype groups under a scenario of selection on a de novo mutation (see Methods), and the right panel is the distribution under a scenario of selection on standing variation (see Methods) for a region of size ~32.7kb. The initial frequency of the selected allele in the SSV model is 1%. Each row of panels corresponds to different selection strength (2Ns) from 200 to 1000. The red lines mark the number of fixed differences observed between the two haplotype classes in the real data for the given window size.
Extended Data Figure 3. Haplotype frequencies for Tibetans, our Han samples and the populations from the 1000 genomes project for the 5-SNP motif in the EPAS1 region. The y-axis is the haplotype frequency. The legend shows all the possible haplotypes for the region considered among these populations: YRI, Yoruban; LWK, Luhya; ASW, African American from the South West; IBS, Iberian; TSI, Toscani; CEU, Utah Residents with Northern and Western European ancestry; GBR, British; FIN, Finnish; JPT, Japanese; CHS, Southern Han Chinese; CHB, Han Chinese from Beijing; HAN, Han Chinese from Beijing; TIB, Tibetan; MXL, Mexican; PUR, Puerto Rican; CLM, Colombian (see Methods for Extended Data Figure 3).
Extended Data Figure 4. Derived allele frequency of a subset of SNPs from Table S3 in the 1000 Genomes Project populations. At these SNPs, the frequency difference between Tibetans and the 1000 Genomes project populations is 0.65 or larger. Positions 46571435, 46579689, 46584859 and 46600358 were not called as SNPs in the 1000 Genomes data, so we assume these positions were fixed for the human reference allele. Note that even though position 46577251, 46588331, 46594122 and 46598025 appear to have a frequency of 0.0 for the populations in the 1000 Genomes data, the derived allele in these SNPs are observed at very low frequency in at least one population (e.g. CHB).
Extended Data Figure 5. Differences between haplotypes. a The full matrix of pairwise differences between all the unique haplotypes in Figure 3, for the 40 most common haplotypes identified in the 1000 Genomes and the Tibetan samples in the 32.7kb region of EPAS1. The Denisovan haplotype (of frequency two) was added afterwards for comparison. The unique haplotypes are labeled with Roman numerals (here and in Figure 3 of the main text), and the Denisovan haplotype is the first column, haplotype I. Refer to Figure 3 in the main text and the supplementary material for the representation of populations for each haplotype. b, Illustration of the genealogical structure in a model with gene-flow from Denisovans to Tibet. Letters (a–k) represent the lengths of the branches of the tree. The divergence between modern human haplotypes and the introgressed haplotype in Tibetans would be larger than the haplotypes in other modern human populations and the Denisovan haplotype (see Methods Section for Figure 3 and Supplementary Information titled “Extended Data Figure 5b”). Tib, CEU and YRI denote Tibetan, European and Yoruban populations. Note that the lengths i and k are unknown since we do not know when these populations went extinct.
Extended Data Figure 6. Other haplotype netwoks. a, A haplotype network based on the number of pairwise differences between 43 unique haplotypes defined from the 20 most differentiated SNPs between Tibetans and the 14 populations from the 1000 Genomes Project. The R software package pegas22 was used to generate the figure. The haplotype distances are from pairwise differences. Each pie chart represents one unique haplotype and the size of the pie chart is proportional to log2(number of chromosomes with that haplotype). The sections in the pie provide the breakdown of the haplotypes amongst populations. The width of the edges is proportional to the number of pairwise differences between the joined haplotypes; the thinnest edge width represents a difference of 1 mutation. The number 57 next to a Tibetan haplotype is the number of Tibetan chromosomes with that haplotype. Similarly, the number 1912 is the number of chromosomes (across several populations) with that haplotype. b, The number of pairwise differences between the Denisovan haplotype and the 43 unique haplotypes defined from the 20 most differentiated SNPs between Tibetans and the 14 populations from the 1000 Genomes Project (same haplotypes as in Panel a). Each bar is a unique haplotype, and they are sorted in increasing order of pairwise differences. The colors within each bar represent the proportion of chromosomes with that haplotype broken down by populations. The numbers on top of each bar represent the total number of chromosomes within the 1000 Genomes dataset and Tibetans that have the haplotype. Note this is the same data set used to create the haplotype network in panel a. Tables S5 and S6 contain the 43 haplotypes and the frequencies within each of the populations.
