SUMMARY
Extensive chromatin reprogramming occurs at fertilization and is thought to be under the control of maternal factors, but the underlying mechanisms remain poorly understood. We report that maternal Hira, a chaperone for the histone variant H3.3, is required for mouse development past the zygote stage. Male pronucleus formation is inhibited upon deletion of Hira due to a lack of nucleosome assembly in the sperm genome. Hira mutant oocytes are incapable of developing parthenogenetically, indicative of a role for Hira in the female genome. Both parental genomes show highly reduced levels of DNA replication and transcription in the mutants. It has long been thought that transcription is not required for zygote development. Surprisingly, we found that Hira/H3.3-dependent transcription of ribosomal RNA is required for first cleavage. Our results demonstrate that Hira-mediated H3.3 incorporation is essential for parental genome reprogramming, and reveal an unexpected role for rRNA transcription in the mouse zygote.
INTRODUCTION
A successful fertilization event occurs when a sperm cell fuses with an oocyte to form a totipotent zygote and initiates embryogenesis (Clift and Schuh, 2013). Sperm DNA is delivered to the oocyte at fertilization depleted of histones and highly packaged by protamines, and therefore needs to reacquire a nucleosomal organization to support development. Genome-wide chromatin reprogramming occurs at fertilization and is thought to center on the paternal genome, under the control of largely unknown maternal factors (Gu et al., 2011). This property of the oocyte is harnessed using Somatic Cell Nuclear Transfer (SCNT) to reprogram somatic cells to totipotency (Yamanaka and Blau, 2010) . In both mouse and Drosophila, the histone variant H3.3 is incorporated into the paternal pronucleus (Loppin et al., 2005; Torres-Padilla et al., 2006; van der Heijden et al., 2005). H3.3 is generally found associated with active chromatin (Ahmad and Henikoff, 2002), and we recently reported that it maintains a decondensed chromatin state essential during mouse embryo cleavage stages (Lin et al., 2013). The histone chaperone histone cell cycle regulation defective homolog A (Hira) (Ray-Gallet et al., 2002; Tagami et al., 2004) is required in the Drosophila oocyte for incorporation of H3.3 into the paternal genome, although maternal Hira mutants can develop to late embryogenesis (Loppin et al., 2005). We therefore sought to investigate the maternal role of Hira during early pre-implantation mouse development.
RESULTS
Maternal Hira is Strictly Required for Zygote Development to the 2-Cell Stage
Of all the known H3.3 chaperones, we found that Hira is the only one that is incorporated broadly into decondensed sperm DNA at fertilization, while Atrx and Daxx show restricted patterns of incorporation and Dek is undetectable (data not shown). These data suggested to us that other H3.3 chaperones might not be able to compensate for the loss of Hira. We therefore used a genetic approach to specifically delete Hira during oogenesis using Zp3-Cre (de Vries et al., 2000) and a conditional (“floxed”) allele of Hira derived from the KOMP repository (Figures 1A and S1A). For simplicity, from here on we refer to littermate control females whose oocytes carry one functional copy of Hira (see Figure S1A) as heterozygotes.
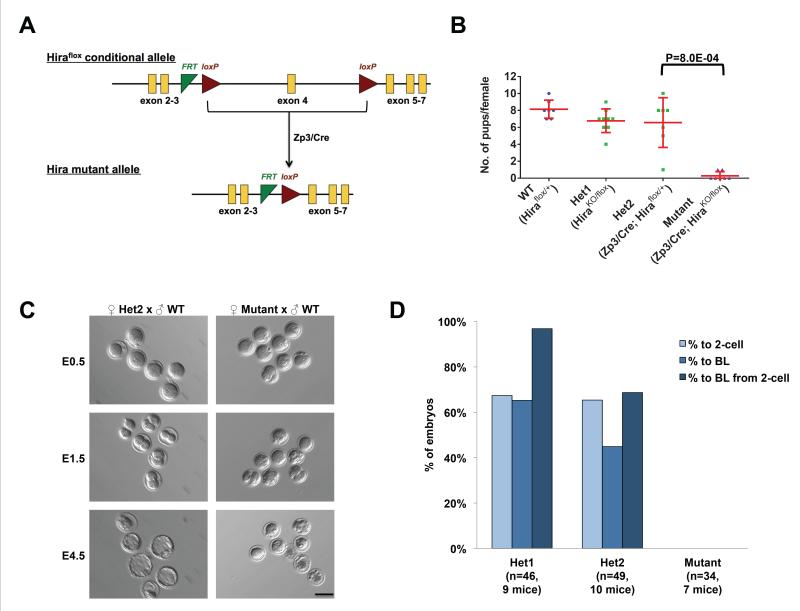
Figure 1. Maternal Hira is essential for zygote development.
