Abstract
Elevated levels of homocysteine (Hcy), known as hyperhomocysteinemia (HHcy), is an independent risk factor of various diseases. Clinical studies report that people born with severe HHcy develop skeletal malformations with weaker bone. Studies also report that altered mitochondrial dynamics and altered epigenetics contribute to weaker bones and bone diseases. Although Hcy-induced mitochondrial dysfunction has been shown to affect bone metabolism, the role of mitochondrial epigenetics (mito-epigenetics) has not been studied in bones. The epigenetics in mitochondria is interesting as the mitochondrial genome size is small (16 kb) with fewer CpG, and without histones and introns. Recently, fascinating works on epigenetics along with the discovery of histone-like proteins in mitochondria are giving exciting areas for novel studies on mitochondria epigenetics. There are mutual cause and effect relationships between bone, mitochondria, Hcy, and epigenetics but unfortunately, studies are lacking which describe the involvement of all these together in bone disease progression. This review describes the reciprocal relationships and mechanisms of Hcy-bone-mitochondria-epigenetics along with a short discussion of techniques which could be employed to assess Hcy-induced anomaly in bone, mediated through alterations in mitochondrial epigenetics.
Keywords: Bone, DNA methylation, Epigenetics, Homocysteine, Mitochondria
1. Introduction
Many clinical and basic studies have shown the relationship between high homocysteine (Hcy) levels in serum and impaired bone health in children, adolescents, postmenopausal woman and the elderly. Hcy is being proved as an independent risk factor of various diseases including bone diseases (osteoporosis, arthritis, fracture risk, and bone deformity) [1–3]. Studies also report that hyperhomocysteinemia (HHcy), an elevated level Hcy, affect mitochondria dysfunction and dynamics and that this process confirms the association between mitochondrial DNA, oxidative stress and premature bone fragility [4, 5]. Mitochondria are the power house of the cells; they generate energy in the form of adenosine triphosphates (ATPs), and they are five times more prone to oxidative stress [6]. Mitochondria contain their own genome, also called as nucleoid, which is around 16 kb in size and does not contain histones and introns. Despite this, reports suggest that epigenetics play an important role in mitochondrial metabolism as well as other mechanisms. Interestingly, recent studies determined the role of DNA methylation, hydroxymethylation, and microRNA based mechanisms which regulate mitochondrial functional integrity. The discovery of histone-like bodies in mitochondria also raises interest towards hidden molecules and mechanisms regulating mitochondria dynamics [7]. Bone has been studied with respect to mitochondrial aspects but unfortunately these studies are not directed towards epigenetic alterations induced with HHcy. These studies could certainly benefit from the discovery of novel mitochondrial epigenetic mechanisms and biomarkers for bone diseases. Hence, the current review describes the mutual associations of Hcy, mitochondria, epigenetics and bone. This review also suggests the possible mitochondrial epigenetic (mito-epigenetic) mechanisms that impart bone weakness, disease progression and affect bone metabolism. At last, potential techniques are discussed which could be employed finding epigenetic mechanisms in impaired bone health.
2. Historical perspective and effect of homocystinuria on bone
The first study that reported the link between Hcy and pathological changes in bones, in patients with homocystinuria, was done by McKusick (1966). Homocysteinuria is a genetic disorder caused by mutation in cystathionine-beta-synthase (CBS) gene [8]. Deficiency of the gene led to high level of Hcy in plasma. Homocystinuria manifests as a distinctive spondylo-epimetaphyseal dysplasia which is also characterized by skeletal deformities, osteopaenia, elongated appendicular skeleton and flattening of the vertebral bodies [9–11]. A recent study described that patients with good metabolic control of homocystinuria with a low-methionine diet prevented bone anomalies with an adequate BMD gain [12]. A meta-analysis study provided prospective evidence of Hcy association with significant increase of fracture risk. The study demonstrated elevated Hcy increased risk of all fractures (RR 1.59, 95% CI 1.30–1.96) and hip fracture (RR 1.67, 95% CI 1.17–2.38). The same study also described that the risk was more pronounced in male subjects [3]. The study done with 3502 subjects-years of follow-up determined 103 individuals with at least one osteoporotic fracture and the association of osteoporotic fractures with quartiles of Hcy (p=0.047) [13]. A study with 1,475 post-menopausal women concluded significant risk to vertebral fracture with high plasma Hcy levels (OR = 1.27, 95 % CI 1.04–1.58, p = 0.021) [2]. Herrmann et al. have explored the role of HHcy on bone remodeling primarily by decrease in osteoblast activity, increase in osteoclast activity, alteration in extracellular matrix, and decrease in blood flow [10, 14, 15]. There have been various mechanisms described for the effect of Hcy on bone. Mice fed on high Hcy diet showed high serum Hcy concentration (102.2 +/− 64.5 micromol/l) as compared to normal standard diet fed mice (2.8 +/− 1.5 micromol/l) and appear to have impaired fracture healing [16]. The study from our lab on genetic hyperhomocysteinemic mice (CBS+/−) showed decrease in bone-blood flow and increase in NOX-4, iNOS (inducible niric oxide synthase), MMP-9 (matrix metalloproteinase) protein, MMP9 activity along with decrease in Trx-1 (thioredoxin), eNOS (endothelial nitric oxide synthase) and NO (nitric oxide) bioavailability [17]. HHcy has been described as altering the redox regulatory mechanism in the osteoblast by activating PP2A and deranging FOXO1 and MAPK signalling cascades that eventually shifted the OPG:RANKL (Receptor activator of nuclear factor kappa-B ligand) ratio toward increased osteoclast activity and decreased bone quality [18]. Hcy lowering treatment, folate and vitamin B12, have shown benefits, with reduced risk of hip fracture [19]. Elevated homocysteine also activated JNK signal via ROS and induced the apoptosis of bone marrow mesenchymal stem cells (BMSCs) [20]. Collagen cross-linkers are determinants of bone quality and Hcy is known to affect collagen cross link by decreasing enzymatic cross link and increasing non-enzymatic cross link [21]. A study with 10 homocystinuric patients determined significantly lower collagen I C terminal telopeptides (ICTP) which is a cross-linked telopeptide from the C-terminal non-helical part of collagen I [22]. Treatment of Hcy on MC3T3-E1 (pre-osteoblast cells) for 21 days led to significant decreases in collagen cross-linking along with activation of PTK2-PXN-CTNNB1 pathway [23]. Hcy treatment to osteoblastic cells led to an increase in release of mitochondrial cytochrome c. Treatment also triggered ER stress by increased expression of glucose-regulated protein 78, inositol-requiring transmembrane kinase and endonuclease 1α (IRE-1α), spliced X-box binding protein, activating transcription factor 4, and C/EBP homologous protein [24]. There is a significant correlation found between serum and bone homocysteine level and increased level of homocysteine in bone was also found to affect bone morphology [1].
Bone mineral density (BMD), a diagnostic parameter of osteoporosis, is significantly decreased with elevated levels of Hcy. Hence, homocysteinuria increase the risk of osteoporosis by decreasing BMD. Numerous reports also suggest that elevated plasma Hcy reduces BMD and increases fracture risk [25–28]. On the contrary, a few epidemiological studies have determined that no significant relationship exists between plasma Hcy and fracture risk [26, 29]. Because of contradictory results with the studies, more studies are required to come to a final conclusion. The Hcy-induced fracture risk could be assessed by analyzing bone metabolism markers which are divided into bone formation (alkaline phosphatase, pro-collagen type I N and C-terminal peptides, osteocalcin) [15, 25, 29–31], and bone resorption (collagen I, beta-crosslaps, urinary deoxypyridinoline crosslinks, tartrate resistant acid phosphatase 5b) [30, 32, 33]. However, metabolism anomalies arise after disturbance of normal axis between bone formation and resorption. Existing clinical and animal studies report the action of Hcy is through shifting the bone metabolism to bone resorption. Kim et al. have shown the effect of Hcy on primary human osteoclasts collected from 10 healthy male donors. They determined significant stimulation of resorption activity. Significant increase in tartrate-resistant acid phosphatase (TRAP; maximum increase of 24%) and cathepsin K (CK; maximum increase of 24%) activity confirmed increase rate of bone resorption with exogenous Hcy administration with decreasing concentrations of folate, B6 and B12 markers [14].