Extended Data Figure 7. Number of pairwise differences. Red bars are the histograms of the number of pairwise differences between Denisovan and Tibetans. Blue bars are the histograms of the number of pairwise differences between Denisovan and (GBR, CHS, FIN, PUR, CLM, IBS, CEU, YRI, CHB, JPT, LWK, ASW, MXL or TSI). All comparisons are within the 32.7kb region of high differentiation (green box in Extended Data Fig. 1).
Extended Data Figure 8. Divergence distributions. Modern human/Denisovan divergence (see Methods for Extended Data Fig. 8) for intronic regions of size 32.7kb is plotted in red. Modern human/modern human divergence for the same intronic regions is plotted in blue. At the EPAS1 32.7kb region, in green, we plot the Tibetan-Han divergence. The black arrow points to the number of nucleotide differences between the Denisovan and the most common Tibetan haplotype (0.0038). This value is significantly lower than what we observe between modern human/Denisovan (red curve, p-value =0.0028).
Extended Data Figure 9. Null distributions of D for an assumed Tibet-Han divergence of 3000 years. Each histogram corresponds to the D values obtained under null models without gene flow, and the red vertical bar corresponds to the D values observed in the real data. The observed D values are significant (p-value < 0.001) even when we assume Tibet-Han divergence of 5000 or 10000 years (see Methods section for Extended Data Figure 9 and Tables S8–S10). Model abbreviations are given in the supplementary section titled “D statistics under models of no gene flow.”
Extended Data Figure 10. S statistics and PCA plot. a, A measure of introgression, S*, from Vernot et al. 201423. Distributions are for 1000 simulations under the 4 demographic models described in the supplementary section titled “D statistics under models of no gene flow”. S* for the Tibetan individuals is shown as vertical grey line. For all models, the empirical p-values are 0.035, 0.028, 0.019 and 0.017 respectively for each model (top to bottom). b, Plots the first and second principal components using all the CHS (100 individuals) and the CHB (97 individuals) from the 1000 Genomes and the 77 Tibetan individuals from Simonson et al. 201045 (see Methods for Extended Data Fig. 10b). The black circle and the black triangle represent the single CHB and the CHS individuals carrying the 5-SNP Tibetan/Denisovan-haplotype (Extended Data Fig. 3). All SNPs in the intersection between the 1000 Genomes populations and the 77 Tibetan individuals from chromosome 2 were used for this analysis.
Acknowledgments
This research was funded by the State Key Development Program for Basic Research of China, 973 Program (2011CB809203, 2012CB518201, 2011CB809201, 2011CB809202), China National GeneBank-Shenzhen and Shenzhen Key Laboratory of Transomics Biotechnologies (NO.CXB201108250096A). This work was also supported by research grants from the US NIH (R01HG003229) to R.N. and to E.H.S (R01HG003229-08S2). We thank Flora Jay, Mason Liang and Fergal Casey for useful discussions.
Footnotes
Author Contributions
RN, Jian W and Jun W supervised the project. XJ, Asan, ZB, YL, XY, MH, PN, BW, XO, Huasang, JL, ZXPC, GG, YY, WW, XZ, XX, HY, YL, Jian W and Jun W collected and generated the data, and performed the preliminary bioinformatic analyses to call SNPs and indels from the raw data. EHS and NV filtered the data and BP phased the data. EHS performed the majority of the population genetic analysis with some contributions from BP and MS. EHS and RN wrote the manuscript with critical input from all the authors.