(A) Strategy for generation of the maternal Hira mutants: Zp3/Cre induces excision of exon 4 specifically in oocytes.
(B) Hira mutant females are infertile. The size of litters derived from natural matings of experimental females to wild-type males was recorded. Hira mutant females were defined as infertile if no pups were born after three months of breeding. The two pups born to Hira mutant females were perinatal lethal. Two-tailed t-test with unequal variance was used and error bars indicate s.d..
(C) Embryos obtained from Hira mutant females naturally mated to wild-type males undergo developmental arrest at the zygote stage. Embryos were flushed on the day of vaginal plug detection and cultured in vitro for 4 days. Scale bar: 100 m.
(D) Quantification of the developmental potential of Hira mutant and heterozygous zygotes. 1st bar – % development from zygote to 2-cell stage; 2nd bar – % development from zygote to blastocyst stage; 3rd bar – % development from 2-cell stage to blastocyst. No maternally-deleted Hira zygotes developed to the 2-cell stage.
See also Figure S1.
We first validated the complete loss of Hira mRNA and protein in mutant Germinal Vesicle (GV) stage oocytes by qRT-PCR and immunofluoresence (Figures S1B and S1C). Fully-grown Hira mutant oocytes show no significant difference in diameter (89.5±4.6 μm in heterozygotes versus 91.8±4.9 μm in mutants, P=0.41), and chromatin appears normally condensed at the GV stage (Figures S1C). In addition, mutant females can ovulate naturally, produce similar numbers of metaphase II (MII) oocytes compared to heterozygotes and show proper spindle and chromosomal alignment. These data indicate that the loss of Hira during oogenesis does not appear to affect oocyte development through meiosis.
Mutant females were mated to wild-type males and found to be infertile (Figure 1B). There is no significant difference in litter size between wild-type and heterozygous controls. To identify the stage at which maternal Hira mutant embryos arrest, we collected embryonic stage (E) 0.5 embryos from natural matings and assessed the in vitro development of heterozygotes versus mutants. While the majority of fertilized embryos from heterozygous oocytes develop to blastocysts, no mutant zygotes progress to the 2-cell stage (Figures 1C and 1D). These data indicate that maternal Hira is strictly required for development past the zygote stage.
Hira-Mediated H3.3 Incorporation Underlies Paternal Chromatin Assembly