3. Bone and mitochondrial metabolism
The number and activity of mitochondria differs in different cells dependent upon energy demand of the cell type. Bone cells contain a large number of mitochondria and mitochondrial dysfunction may contribute to bone impairment. It was reported that the content of mitochondria was found to be decreasing in periosteum osteoblasts during the life span of the rat from weeks 1 to 104 [34]. In another study authors have measured the levels of mRNA derived from mitochondrial genes in healing fractures of young, adult, and old rats and found that older rats exhibit reduced mitochondrial gene expression during fracture repair [35]. In humans, few studies have addressed association between mitochondrial DNA, oxidative stress and premature bone fragility [4, 5]. At the tissue level bone metabolism is linked to coordinated function of osteocytes, osteoblasts and osteoclasts. Osteoblasts have been referred to as ‘bone builders’ and their increased activity was found to increase number of mitochondria during bone formation. Cultured osteoblasts can produce energy via glycolysis, citric acid cycle and beta oxidation pathways [36, 37]. Besides energy regulation mitochondria also regulate life of bone cells and thus mitochondria health is important as well for proper bone functioning.
Osteoblast metabolism is based on the generation of products of bone matrix production and alkaline phosphatase activity. It has been reported that glucose utilization is proportional to alkaline phophatase activity as measured by using [(18)F]-fluorodeoxyglucose in human osteoblast-like cells in vitro [38]. The primary osteoblast cells derived from fetal rat calvaria were kept on differentiation with ascorbate treatment and bioenergetics was measured. The rate of respiration, ATP production and ATP content were found to be increased after 14 days of osteoblast differentiation as compared to immature cells. Likewise as compared to immature cells, mature cells were reported to have high-transmembrane-potential mitochondria as measured by using JC-1 (5,5′,6,6′-tetrachloro-1,1′,3,3′-tetraethylbenzimidazolylcarbocyanineiodide) dye uptake [39]. Proper mitochondrial function is linked to the maintenance of the phosphocreatinine pool which is generated from creatinine by mitochondrial creatine kinase. Phosphocreatinine then diffuses from mitochondria via voltage dependent anion channel to the cytosol where creatinine kinase can generate ATP for utilization. Creatinine kinase is therefore a pivotal enzyme for cellular energy metabolism for of osteoblasts and it has been seen that supplementation of creatinine to cultured osteoblast not only increases metabolic activity but also enhances their survival [40–42]. Mitochondria also produce citrate which is important to maintain bone nanostructure since citrate covers approximately one-sixth of the hydroxyapatite surface [43]. Another important enzyme which is produced by mitochondria of osteoblasts, is cytochrome c oxidase which determines apoptotic events in osteoblast cells [44–46].
Mitochondrial quality is an important determinant of osteogenic differentiation capacity and viability of human mesenchymal stem cells (hMSCs) [47]. Different reports suggest the metabolic status of cells during differentiation including few studies on mitochondrial bioenergetics and regulation at the time of bone cells differentiation. Chen et al. reported that undifferentiated hMSCs showed high level of glycolytic enzymes and lactate production while differentiating hMSCs exhibited more mitochondrial DNA copy number, protein subunits of respiratory enzyme, rate of oxygen consumption and intracellular ATP content. The increase in mitochondrial bioenergetics indicated upregulation of aerobic mitochondrial metabolism [48]. An et al. showed increase in mitochondrial biogenesis during Wnt-induced osteblastic differentiation of murine mesenchymal C3H10T1/2 cells. However, the biogenesis was significantly attenuated with zidovudane which is an inhibitor of mitochondrial biogenesis [49]. Peroxisome proliferator-activated receptor gamma coactivator-1alpha (PGC-1α) is a known regulator of mitochondrial biogenesis and together with NR4A/NGF-1β orphan nuclear receptor (NR) regulated primary mouse osteoblasts gene expression upon treatment with parathyroid hormone [50]. Osteoblast also differentiated into osteocytes which are relatively inert cells but capable of molecular synthesis and modification, as well as transmission of signals over long distances [51]. Osteoclasts also have high energy demands and therefore, they are rich in mitochondria [52]. The differentiation of osteoblasts to osteocytes led to increased expression of glycolytic enzymes and mitochondrial heat shock protein 70 (HSP 70). Osteoclasts are prone to mitochondrial dysfunction due to their high susceptibility for ROS.
3.1. Homocysteine, bone and mitochondria
One study reported that HHcy contributes to the development of osteoporosis by reducing bone formation. This study demonstrated that Hcy induced apoptosis in primary human bone marrow stromal cells (hBMSC) and the human bone marrow stromal cell line (HS-5) was mitochondrial dependent since caspase-3 and caspase-9 were involved. The same study confirmed that Hcy increased cytochrome c, ROS and activated the NF-κB pathway [53]. We earlier proposed that Hcy increases ROS that directs mitochondrial membrane permeability transition (MPT) that further allows the extrusion of proteins through mitochondria in osteocytes [54]. We also proposed the pathway of MMPs activation in bone remodeling during HHcy [54]. MMPs play a very important role in bone turnover processes. Different MMPs, for example MMP-1, -2, -3, -9, -10, 12, -13, 14, are expressed in different bone processing and execute specific functions [55]. Hcy is well known to activate MMPs and thus probably involve proteases in mediating bone pathogenesis [54, 56]. Moreover, absence of enough studies in Hcy mediated bone remodeling through mitochondria warranted important studies to fill out the knowledge gap in the proposed area.
4. Epigenetics underlying bone metabolism
Epigenetics is the study of heritable changes in gene activity. The study does not involve changes in the DNA nucleotide sequence but affects on gene expression. Tight control of genetic mechanisms occur under the influence of several important epigenetic mechanisms, for example; DNA methylation, non-coding RNA (microRNA), histone modification for example ubiquitylation, ADP ribosylation, biotinylation, and sumoylation. These processes not only control the genetic regulation but also control development, cell differentiation and aging. Besides, these modifications can also be treated as marker for the disease event and termed as epigenetic biomarker.
4.1. Role of DNA methylation
There have been several studies reported for these modifications in bone and bone cells. Investigators have determined that in case of osteoarthritis (OA) the level of MMP-13 is increased in OA chondrocytes which is associated with demethylation of CpG sites in the MMP-13 promoter [57]. In another study in human chondrocytes, authors have find out demethylation of MMP13 at −110 bp and −299 bp which was associated with increased levels of MMP-13 gene expression [58]. Another gene which is found to be associated with catabolic activity in cartilage is the inducible nitric oxide synthase (iNOS). The association between demethylation of specific NF-kappaB-responsive enhancer elements and the activation of iNOS transactivation in human OA chondrocytes was seen which was consistent with the differences in methylation status observed in vivo in normal and human OA cartilage and, importantly, show association with the OA process [59]. The pattern of methylation (hypo-, or hyper-mehylation) have earlier been seen in rheumatoid arthritis synovial fibroblast (RASF) by Nakano et al [60]. Besides that an altered methylation status in rheumatoid arthritis have been found for many genes such as; DR3 [61], IL6 [62, 63], IL10 [64, 65], IL1R2 [65] and CXCL12 [66]. The functional consequence of altered DNA methylation is also evident by one of the earlier studies that explained the increased risk to develop RA (rheumatoid arthritis) in women. Authors of this study observed the pattern of X chromosome inactivation and found that female RA patients exhibited increased skewed X chromosome inactivation pattern as compared to controls [67]. Few recent studies in systemic lupus erythematosus (SLE) enlighten the significance of hypomethylated genes that were concerned with various pathways of SLE pathogenesis such as; defence response, cell activation and proliferation, immune response and cytokine production [65]. Besides that genome wide DNA methylation arrays identified global DNA methylation changes in SLE T-cells as compared to healthy T cells [68]. In addition, differentially methylated genes were found to be having potential to be used as epigenetic biomarkers for SLE pathogenesis [69].