References
- 1.Moore LG, Young D, McCullough RE, Droma T, Zamudio S. Tibetan protection from intrauterine growth restriction (IUGR) and reproductive loss at high altitude. Am J Hum Biol. 2001;13:635–44. doi: 10.1002/ajhb.1102. [DOI] [PubMed] [Google Scholar]
- 2.Niermeyer S, et al. Child health and living at high altitude. Arch Dis Child. 2009;94:806–811. doi: 10.1136/adc.2008.141838. [DOI] [PubMed] [Google Scholar]
- 3.Wu T, et al. Hemoglobin levels in Quinghai-Tibet: different effects of gender for Tibetans vs. Han J Appl Physiol. 2005;98:598–604. doi: 10.1152/japplphysiol.01034.2002. [DOI] [PubMed] [Google Scholar]
- 4.Yi X, et al. Sequencing of 50 human exomes reveals adaptation to high altitude. Science. 2010;329:75–78. doi: 10.1126/science.1190371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bigham A, et al. Identifying signature of natural selection in Tibetan and Andean populations using dense genome scan data. PLoS Genet. 2010;6:e1001116. doi: 10.1371/journal.pgen.1001116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Simonson ST, et al. Genetic evidence for high-altitude adaptation in Tibet. Science. 2010;329:72–75. doi: 10.1126/science.1189406. [DOI] [PubMed] [Google Scholar]
- 7.Beall MC, et al. Natural selection on EPAS1 (HIF2a) associated with low hemoglobin concentration in Tibetan highlanders. Proc Natl Acad Sci USA. 2010;107:11459–11464. doi: 10.1073/pnas.1002443107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Peng Y, et al. Genetic variations in Tibetan populations and high-altitude adaptation at the Himalayas. Mol Biol Evol. 2011;28:1075–1081. doi: 10.1093/molbev/msq290. [DOI] [PubMed] [Google Scholar]
- 9.Xu S, et al. A genome-wide search for signals of high-altitude adaptation in Tibetans. Mol Biol Evol. 2011;28:1003–1011. doi: 10.1093/molbev/msq277. [DOI] [PubMed] [Google Scholar]
- 10.Wang B, et al. On the origin of Tibetans and their genetic basis in adapting high-altitude environments. PLoS One. 6:e17002. doi: 10.1371/journal.pone.0017002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moore LG, et al. Maternal adaptation to high-altitude pregnancy: an experiment of nature—a review. Placenta. 2004;25:S60–S71. doi: 10.1016/j.placenta.2004.01.008. [DOI] [PubMed] [Google Scholar]
- 12.Vargas EP, Spielvogel H. Chronic mountain sickness, optimal hemoglobin, and heart disease. High Alt Med Biol. 2006;7:138–49. doi: 10.1089/ham.2006.7.138. [DOI] [PubMed] [Google Scholar]
- 13.Yip R. Significance of an abnormally low or high hemoglobin concentration during pregnancy: special consideration of iron nutrition1′2′3. Am J Clin Nutr. 2000;72:272–279. doi: 10.1093/ajcn/72.1.272S. [DOI] [PubMed] [Google Scholar]
- 14.Meyer M, et al. A high-coverage genome sequence from an archaic Denisovan individual. Science. 2012;388:222–226. doi: 10.1126/science.1224344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li JZ, et al. Worldwide human relationships inferred from genome-wide patterns of variation. Science. 2008;319:1100–1104. doi: 10.1126/science.1153717. [DOI] [PubMed] [Google Scholar]
- 16.Rosenberg NA. Standardized subsets of the HGDP-CEPH Human Genome Diversity Cell Line Panel, accounting for atypical and duplicated samples and pairs of close relatives. Ann Hum Genet. 2006;70:841–847. doi: 10.1111/j.1469-1809.2006.00285.x. [DOI] [PubMed] [Google Scholar]
- 17.Soejima M, Koda Y. Population differences of two coding SNPs in pigmentation-related genes SLC24A5 and SLC45A2. Int J Legal Med. 2007;121:36–39. doi: 10.1007/s00414-006-0112-z. [DOI] [PubMed] [Google Scholar]
- 18.Sulem P, et al. Genetic determinants of hair, eye and skin pigmentation in Europeans. Nat Genet. 2007;39:1443–1452. doi: 10.1038/ng.2007.13. [DOI] [PubMed] [Google Scholar]
- 19.Coop G, et al. The role of geography in human adaptation. PLoS Genetics. 2009;5:e1000500. doi: 10.1371/journal.pgen.1000500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pickrell JK, et al. Signals of recent positive selection in a worldwide sample of human populations. Genome Res. 2009;19:826–37. doi: 10.1101/gr.087577.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McVean G, et al. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Paradis E. Pegas: an R package for population genetics with an integrated–modular approach. Bioinformatics. 2010;26:419–420. doi: 10.1093/bioinformatics/btp696. [DOI] [PubMed] [Google Scholar]