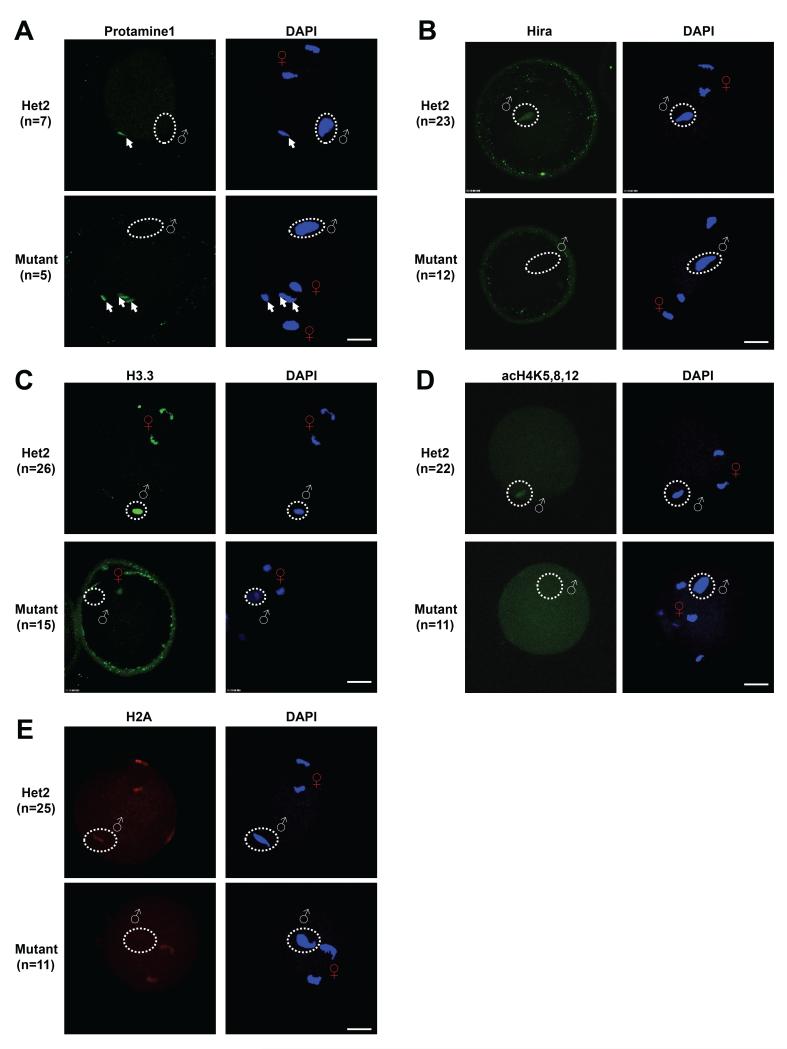
Mutant oocytes resulting from superovulated females appear morphologically normal at the MII stage with proper chromatin alignment and a first polar body (Figure S1D). Proper second meiotic division was also observed when these oocytes were fertilized in vitro (Figures 2 and S2). We went on to characterize the early stages of zygote development (PN0 stage), and found that protamines are still removed from the sperm genome in the mutants (Figure 2A). In both heterozygous control and mutant oocytes, only unfused sperm heads showed positive protamine signal. We thus conclude that Hira is not involved in the first step of paternal genome reprogramming, that of protamine removal. However, in the absence of Hira (Figure 2B), H3.3 cannot be incorporated into the decondensed sperm head (Figure 2C). The lack of H3.3 incorporation in the mutants coincides with a failure of other nucleosome components such as acetyl-H4 (Adenot et al., 1997) and H2A to incorporate into the paternal genome (Figure 2D and 2E). Interestingly, H3.3 levels also appear to be reduced in the maternal chromosomes (Figure 2C, see below).
Figure 2. Hira is required for chromatin assembly at the paternal genome, but not for protamine removal.
(A) Protamine1 is undetectable in decondensed sperm heads in both mutant and heterozygous zygotes. Arrows indicate protamine1 signal in unfused sperm heads.
(B-E) Hira, H3.3, acetylated H4 and H2A are present in decondensed sperm heads of Hira heterozygous zygotes, but are completely undetectable in that of mutant zygotes. Scale bar: 20 m. White “♂” symbol or white dashed circle indicates decondensed sperm head; Red “♀” symbol indicates female chromatin.
See also Figure S2.
When cultured to late zygote stages (PN3-4), the majority of the mutants (82%) form embryos with an abnormal single pronucleus (1PN) instead of the normal two pronuclei (2PN) (Figure S3A). Here again, we could not detect any histones at the paternal genome in PN3-4 mutant zygotes, including H3.3, panH3, acetyl-H4 or H2A (Figures S2B-E). We confirmed that the 1PN present in mutant zygotes is derived from the maternal and not the paternal genome by using the maternal-specific marker H3K9me2 (Nakamura et al., 2012) (Figure S2F). Therefore, Hira has a key role in chromatinization of the paternal genome at fertilization. These results strongly suggest that nucleosomes do not assemble at the paternal genome in maternal Hira mutant zygotes.