4.2. Role of histone modifications
Histone modification is a dynamic process that leads to chromatin remodeling and modulate gene expression. Histones (H2A, H2B, H3 and H4) form an octamer complex and connected through H1 and around this DNA wraps and packaged. The histones could be modified by several post translational modifications, such as: acetylation, methylation, phosphorylation, ADP ribosylation, sumoylation and biotinylation. The central mechanism of histone modification is controlled by histone acetylation/deacetylation which further control gene expression. Histone acetyltransferases (HAT) loosens the histone-DNA interactions, relaxes the chromatin and favours transcription of genes while histone deacetylases (HDAC) increases histone-DNA interaction, chromatin condensation and attenuates transcription machinery [70]. HDAC inhibitors were profoundly used to alleviate osteoblast and osteoclast differentiation such as sodium butyrate, valproic acid, MS-275, and TSA [71, 72]. These agents were found to influence differentiation by influencing genes associated with osteoblast/osteoclast differentiation (e.g., type I collagen, bone sialoprotein, osteopontin, osteocalcin, ALPL, OSX and RUNX2 cathepsin K, calcitonin receptor and OSCAR).
4.3. Role of microRNA
MicroRNA (miRNA) are 20–22 nucleotides long, processed from stem loop pre-miRNAs to mature miRNAs, and regulate a large number of target genes by binding to the 3′ region of their corresponding messenger RNA (mRNA). There are many reports available for many miRs regulating osteoblast differentiation [73]. Some of them are miR-30, mir433, and miR-335 by directly targeting osteogenic transcription factor RUNX2 [74–76]. Likewise miRs also regulate osteoclast differentiation for example miR-21 by targeting programmed cell death 4 (PDCD4) protein levels [77] and miR-155 by targeting microphthalmia associated transcription factor (MITF) [78].
5. Epigenetics and Mitochondria
The mitochondria genome size is 16,569bp, represents less than 1% of total cellular DNA, encodes 13 proteins in the electron transport chain (ETC) and the displacement loop (D-loop), two rRNA (rRNAs) and 22 tRNAs necessary for the translation of mitochondrial genes. Mitochondrial gene products are essential for normal cellular function. More than 200 different molecular defects have been reported in patients with mitochondrial diseases. The number of mitochondria varies from tissue to tissue. It contains fewer CpG dinucleotides (435) than genomic DNA and lacks the conventional CpG islands that are found in genomic DNA. Furthermore, mitochondrial genome doesn’t contain retrotransposons (LINE-1 and Alus), which are abundantly (~40%) present in human genome. The lack of histones in mitochondrial genome also suggests an important role for DNA methylation in mitochondrial DNA (mtDNA) stability and function. It is earlier reported that mtDNA was less methylated, as compared to nuclear DNA, in loach embryos and the same study reported presence of DNMT activity in mitochondria [79]. After that mtDNA methylation was reported in human [80] and mammals [81]. The mitochondrial DNMT was found different than nuclear one [82]. MtDNA methylation is done by means of 5-methylcytosine and 5-hydroxymethylation and has been reported in many disease related studies [83].
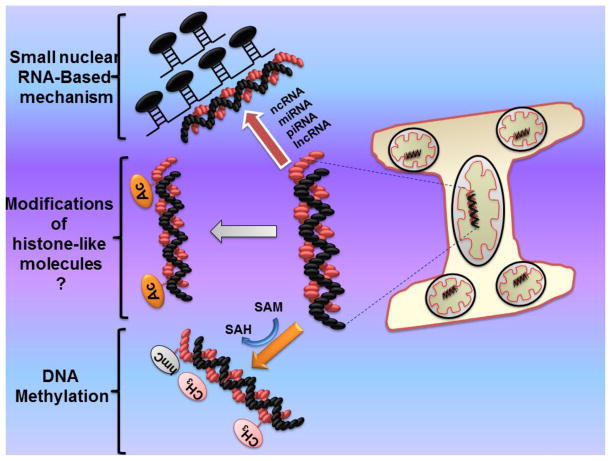
Apart from this many studies report role of mtDNA methylation in different disease progression. The absence of histones in mitochondria did not open the epigenetic area of histone modifications; however a report suggests the presence of histone family members in mitochondria [7]. More studies are warranted to come with existing contradictory reports and explore histone-based modifications in mitochondria. Likewise there is no report, to date, that identifies mitochondria-allied disorders by directly affecting microRNAs. Nevertheless microRNA have been seen to be involved in mitochondrial metabolism regulations, for example miR-210 is shown to regulate electron transport chain and tri-carboxylic acid cycles [84], and miR-30 is observed for its role in mitochondrial fission targeted through p53 and dynamin-related protein-1 (DRP-1) [12]. In rats, a small pool of microRNA, identified in highly purified liver-derived mitochondria, appears to regulate the expression of genes related to apoptosis, cell proliferation, and cellular differentiation [85]. Fig. 1 describes the several mechanisms associated to mitochondria epigenetic regulations.
Fig. 1.
Possible epigenetic mechanisms in bone mitochondria. Here; ncRNA, non-coding RNA; miRNA, micro RNA; piRNA, piwi RNA; lnc, long non-coding RNA; SAM, s-adenosylmethionine; SAH, s-adenosylhomocysteine; hmC, hydroxymethycytosine
5.1. Hcy-bone-mitochondria-epigenetics
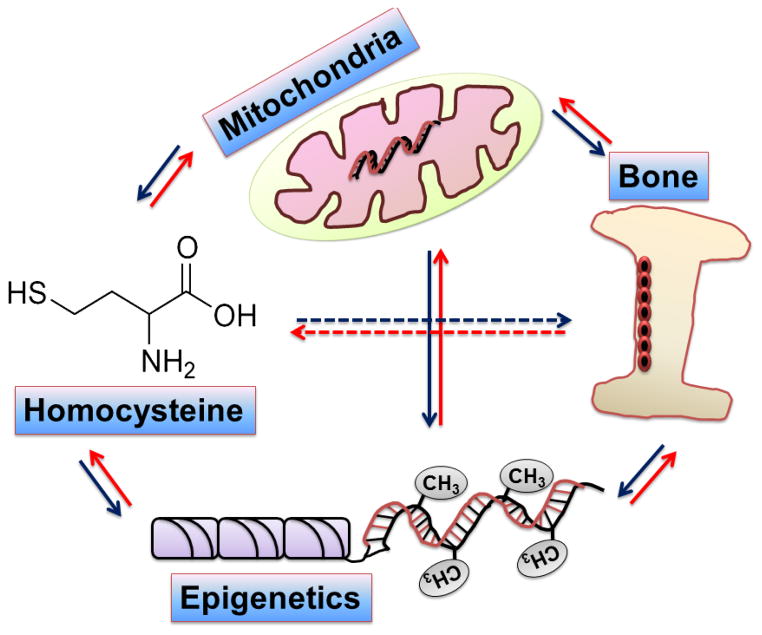
The previous studies showed the relations in between Hcy, epigenetics, bone, and mitochondria (connection showed in fig. 2), but the reports are lacking which unite these together to direct research in bone anomaly-induced by homocysteinuria and mediates through alterations of mitochondrial epigenetics. We and others have already established that Hcy influence epigenetic mechanisms along with mitochondrial dysfunctions and therefore; research is required to find out their role in bone weakness during homocysteinuria. The research will provide a novel milestone in the bone science.