- 23.Vernot B, Akey J. Resurrecting Surviving Neandertal Lineages from Modern Human Genomes. Science. 2014 doi: 10.1126/science.1245938. [DOI] [PubMed] [Google Scholar]
- 24.Plagnol V, Wall JD. Possible Ancestral Structure in Human Populations. PLoS Genet. 2006;2(7):e105. doi: 10.1371/journal.pgen.0020105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reich D, et al. Genetic history of an archaic hominin group from Denisova cave in Siberia. Nature. 2010;468:1053–1060. doi: 10.1038/nature09710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Prufer K, et al. The complete genome sequence of a Neanderthal from the Altai Mountains. Nature. 2014;505:43–49. doi: 10.1038/nature12886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Skoglund P, Jakobsson M. Archaic human ancestry in East Asia. Proc Natl Acad Sci USA. 2011;108:18301–18306. doi: 10.1073/pnas.1108181108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Abi-Rached L, Jobin MJ, Kulkarni S, McWhinnie A, Dalva K, et al. The shaping of modern human immune systems by multiregional admixture with archaic humans. Science. 2011;334:89–94. doi: 10.1126/science.1209202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mendez FL, Watkins JC, Hammer MF. A haplotype at STAT2 introgressed from Neanderthals and serves as a Candidate of Positive selection in Papua New Guinea. Am J Hum Genet. 2012;91:265–274. doi: 10.1016/j.ajhg.2012.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sankararaman S, et al. The genomic landscape of Neanderthal ancestry in present-day humans. Nature. 2014 doi: 10.1038/nature12961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li R, Li Y, Kristiansen K, Wang J. SOAP: short oligonucleotide alignment program. Bioinformatics. 2008;24:713–714. doi: 10.1093/bioinformatics/btn025. [DOI] [PubMed] [Google Scholar]
- 32.Li R, et al. SNP detection for massively parallel whole-genome resequencing. Genome Res. 2009;19:1124–1132. doi: 10.1101/gr.088013.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Browning BL, Browning SR. A fast, powerful method for detecting identity by descent. Am J Hum Genet. 2011;88:173–182. doi: 10.1016/j.ajhg.2011.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Coop G, et al. The role of geography in human adaptation. PLoS Genetics. 2009;5:e1000500. doi: 10.1371/journal.pgen.1000500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Reynolds J, Weir BS, Cockerham CC. Estimation of the coancestry coefficient: basis for a short-term genetic distance. Genetics. 1983;105:767–779. doi: 10.1093/genetics/105.3.767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.R (http://r-cran.org)
- 37.Ewing G, Hermisson J. MSMS: a coalescent simulation program including recombination, demographic structure, and selection at a single locus. Bioinformatics. 2010;26:2064–2065. doi: 10.1093/bioinformatics/btq322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Myers S, et al. A Fine-Scale Map of Recombination Rates and Hotspots Across the Human Genome. Science. 2005;310:321–324. doi: 10.1126/science.1117196. [DOI] [PubMed] [Google Scholar]
- 39.Hinch, et al. The landscape of recombination in African Americans. Nature Genetics. 2011;476:170–177. doi: 10.1038/nature10336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Scally A, Durbin R. Revising the human mutation rate: implications for understanding human evolution. Nat Rev Genet. 2012;13:745–53. doi: 10.1038/nrg3295. [DOI] [PubMed] [Google Scholar]
- 41.Teshima KM, Innan H. mbs: modifying Hudson’s ms software to generate samples of DNA sequences with a biallelic site under selection. BMC bioinformatics. 2009;10:166. doi: 10.1186/1471-2105-10-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hudson RR. Generating samples under a Wright-Fisher neutral model of genetic variation. Bioinformatics (Oxford, England) 2002;18:337–338. doi: 10.1093/bioinformatics/18.2.337. [DOI] [PubMed] [Google Scholar]