Loss of DNA Replication and RNA Transcription in both Pronuclei in Hira-Mutant Zygotes
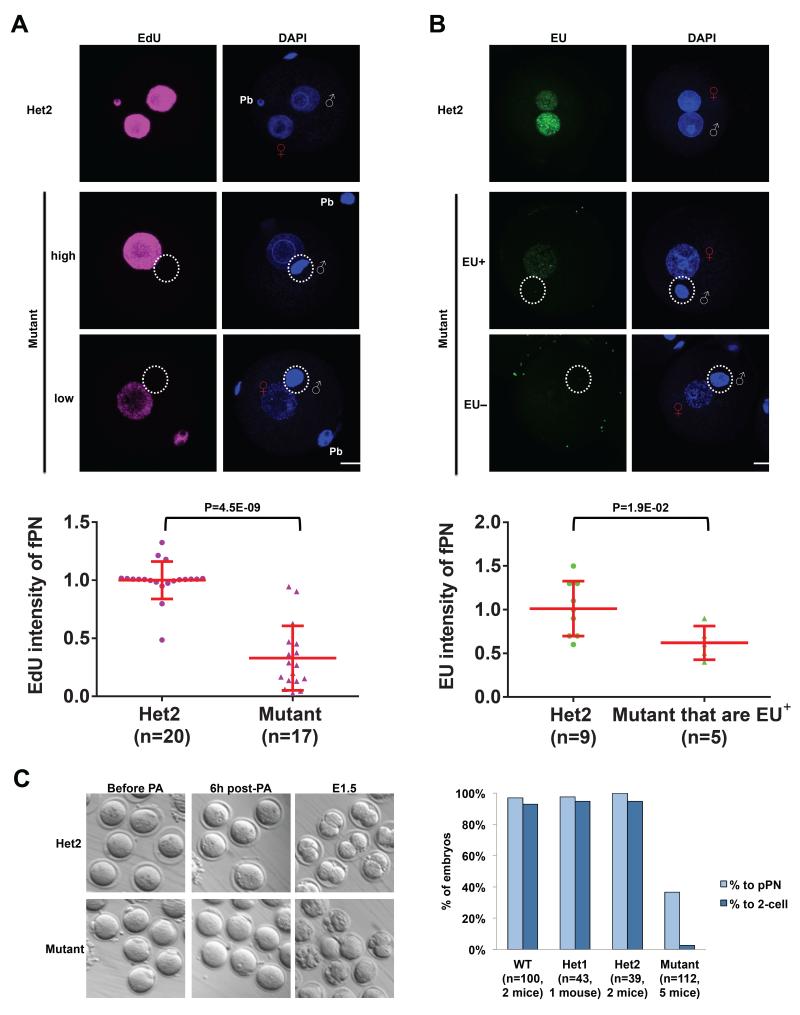
Subsequent to paternal genome reprogramming, DNA replication is believed to be the first key cellular event in the zygote in preparation for the first cleavage. We therefore examined DNA synthesis by 5-ethynyl-2′-deoxyuridine (EdU) labeling. Zygotes were incubated 5 hours post-in vitro fertilization (IVF) (hpi) with EdU for another 5 hours (h) to label all newly synthesized DNA in both paternal and maternal genomes. In contrast to the strong staining in controls, we could not detect any EdU signal at the paternal genome in the mutants (n=20, Figure 3A). These results suggest that chromatinization of the paternal genome is a prerequisite for DNA replication, possibly due to the role of histone marks in productive DNA synthesis (Dorn and Cook, 2011). Surprisingly, the EdU intensity in the mutant female PN is also highly reduced (33% compared with heterozygotes, P=4.5E-09, Figure 3A). Therefore, DNA replication is absent in the paternal genome and significantly reduced in the maternal genome in Hira mutants. These results were the first indication that the maternal genome also requires Hira for developmental progression. Interestingly, the fPN in mutant zygotes is significantly larger than in the heterozygotes (diameter of 15.1±1.2 μm in heterozygotes versus 20.9±2.5 μm in mutants, P=4.1E-12).
Figure 3. Maternal Hira regulates DNA replication and RNA transcription in both parental genomes.
(A) DNA replication as detected by EdU staining is consistently absent in the paternal genome, while it is reduced in the maternal genome in Hira mutant zygotes. Representative EdU labeling images in heterozygous zygotes and high- or low-EdU-intensity in Hira mutants are shown (upper panel). Quantification of EdU intensity of female pronucleus (fPN) in Hira mutant and heterozygous zygotes (lower panel).