Fig. 2.
Figure showing mutual relations between homocysteine, bone, mitochondria and epigenetics.
6. Supporting techniques
Performing epigenetics studies in bone mitochondrial genome is not easy as in the nuclear genome. One should have to be very careful from isolating purified mitochondria to perform epigenetic studies in bone mitochondria. Another difficulty is the mitochondria genome size which is reported to consist of fewer CpG and is devoid of histones and introns. There are several techniques which can be used to perform Hcy-induced epigenetic modifications in bone mitochondria:
6.1. Bisulphite sequencing
This technique is used to check bisulphite modification of DNA and used to differentiate between cytosine bases in methylated CpG and unethylated CpG sites. Methylated cytosine bases have a methyl group bound to the carbon-5 position of their pyrimidine ring. With bisulphite treatment 5-mC (5-methylcytosine) residues are protected but cytosine residues are converted to uracil. A number of downstream applications can be employed to estimate the level of methylation in the given sample such as sequencing of methylated DNA and Polymerase chain reaction specific to methylated region. The technique is advantageous in order to assess even a single CpG site and is relatively quick to perform. Another advantage of bisulphite modification is that it does not differentiate between 5-mC and 5-hydroxymethyl cytosine (5-hmC). The drawback of bisulphite sequencing is that the technique does not estimate the percentage of methylation at a given site.
6.2. Methylation-based arrays
For high throughput analysis of methylated DNA, the arrays specific for methylated DNA are available. Although these arrays are specific to bind nuclear targets but those targets may be specific to bone mitochondrial gene functions.
6.3. Biochemical epigenetic assays
Epigentek (Epigentek, Farmingdale, NY, USA) has launched several kits for determining DNA, RNA methylation, DNA methyltransferase assay, DNA demethyase assay, methylated DNA immunoprecipitation, mehylated DNA quantification (5-hydroxymethylation assay, 5-mthylacytosine estimation), and methyl-DNA binding protein Chip estimation. There are several other kits available to quantify histone modification too. Depending upon the need these kits could be explored for their use in bone mitochondrial epigenetics projects.
6.4. MicroRNA profiling
The profiling of microRNA in test sample can be performed through array-based methods and compared to control sample. This will give some microRNA which could be associated to mitochondrial epigenetics. Finding individual microRNA and their downstream targets could further throw light for individual important microRNA for the research projects. After that quantitative PCR methods would benefit for assessing individual microRNA role in bone disease progression. The use of mimics (microRNA) and anti-microRNA (antagomiR or miR inhibitors) designed for individual microRNA would further confirm the regulation of microRNA for mitochondria-related bone diseases.
6.5. Chromatin modifications
Although mitochondrial DNA lacks histones, but packed with proteins to form nucleoids. A Chip assay can be designed to analyze protein-DNA interactions followed by downstream analysis of the purified DNA/protein and could be useful for mitochondrial epigenetics in bone.
6.6. Transcriptome sequencing
The purified bone mitochondrial RNA can be explored by next generation sequencing to identify relative transcript abundance, RNA variation and post-translational modifications. A relatively small genome could further be analyzed for global schema of polycisron cleavage using parallel analysis of RNA ends (PARE).
7. Summary and future directions
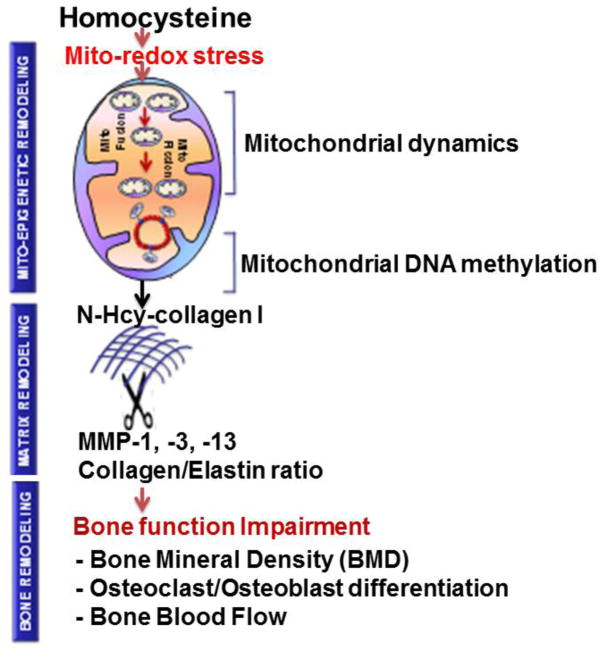
In summary, we have discussed the relationship of Hcy, bone, mitochondria and epigenetics. Although it is reported that homocystenuria leads to dysfunction of bone, the epigenetic pathways in bone mitochondria that lead to bone pathology have not been discussed. This suggests that studies should focus in the direction of Hcy-induced mitochondrial epigenetics in bone and these studies could explore novel mechanisms during disease pathology. For example, we are proposing a mechanism of mito-epigenetic remodeling that lead to bone function impairments during HHcy as shown in fig. 3. HHcy causes mito-redox stress that alters mitochondrial dynamics and mito-epigenetics (DNA methylation, DNA hydroxymethylation) concomitant with N-homocysteinylation of collagen 1. These altered events affect MMPs/TIMPs axis along with collagen/elastin ratio and eventually impairs bone structurally and functionally.
Fig. 3.
Schematic representation of proposed mechanism of mito-epigenetic remodeling and change in bone structure/function during genetic hyperhomocysteinemia. The central hypothesis of this proposal is that HHcy contributes to mito-epigenetic remodeling (DNA methylation, DNA hydroxymethylation) mediated bone-matrix feebler through N-Hcy-collagen 1, in part, by altering mitochondrial remodeling. These events disturb normal MMPs/TIMPs axis, collagen/elastin ratio, and eventually led to bone function impairments which could be determined by bone mineral density, assessing osteoclast/osteoblast differentiation and bone blood flow.
Here, MMP, matrix metalloproteinases; TIMP, tissue inhibitor of matrix metalloproteinases; mito-fusion, mitochondrial fusion; mito-fission, mitochondrial fission.
In conclusion, we highlighted interactions between Hcy, bone, mitochondria, and epigenetics and presented possible mechanisms for disease progression. We also discussed in short epigenetics-related techniques which can be employed to find out epigenetic alterations in mitochondria during Hcy-induced bone anomaly. Identifying these important pathways and epigenetic molecules would certainly benefit epigenetic molecular targets that could be used to attenuate disease pathology or to be used as biomarker for bone diseases. In our opinion, epigenetic approaches with bone mitochondria during HHcy are very promising and further studies are required to determine pathways/molecules that would improve bone disease and alleviate therapeutic directions.
Acknowledgments
A part of this work was supported by National Institutes of Health grants HL71010-NT and NS-084823-SCT.