- 43.Sankararaman S, et al. The Date of Interbreeding between Neandertals and Modern Humans. PLoS Genet. 2012;8(10):e1002947. doi: 10.1371/journal.pgen.1002947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Durand EY, et al. Testing for ancient admixture between closely related populations. Molecular Biology and Evolution. 2011;28:2239–2252. doi: 10.1093/molbev/msr048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Simonson ST, et al. Genetic evidence for high-altitude adaptation in Tibet. Science. 2010;329:72–75. doi: 10.1126/science.1189406. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Extended Data Figure 1. FST calculated for each SNP between Tibetan and Han populations. Each dot represents the FST value for each SNP in EPAS1. The x-axis is the physical position in the gene. Positions are based on the hg18 build of the human genome. The green box defines a 32.7kb region where we observe the largest genetic differentiation between Han and Tibetans. The first and last positions of this 32.7kb region correspond to the first and last position of the SNPs listed in Table S3. For comparison, from Yi et al. 20104, the genome-wide FST between Han and Tibetans is 0.02. The site with the largest frequency difference (and therefore largest FST) is circled.
Extended Data Figure 2 Distribution of fixed differences. The left panel is the distribution of fixed differences between two haplotype groups under a scenario of selection on a de novo mutation (see Methods), and the right panel is the distribution under a scenario of selection on standing variation (see Methods) for a region of size ~32.7kb. The initial frequency of the selected allele in the SSV model is 1%. Each row of panels corresponds to different selection strength (2Ns) from 200 to 1000. The red lines mark the number of fixed differences observed between the two haplotype classes in the real data for the given window size.
Extended Data Figure 3. Haplotype frequencies for Tibetans, our Han samples and the populations from the 1000 genomes project for the 5-SNP motif in the EPAS1 region. The y-axis is the haplotype frequency. The legend shows all the possible haplotypes for the region considered among these populations: YRI, Yoruban; LWK, Luhya; ASW, African American from the South West; IBS, Iberian; TSI, Toscani; CEU, Utah Residents with Northern and Western European ancestry; GBR, British; FIN, Finnish; JPT, Japanese; CHS, Southern Han Chinese; CHB, Han Chinese from Beijing; HAN, Han Chinese from Beijing; TIB, Tibetan; MXL, Mexican; PUR, Puerto Rican; CLM, Colombian (see Methods for Extended Data Figure 3).
Extended Data Figure 4. Derived allele frequency of a subset of SNPs from Table S3 in the 1000 Genomes Project populations. At these SNPs, the frequency difference between Tibetans and the 1000 Genomes project populations is 0.65 or larger. Positions 46571435, 46579689, 46584859 and 46600358 were not called as SNPs in the 1000 Genomes data, so we assume these positions were fixed for the human reference allele. Note that even though position 46577251, 46588331, 46594122 and 46598025 appear to have a frequency of 0.0 for the populations in the 1000 Genomes data, the derived allele in these SNPs are observed at very low frequency in at least one population (e.g. CHB).
Extended Data Figure 5. Differences between haplotypes. a The full matrix of pairwise differences between all the unique haplotypes in Figure 3, for the 40 most common haplotypes identified in the 1000 Genomes and the Tibetan samples in the 32.7kb region of EPAS1. The Denisovan haplotype (of frequency two) was added afterwards for comparison. The unique haplotypes are labeled with Roman numerals (here and in Figure 3 of the main text), and the Denisovan haplotype is the first column, haplotype I. Refer to Figure 3 in the main text and the supplementary material for the representation of populations for each haplotype. b, Illustration of the genealogical structure in a model with gene-flow from Denisovans to Tibet. Letters (a–k) represent the lengths of the branches of the tree. The divergence between modern human haplotypes and the introgressed haplotype in Tibetans would be larger than the haplotypes in other modern human populations and the Denisovan haplotype (see Methods Section for Figure 3 and Supplementary Information titled “Extended Data Figure 5b”). Tib, CEU and YRI denote Tibetan, European and Yoruban populations. Note that the lengths i and k are unknown since we do not know when these populations went extinct.