Abbreviations
- Hcy
homocysteine
- HHcy
hyperhomocysteinemia
- Mito-epigenetics
mitochondrial epigenetics
- ATP
adenosine triphosphate
- CBS
cystathionine-beta-synthase
- BMD
bone mineral density
- BMSCs
bone marrow mesenchymal stem cells
- ICTP
collagen I C terminal telopeptides
- TRAP
tartrate-resistant acid phosphatase
- CK
cathepsin K
- JC-1
5,5′,6,6′-tetrachloro-1,1′,3,3′-tetraethylbenzimidazolylcarbocyanineiodide
- hMSCs
human mesenchymal stem cells
- MPT
membrane permeability transition
- MMP
matrix metalloproteinase
- DNMT
DNA methyltransferase
- iNOS
inducible nitric oxide synthase
- eNOS
endothelial nitric oxide synthase
- RASF
rheumatoid arthritis synovial fibroblast
- SLE
systemic lupus erythematosus
- HAT
Histone acetyltransferases
- HDAC
histone deacetylases
- PDCD4
programmed cell death 4
- MITF
microphthalmia associated transcription factor
- D-loop
displacement loop
- mtDNA
mitochondrial DNA
- DRP-1
dynamin-related protein-1
- PARE
parallel analysis of RNA ends
- 5-hmC
5-hydroxymethycytosine
- 5-mC
5-methylcytosine
Footnotes
Conflict of Interest: The authors declared no conflict of interest.
References
- 1.Holstein JH, Herrmann M, Splett C, Herrmann W, Garcia P, Histing T, Klein M, Kurz K, Siebel T, Pohlemann T, Menger MD. High bone concentrations of homocysteine are associated with altered bone morphology in humans. Br J Nutr. 2011;106:378–382. doi: 10.1017/S0007114511000304. S0007114511000304 [pii] [DOI] [PubMed] [Google Scholar]
- 2.Kuroda T, Tanaka S, Saito M, Shiraki Y, Shiraki M. Plasma level of homocysteine associated with severe vertebral fracture in postmenopausal women. Calcif Tissue Int. 2013;93:269–275. doi: 10.1007/s00223-013-9754-2. [DOI] [PubMed] [Google Scholar]
- 3.Yang J, Hu X, Zhang Q, Cao H, Wang J, Liu B. Homocysteine level and risk of fracture: A meta-analysis and systematic review. Bone. 2012;51:376–382. doi: 10.1016/j.bone.2012.05.024. S8756-3282(12)00942-8 [pii] [DOI] [PubMed] [Google Scholar]
- 4.Papiha SS, Rathod H, Briceno I, Pooley J, Datta HK. Age related somatic mitochondrial DNA deletions in bone. J Clin Pathol. 1998;51:117–120. doi: 10.1136/jcp.51.2.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Varanasi SS, Francis RM, Berger CE, Papiha SS, Datta HK. Mitochondrial DNA deletion associated oxidative stress and severe male osteoporosis. Osteoporos Int. 1999;10:143–149. doi: 10.1007/s001980050209. 90100143.198 [pii] [DOI] [PubMed] [Google Scholar]
- 6.Byun HM, Baccarelli AA. Environmental exposure and mitochondrial epigenetics: study design and analytical challenges. Hum Genet. 2014;133:247–257. doi: 10.1007/s00439-013-1417-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Choi YS, Hoon JJ, Min HK, Jung HJ, Hwang D, Lee SW, Kim PY. Shotgun proteomic analysis of mitochondrial D-loop DNA binding proteins: identification of mitochondrial histones. Mol Biosyst. 2011;7:1523–1536. doi: 10.1039/c0mb00277a. [DOI] [PubMed] [Google Scholar]
- 8.Kraus JP, Janosik M, Kozich V, Mandell R, Shih V, Sperandeo MP, Sebastio G, de FR, Andria G, Kluijtmans LA, Blom H, Boers GH, Gordon RB, Kamoun P, Tsai MY, Kruger WD, Koch HG, Ohura T, Gaustadnes M. Cystathionine beta-synthase mutations in homocystinuria. Hum Mutat. 1999;13:362–375. doi: 10.1002/(SICI)1098-1004199913:5<362::AID-HUMU4>3.0.CO;2-K. [pii] [DOI] [PubMed] [Google Scholar]
- 9.Brenton DP. Skeletal abnormalities in homocystinuria. Postgrad Med J. 1977;53:488–494. doi: 10.1136/pgmj.53.622.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Herrmann M, Widmann T, Herrmann W. Homocysteine--a newly recognised risk factor for osteoporosis. Clin Chem Lab Med. 2005;43:1111–1117. doi: 10.1515/CCLM.2005.194. [DOI] [PubMed] [Google Scholar]
- 11.Schedewie H, Willich E, Grobe H, Schmidt H, Muller KM. Skeletal fingings in homocystinuria: a collaborative study. Pediatr Radiol. 1973;1:12–23. doi: 10.1007/BF00972819. [DOI] [PubMed] [Google Scholar]
- 12.Lim JS, Lee DH. Changes in bone mineral density and body composition of children with well-controlled homocystinuria caused by CBS deficiency. Osteoporos Int. 2013;24:2535–2538. doi: 10.1007/s00198-013-2351-4. [DOI] [PubMed] [Google Scholar]
- 13.Enneman AW, van dV, de JR, Heil SG, Stolk L, Hofman A, Rivadeneira F, Zillikens MC, Uitterlinden AG, van Meurs JB. The association between plasma homocysteine levels, methylation capacity and incident osteoporotic fractures. Bone. 2012;50:1401–1405. doi: 10.1016/j.bone.2012.03.013. S8756-3282(12)00731-4 [pii] [DOI] [PubMed] [Google Scholar]
- 14.Herrmann M, Umanskaya N, Wildemann B, Colaianni G, Widmann T, Zallone A, Herrmann W. Stimulation of osteoblast activity by homocysteine. J Cell Mol Med. 2008;12:1205–1210. doi: 10.1111/j.1582-4934.2008.00104.x. JCMM104 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Herrmann M, Kraenzlin M, Pape G, Sand-Hill M, Herrmann W. Relation between homocysteine and biochemical bone turnover markers and bone mineral density in peri- and post-menopausal women. Clin Chem Lab Med. 2005;43:1118–1123. doi: 10.1515/CCLM.2005.195. [DOI] [PubMed] [Google Scholar]
- 16.Claes L, Schmalenbach J, Herrmann M, Olku I, Garcia P, Histing T, Obeid R, Schorr H, Herrmann W, Pohlemann T, Menger MD, Holstein JH. Hyperhomocysteinemia is associated with impaired fracture healing in mice. Calcif Tissue Int. 2009;85:17–21. doi: 10.1007/s00223-009-9262-6. [DOI] [PubMed] [Google Scholar]
- 17.Tyagi N, Kandel M, Munjal C, Qipshidze N, Vacek JC, Pushpakumar SB, Metreveli N, Tyagi SC. Homocysteine mediated decrease in bone blood flow and remodeling: role of folic acid. J Orthop Res. 2011;29:1511–1516. doi: 10.1002/jor.21415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vijayan V, Khandelwal M, Manglani K, Singh RR, Gupta S, Surolia A. Homocysteine alters the osteoprotegerin/RANKL system in the osteoblast to promote bone loss: pivotal role of the redox regulator forkhead O1. Free Radic Biol Med. 2013;61C:72–84. doi: 10.1016/j.freeradbiomed.2013.03.004. S0891-5849(13)00098-1 [pii] [DOI] [PubMed] [Google Scholar]
- 19.Sato Y, Honda Y, Iwamoto J, Kanoko T, Satoh K. Effect of folate and mecobalamin on hip fractures in patients with stroke: a randomized controlled trial. JAMA. 2005;293:1082–1088. doi: 10.1001/jama.293.9.1082. 293/9/1082 [pii] [DOI] [PubMed] [Google Scholar]
- 20.Cai B, Li X, Wang Y, Liu Y, Yang F, Chen H, Yin K, Tan X, Zhu J, Pan Z, Wang B, Lu Y. Apoptosis of bone marrow mesenchymal stem cells caused by homocysteine via activating JNK signal. PLoS One. 2013;8:e63561. doi: 10.1371/journal.pone.0063561. PONE-D-12-32530 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Saito M, Marumo K, Soshi S, Kida Y, Ushiku C, Shinohara A. Raloxifene ameliorates detrimental enzymatic and nonenzymatic collagen cross-links and bone strength in rabbits with hyperhomocysteinemia. Osteoporos Int. 2010;21:655–666. doi: 10.1007/s00198-009-0980-4. [DOI] [PubMed] [Google Scholar]
- 22.Lubec B, Fang-Kircher S, Lubec T, Blom HJ, Boers GH. Evidence for McKusick’s hypothesis of deficient collagen cross-linking in patients with homocystinuria. Biochim Biophys Acta. 1996;1315:159–162. doi: 10.1016/0925-4439(95)00119-0. [DOI] [PubMed] [Google Scholar]
- 23.Thaler R, Zwerina J, Rumpler M, Spitzer S, Gamsjaeger S, Paschalis EP, Klaushofer K, Varga F. Homocysteine induces serum amyloid A3 in osteoblasts via unlocking RGD-motifs in collagen. FASEB J. 2013;27:446–463. doi: 10.1096/fj.12-208058. fj.12-208058 [pii] [DOI] [PubMed] [Google Scholar]
- 24.Park SJ, Kim KJ, Kim WU, Oh IH, Cho CS. Involvement of endoplasmic reticulum stress in homocysteine-induced apoptosis of osteoblastic cells. J Bone Miner Metab. 2012;30:474–484. doi: 10.1007/s00774-011-0346-9. [DOI] [PubMed] [Google Scholar]
- 25.Dhonukshe-Rutten RA, Pluijm SM, de Groot LC, Lips P, Smit JH, van Staveren WA. Homocysteine and vitamin B12 status relate to bone turnover markers, broadband ultrasound attenuation, and fractures in healthy elderly people. J Bone Miner Res. 2005;20:921–929. doi: 10.1359/JBMR.050202. [DOI] [PubMed] [Google Scholar]
- 26.Gjesdal CG, Vollset SE, Ueland PM, Refsum H, Drevon CA, Gjessing HK, Tell GS. Plasma total homocysteine level and bone mineral density: the Hordaland Homocysteine Study. Arch Intern Med. 2006;166:88–94. doi: 10.1001/archinte.166.1.88. 166/1/88 [pii] [DOI] [PubMed] [Google Scholar]
- 27.Gjesdal CG, Vollset SE, Ueland PM, Refsum H, Meyer HE, Tell GS. Plasma homocysteine, folate, and vitamin B 12 and the risk of hip fracture: the hordaland homocysteine study. J Bone Miner Res. 2007;22:747–756. doi: 10.1359/jbmr.070210. [DOI] [PubMed] [Google Scholar]
- 28.McLean RR, Jacques PF, Selhub J, Tucker KL, Samelson EJ, Broe KE, Hannan MT, Cupples LA, Kiel DP. Homocysteine as a predictive factor for hip fracture in older persons. N Engl J Med. 2004;350:2042–2049. doi: 10.1056/NEJMoa032739. 350/20/2042 [pii] [DOI] [PubMed] [Google Scholar]
- 29.Perier MA, Gineyts E, Munoz F, Sornay-Rendu E, Delmas PD. Homocysteine and fracture risk in postmenopausal women: the OFELY study. Osteoporos Int. 2007;18:1329–1336. doi: 10.1007/s00198-007-0393-1. [DOI] [PubMed] [Google Scholar]
- 30.Gerdhem P, Ivaska KK, Isaksson A, Pettersson K, Vaananen HK, Obrant KJ, Akesson K. Associations between homocysteine, bone turnover, BMD, mortality, and fracture risk in elderly women. J Bone Miner Res. 2007;22:127–134. doi: 10.1359/jbmr.061003. [DOI] [PubMed] [Google Scholar]
- 31.Kim DJ, Park BL, Koh JM, Kim GS, Kim LH, Cheong HS, Shin HD, Hong JM, Kim TH, Shin HI, Park EK, Kim SY. Methionine synthase reductase polymorphisms are associated with serum osteocalcin levels in postmenopausal women. Exp Mol Med. 2006;38:519–524. doi: 10.1038/emm.2006.61. 200610308 [pii] [DOI] [PubMed] [Google Scholar]
- 32.Bode MK, Laitinen P, Risteli J, Uusimaa P, Juvonen T. Atherosclerosis, type 1 collagen cross-linking and homocysteine. Atherosclerosis. 2000;152:531–532. doi: 10.1016/s0021-9150(00)00548-7. S0021915000005487 [pii] [DOI] [PubMed] [Google Scholar]
- 33.Stanger O, Herrmann W, Pietrzik K, Fowler B, Geisel J, Dierkes J, Weger M. DACH-LIGA homocystein (german, austrian and swiss homocysteine society): consensus paper on the rational clinical use of homocysteine, folic acid and B-vitamins in cardiovascular and thrombotic diseases: guidelines and recommendations. Clin Chem Lab Med. 2003;41:1392–1403. doi: 10.1515/CCLM.2003.214. [DOI] [PubMed] [Google Scholar]
- 34.TONNA EA, PILLSBURY N. Changes in the osteoblastic and mitochondrial population of aging periosteum. Nature. 1959;183:337–338. doi: 10.1038/183337a0. [DOI] [PubMed] [Google Scholar]
- 35.Meyer MH, Meyer RA., Jr Altered expression of mitochondrial genes in response to fracture in old rats. Acta Orthop. 2006;77:944–951. doi: 10.1080/17453670610013277. 770235899 [pii] [DOI] [PubMed] [Google Scholar]
- 36.Adamek G, Felix R, Guenther HL, Fleisch H. Fatty acid oxidation in bone tissue and bone cells in culture. Characterization and hormonal influences. Biochem J. 1987;248:129–137. doi: 10.1042/bj2480129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.WALKER DG. Citric acid cycle in osteoblasts and osteoclasts. A histochemical study of normal and parathor-mone-treated rats. Bull Johns Hopkins Hosp. 1961;108:80–99. [PubMed] [Google Scholar]
- 38.Rosenberg N, Bettman L, Rosenberg O, Soudry M, Gavish M, Bar-Shalom R. Measurement of [(18)F]-fluorodeoxyglucose incorporation into human osteoblast-An experimental method. Cytotechnology. 2007;54:1–4. doi: 10.1007/s10616-007-9066-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Komarova SV, Ataullakhanov FI, Globus RK. Bioenergetics and mitochondrial transmembrane potential during differentiation of cultured osteoblasts. Am J Physiol Cell Physiol. 2000;279:C1220–C1229. doi: 10.1152/ajpcell.2000.279.4.C1220. [DOI] [PubMed] [Google Scholar]
- 40.Gerber I, ap GI, Alini M, Wallimann T. Stimulatory effects of creatine on metabolic activity, differentiation and mineralization of primary osteoblast-like cells in monolayer and micromass cell cultures. Eur Cell Mater. 2005;10:8–22. doi: 10.22203/ecm.v010a02. vol010a02 [pii] [DOI] [PubMed] [Google Scholar]
- 41.Hobson GM, Funanage VL, Elsemore J, Yagami M, Rajpurohit R, Perriard JC, Hickok NJ, Shapiro IM, Tuan RS. Developmental expression of creatine kinase isoenzymes in chicken growth cartilage. J Bone Miner Res. 1999;14:747–756. doi: 10.1359/jbmr.1999.14.5.747. [DOI] [PubMed] [Google Scholar]