Extended Data Figure 6. Other haplotype netwoks. a, A haplotype network based on the number of pairwise differences between 43 unique haplotypes defined from the 20 most differentiated SNPs between Tibetans and the 14 populations from the 1000 Genomes Project. The R software package pegas22 was used to generate the figure. The haplotype distances are from pairwise differences. Each pie chart represents one unique haplotype and the size of the pie chart is proportional to log2(number of chromosomes with that haplotype). The sections in the pie provide the breakdown of the haplotypes amongst populations. The width of the edges is proportional to the number of pairwise differences between the joined haplotypes; the thinnest edge width represents a difference of 1 mutation. The number 57 next to a Tibetan haplotype is the number of Tibetan chromosomes with that haplotype. Similarly, the number 1912 is the number of chromosomes (across several populations) with that haplotype. b, The number of pairwise differences between the Denisovan haplotype and the 43 unique haplotypes defined from the 20 most differentiated SNPs between Tibetans and the 14 populations from the 1000 Genomes Project (same haplotypes as in Panel a). Each bar is a unique haplotype, and they are sorted in increasing order of pairwise differences. The colors within each bar represent the proportion of chromosomes with that haplotype broken down by populations. The numbers on top of each bar represent the total number of chromosomes within the 1000 Genomes dataset and Tibetans that have the haplotype. Note this is the same data set used to create the haplotype network in panel a. Tables S5 and S6 contain the 43 haplotypes and the frequencies within each of the populations.
Extended Data Figure 7. Number of pairwise differences. Red bars are the histograms of the number of pairwise differences between Denisovan and Tibetans. Blue bars are the histograms of the number of pairwise differences between Denisovan and (GBR, CHS, FIN, PUR, CLM, IBS, CEU, YRI, CHB, JPT, LWK, ASW, MXL or TSI). All comparisons are within the 32.7kb region of high differentiation (green box in Extended Data Fig. 1).
Extended Data Figure 8. Divergence distributions. Modern human/Denisovan divergence (see Methods for Extended Data Fig. 8) for intronic regions of size 32.7kb is plotted in red. Modern human/modern human divergence for the same intronic regions is plotted in blue. At the EPAS1 32.7kb region, in green, we plot the Tibetan-Han divergence. The black arrow points to the number of nucleotide differences between the Denisovan and the most common Tibetan haplotype (0.0038). This value is significantly lower than what we observe between modern human/Denisovan (red curve, p-value =0.0028).
Extended Data Figure 9. Null distributions of D for an assumed Tibet-Han divergence of 3000 years. Each histogram corresponds to the D values obtained under null models without gene flow, and the red vertical bar corresponds to the D values observed in the real data. The observed D values are significant (p-value < 0.001) even when we assume Tibet-Han divergence of 5000 or 10000 years (see Methods section for Extended Data Figure 9 and Tables S8–S10). Model abbreviations are given in the supplementary section titled “D statistics under models of no gene flow.”
Extended Data Figure 10. S statistics and PCA plot. a, A measure of introgression, S*, from Vernot et al. 201423. Distributions are for 1000 simulations under the 4 demographic models described in the supplementary section titled “D statistics under models of no gene flow”. S* for the Tibetan individuals is shown as vertical grey line. For all models, the empirical p-values are 0.035, 0.028, 0.019 and 0.017 respectively for each model (top to bottom). b, Plots the first and second principal components using all the CHS (100 individuals) and the CHB (97 individuals) from the 1000 Genomes and the 77 Tibetan individuals from Simonson et al. 201045 (see Methods for Extended Data Fig. 10b). The black circle and the black triangle represent the single CHB and the CHS individuals carrying the 5-SNP Tibetan/Denisovan-haplotype (Extended Data Fig. 3). All SNPs in the intersection between the 1000 Genomes populations and the 77 Tibetan individuals from chromosome 2 were used for this analysis.