- 42.Katoh R, Iyoda K, Oohira A, Kato K, Nogami H. Zonal and age-related difference in the amounts of creatine kinase subunits in cartilage. Clin Orthop Relat Res. 1991:283–287. [PubMed] [Google Scholar]
- 43.Hu YY, Rawal A, Schmidt-Rohr K. Strongly bound citrate stabilizes the apatite nanocrystals in bone. Proc Natl Acad Sci U S A. 2010;107:22425–22429. doi: 10.1073/pnas.1009219107. 1009219107 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen RM, Liu HC, Lin YL, Jean WC, Chen JS, Wang JH. Nitric oxide induces osteoblast apoptosis through the de novo synthesis of Bax protein. J Orthop Res. 2002;20:295–302. doi: 10.1016/S0736-0266(01)00086-9. [DOI] [PubMed] [Google Scholar]
- 45.Hay E, Lemonnier J, Fromigue O, Marie PJ. Bone morphogenetic protein-2 promotes osteoblast apoptosis through a Smad-independent, protein kinase C-dependent signaling pathway. J Biol Chem. 2001;276:29028–29036. doi: 10.1074/jbc.M011265200. M011265200 [pii] [DOI] [PubMed] [Google Scholar]
- 46.Ho WP, Chen TL, Chiu WT, Tai YT, Chen RM. Nitric oxide induces osteoblast apoptosis through a mitochondria-dependent pathway. Ann N Y Acad Sci. 2005;1042:460–470. doi: 10.1196/annals.1338.039. 1042/1/460 [pii] [DOI] [PubMed] [Google Scholar]
- 47.Pietila M, Lehtonen S, Narhi M, Hassinen IE, Leskela HV, Aranko K, Nordstrom K, Vepsalainen A, Lehenkari P. Mitochondrial function determines the viability and osteogenic potency of human mesenchymal stem cells. Tissue Eng Part C. Methods. 2010;16:435–445. doi: 10.1089/ten.tec.2009.0247. [DOI] [PubMed] [Google Scholar]
- 48.Chen CT, Shih YR, Kuo TK, Lee OK, Wei YH. Coordinated changes of mitochondrial biogenesis and antioxidant enzymes during osteogenic differentiation of human mesenchymal stem cells. Stem Cells. 2008;26:960–968. doi: 10.1634/stemcells.2007-0509. 2007-0509 [pii] [DOI] [PubMed] [Google Scholar]
- 49.An JH, Yang JY, Ahn BY, Cho SW, Jung JY, Cho HY, Cho YM, Kim SW, Park KS, Kim SY, Lee HK, Shin CS. Enhanced mitochondrial biogenesis contributes to Wnt induced osteoblastic differentiation of C3H10T1/2 cells. Bone. 2010;47:140–150. doi: 10.1016/j.bone.2010.04.593. S8756-3282(10)01137-3 [pii] [DOI] [PubMed] [Google Scholar]
- 50.Nervina JM, Magyar CE, Pirih FQ, Tetradis S. PGC-1alpha is induced by parathyroid hormone and coactivates Nurr1-mediated promoter activity in osteoblasts. Bone. 2006;39:1018–1025. doi: 10.1016/j.bone.2006.04.023. S8756-3282(06)00441-8 [pii] [DOI] [PubMed] [Google Scholar]
- 51.Noble BS. The osteocyte lineage. Arch Biochem Biophys. 2008;473:106–111. doi: 10.1016/j.abb.2008.04.009. S0003-9861(08)00194-X [pii] [DOI] [PubMed] [Google Scholar]
- 52.Ishii KA, Fumoto T, Iwai K, Takeshita S, Ito M, Shimohata N, Aburatani H, Taketani S, Lelliott CJ, Vidal-Puig A, Ikeda K. Coordination of PGC-1beta and iron uptake in mitochondrial biogenesis and osteoclast activation. Nat Med. 2009;15:259–266. doi: 10.1038/nm.1910. nm.1910 [pii] [DOI] [PubMed] [Google Scholar]
- 53.Kim DJ, Koh JM, Lee O, Kim NJ, Lee YS, Kim YS, Park JY, Lee KU, Kim GS. Homocysteine enhances apoptosis in human bone marrow stromal cells. Bone. 2006;39:582–590. doi: 10.1016/j.bone.2006.03.004. S8756-3282(06)00373-5 [pii] [DOI] [PubMed] [Google Scholar]
- 54.Vacek TP, Kalani A, Voor MJ, Tyagi SC, Tyagi N. The role of homocysteine in bone remodeling. Clin Chem Lab Med. 2013;51:579–590. doi: 10.1515/cclm-2012-0605. /j/cclm.2013.51.issue-3/cclm-2012-0605/cclm-2012-0605.xml [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pasternak B, Aspenberg P. Metalloproteinases and their inhibitors-diagnostic and therapeutic opportunities in orthopedics. Acta Orthop. 2009;80:693–703. doi: 10.3109/17453670903448257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kalani A, Kamat PK, Givvimani S, Brown K, Metreveli N, Tyagi SC, Tyagi N. Nutri-epigenetics Ameliorates Blood-Brain Barrier Damage and Neurodegeneration in Hyperhomocysteinemia: Role of Folic Acid. J Mol Neurosci. 2013 doi: 10.1007/s12031-013-0122-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bui C, Barter MJ, Scott JL, Xu Y, Galler M, Reynard LN, Rowan AD, Young DA. cAMP response element-binding (CREB) recruitment following a specific CpG demethylation leads to the elevated expression of the matrix metalloproteinase 13 in human articular chondrocytes and osteoarthritis. FASEB J. 2012;26:3000–3011. doi: 10.1096/fj.12-206367. fj.12-206367 [pii] [DOI] [PubMed] [Google Scholar]
- 58.Hashimoto K, Otero M, Imagawa K, de Andres MC, Coico JM, Roach HI, Oreffo RO, Marcu KB, Goldring MB. Regulated transcription of human matrix metalloproteinase 13 (MMP13) and interleukin-1beta (IL1B) genes in chondrocytes depends on methylation of specific proximal promoter CpG sites. J Biol Chem. 2013;288:10061–10072. doi: 10.1074/jbc.M112.421156. M112.421156 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.de Andres MC, Imagawa K, Hashimoto K, Gonzalez A, Roach HI, Goldring MB, Oreffo RO. Loss of methylation in CpG sites in the NF-kappaB enhancer elements of inducible nitric oxide synthase is responsible for gene induction in human articular chondrocytes. Arthritis Rheum. 2013;65:732–742. doi: 10.1002/art.37806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nakano K, Whitaker JW, Boyle DL, Wang W, Firestein GS. DNA methylome signature in rheumatoid arthritis. Ann Rheum Dis. 2013;72:110–117. doi: 10.1136/annrheumdis-2012-201526. annrheumdis-2012-201526 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Takami N, Osawa K, Miura Y, Komai K, Taniguchi M, Shiraishi M, Sato K, Iguchi T, Shiozawa K, Hashiramoto A, Shiozawa S. Hypermethylated promoter region of DR3, the death receptor 3 gene, in rheumatoid arthritis synovial cells. Arthritis Rheum. 2006;54:779–787. doi: 10.1002/art.21637. [DOI] [PubMed] [Google Scholar]
- 62.Ishida K, Kobayashi T, Ito S, Komatsu Y, Yokoyama T, Okada M, Abe A, Murasawa A, Yoshie H. Interleukin-6 gene promoter methylation in rheumatoid arthritis and chronic periodontitis. J Periodontol. 2012;83:917–925. doi: 10.1902/jop.2011.110356. [DOI] [PubMed] [Google Scholar]
- 63.Nile CJ, Read RC, Akil M, Duff GW, Wilson AG. Methylation status of a single CpG site in the IL6 promoter is related to IL6 messenger RNA levels and rheumatoid arthritis. Arthritis Rheum. 2008;58:2686–2693. doi: 10.1002/art.23758. [DOI] [PubMed] [Google Scholar]
- 64.Fu LH, Ma CL, Cong B, Li SJ, Chen HY, Zhang JG. Hypomethylation of proximal CpG motif of interleukin-10 promoter regulates its expression in human rheumatoid arthritis. Acta Pharmacol Sin. 2011;32:1373–1380. doi: 10.1038/aps.2011.98. aps201198 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lin SY, Hsieh SC, Lin YC, Lee CN, Tsai MH, Lai LC, Chuang EY, Chen PC, Hung CC, Chen LY, Hsieh WS, Niu DM, Su YN, Ho HN. A whole genome methylation analysis of systemic lupus erythematosus: hypomethylation of the IL10 and IL1R2 promoters is associated with disease activity. Genes Immun. 2012;13:214–220. doi: 10.1038/gene.2011.74. gene201174 [pii] [DOI] [PubMed] [Google Scholar]
- 66.Karouzakis E, Rengel Y, Jungel A, Kolling C, Gay RE, Michel BA, Tak PP, Gay S, Neidhart M, Ospelt C. DNA methylation regulates the expression of CXCL12 in rheumatoid arthritis synovial fibroblasts. Genes Immun. 2011;12:643–652. doi: 10.1038/gene.2011.45. gene201145 [pii] [DOI] [PubMed] [Google Scholar]
- 67.Chabchoub G, Uz E, Maalej A, Mustafa CA, Rebai A, Mnif M, Bahloul Z, Farid NR, Ozcelik T, Ayadi H. Analysis of skewed X-chromosome inactivation in females with rheumatoid arthritis and autoimmune thyroid diseases. Arthritis Res Ther. 2009;11:R106. doi: 10.1186/ar2759. ar2759 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jeffries MA, Dozmorov M, Tang Y, Merrill JT, Wren JD, Sawalha AH. Genome-wide DNA methylation patterns in CD4+ T cells from patients with systemic lupus erythematosus. Epigenetics. 2011;6:593–601. doi: 10.4161/epi.6.5.15374. 15374 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Furukawa H, Oka S, Matsui T, Hashimoto A, Arinuma Y, Komiya A, Fukui N, Tsuchiya N, Tohma S. Genome, epigenome and transcriptome analyses of a pair of monozygotic twins discordant for systemic lupus erythematosus. Hum Immunol. 2013;74:170–175. doi: 10.1016/j.humimm.2012.11.007. S0198-8859(12)00614-3 [pii] [DOI] [PubMed] [Google Scholar]
- 70.Kalani A, Kamat PK, Tyagi SC, Tyagi N. Synergy of homocysteine, MicroRNA, and epigenetics: a novel therapeutic approach for stroke. Mol Neurobiol. 2013;48:157–168. doi: 10.1007/s12035-013-8421-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Balasubramanian S, Verner E, Buggy JJ. Isoform-specific histone deacetylase inhibitors: the next step? Cancer Lett. 2009;280:211–221. doi: 10.1016/j.canlet.2009.02.013. S0304-3835(09)00112-8 [pii] [DOI] [PubMed] [Google Scholar]
- 72.Khan N, Jeffers M, Kumar S, Hackett C, Boldog F, Khramtsov N, Qian X, Mills E, Berghs SC, Carey N, Finn PW, Collins LS, Tumber A, Ritchie JW, Jensen PB, Lichenstein HS, Sehested M. Determination of the class and isoform selectivity of small-molecule histone deacetylase inhibitors. Biochem J. 2008;409:581–589. doi: 10.1042/BJ20070779. BJ20070779 [pii] [DOI] [PubMed] [Google Scholar]
- 73.Zhang Y, Xie RL, Croce CM, Stein JL, Lian JB, van Wijnen AJ, Stein GS. A program of microRNAs controls osteogenic lineage progression by targeting transcription factor Runx2. Proc Natl Acad Sci U S A. 2011;108:9863–9868. doi: 10.1073/pnas.1018493108. 1018493108 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hassan MQ, Gordon JA, Beloti MM, Croce CM, van Wijnen AJ, Stein JL, Stein GS, Lian JB. A network connecting Runx2, SATB2, and the miR-23a~27a~24-2 cluster regulates the osteoblast differentiation program. Proc Natl Acad Sci U S A. 2010;107:19879–19884. doi: 10.1073/pnas.1007698107. 1007698107 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kim EJ, Kang IH, Lee JW, Jang WG, Koh JT. MiR-433 mediates ERRgamma-suppressed osteoblast differentiation via direct targeting to Runx2 mRNA in C3H10T1/2 cells. Life Sci. 2013;92:562–568. doi: 10.1016/j.lfs.2013.01.015. S0024-3205(13)00043-X [pii] [DOI] [PubMed] [Google Scholar]
- 76.Wu T, Zhou H, Hong Y, Li J, Jiang X, Huang H. miR-30 family members negatively regulate osteoblast differentiation. J Biol Chem. 2012;287:7503–7511. doi: 10.1074/jbc.M111.292722. M111.292722 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sugatani T, Vacher J, Hruska KA. A microRNA expression signature of osteoclastogenesis. Blood. 2011;117:3648–3657. doi: 10.1182/blood-2010-10-311415. blood-2010-10-311415 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Mann M, Barad O, Agami R, Geiger B, Hornstein E. miRNA-based mechanism for the commitment of multipotent progenitors to a single cellular fate. Proc Natl Acad Sci U S A. 2010;107:15804–15809. doi: 10.1073/pnas.0915022107. 0915022107 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Nass MM. Differential methylation of mitochondrial and nuclear DNA in cultured mouse, hamster and virus-transformed hamster cells. In vivo and in vitro methylation. J Mol Biol. 1973;80:155–175. doi: 10.1016/0022-2836(73)90239-8. 0022-2836(73)90239-8 [pii] [DOI] [PubMed] [Google Scholar]
- 80.Shmookler Reis RJ, Goldstein S. Mitochondrial DNA in mortal and immortal human cells. Genome number, integrity, and methylation. J Biol Chem. 1983;258:9078–9085. [PubMed] [Google Scholar]
- 81.Mushkambarov NN, Votrin II, Debov SS. Methylation of endogenous DNA in isolated rat liver nuclei and mitochondria. Vopr Med Khim. 1976;22:659–664. [PubMed] [Google Scholar]
- 82.Kudriashova IB, Kirnos MD, Vaniushin BF. DNA-methylase activities from animal mitochondria and nuclei: different specificity of DNA methylation. Biokhimiia. 1976;41:1968–1977. [PubMed] [Google Scholar]
- 83.Iacobazzi V, Castegna A, Infantino V, Andria G. Mitochondrial DNA methylation as a next-generation biomarker and diagnostic tool. Mol Genet Metab. 2013;110:25–34. doi: 10.1016/j.ymgme.2013.07.012. S1096-7192(13)00257-6 [pii] [DOI] [PubMed] [Google Scholar]
- 84.Chen Z, Li Y, Zhang H, Huang P, Luthra R. Hypoxia-regulated microRNA-210 modulates mitochondrial function and decreases ISCU and COX10 expression. Oncogene. 2010;29:4362–4368. doi: 10.1038/onc.2010.193. onc2010193 [pii] [DOI] [PubMed] [Google Scholar]
- 85.Kren BT, Wong PY, Sarver A, Zhang X, Zeng Y, Steer CJ. MicroRNAs identified in highly purified liver-derived mitochondria may play a role in apoptosis. RNA Biol. 2009;6:65–72. doi: 10.4161/rna.6.1.7534. 7534 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]