(B) EU labeling indicates a complete loss of nascent RNA synthesis in the paternal genome and a severe deficiency in the maternal genome in Hira mutant zygotes (upper panel). 54% of mutant zygotes have maternal genomes with no detectable EU signal (hence quantification not shown), while the remaining 46% show significantly reduced levels of EU-positivity (quantified in the lower panel). Two-tailed t-test with unequal variance was used and error bars indicate s.d..
(C) Hira mutant oocytes fail to undergo cleavage to form 2-cell embryos after parthenogenetic activation (PA) (left panel). Many mutant oocytes show visible cellular fragmentation the day after PA, while almost all control oocytes formed a pseudo-pronucleus (pPN) and proceeded to the 2-cell stage (right panel). Scale bar: 20 m. White “♂” symbol or white dashed circle indicates male chromatin; Red “♀” symbol indicates female chromatin; Pb refers to a polar body.
See also Figure S3.
We next investigated RNA synthesis, which is highly regulated by chromatin architecture. Nascent RNA synthesis was monitored by incubating 5 hpi zygotes for 5 h in 5-ethynyl-uridine (EU). Both PNs show clearly detectable signal in control zygotes that is higher in the male PN (mPN), as previously described (Bouniol et al., 1995). In contrast, no EU signal was detected in the paternal genome in the mutants (Figure 3B). Surprisingly, only 46% of mutant fPN showed EU positive signal compared to 100% of heterozygotes, and the signal intensity is significantly lower in those mutant fPN that are EU-positive when compared to heterozygous controls (63%, P=1.9E-02, Figure 3B). In agreement with the EU data, we observed a complete absence of RNA polymerase II phospho S2 (Pol II pS2), a marker of transcriptional elongation, in mutant paternal genomes (Figure S3). Similarly, there is a significantly lower level of Pol II pS2 in mutant fPN (50% compared with heterozygotes, P=1.9E-03; Figure S3). Taken together, the results above indicate that Hira-mediated chromatin assembly is essential for DNA replication and RNA transcription in both parental genomes, not just the paternal one.
Haploid mouse embryos are capable of developing beyond the 2-cell stage and up to the blastocyst stage (Latham et al., 2002; Modlinski, 1975; Yang et al., 2012). If the role of Hira was strictly to reprogram the male genome, one would expect mutant embryos to progress as haploids through cleavage stages, as is the case in Drosophila maternal Hira mutants (Loppin et al., 2005). The observations that Hira mutants have reduced H3.3 levels in female chromosomes and fPN (Figure 2C and Figure S2B, see quantification in Figure S2G) and reduced DNA replication (Figure 3A) and RNA transcription (Figure 3B) at the fPN suggested that Hira is also required for developmental competence of the female genome. Alternatively, the presence of an abnormal male genome in Hira mutant zygotes might interfere with the function of the fPN. To distinguish between these possibilities, we carried out parthenogenesis, where development is driven exclusively by the maternal genome. We verified that control and mutant oocytes undergo pathenogenetic activation normally as assessed by cortical granule exocytosis using PSA-FITC (data not shown). 93.5% of wild-type oocytes (n=100) and 84.7% of heterozygous oocytes (n=82) develop to morula/blastocysts after parthenogenetic activation (Figure 3C). Strikingly, only 2% of mutant oocytes (n=112) formed 2-cell-like embryos after parthenogenesis. None of them developed beyond the 2-cell stage, even after an extended culture of three additional days. These results demonstrate that, in addition to the role in paternal genome reprogramming, Hira is required for developmental competence of the female genome.
Transcription of rRNA by RNA Pol I is Required for Zygote Development
Given the requirement of DNA replication for first cleavage, the developmental arrest of Hira mutants may well be attributable to the reduction in DNA replication at both PN. Nevertheless, we explored further the concomitant reduction in RNA transcription in the mutants. Transcription is widely accepted to not be required for zygote cleavage, because treatment of zygotes with α-Amanitin, an RNA Pol II inhibitor, leads to an arrest only at the 2-cell stage during zygotic gene activation (Braude et al., 1979; Johnson, 1981; Schultz, 2002). We analyzed two independent published gene expression datasets (Wang et al., 2004; Zeng et al., 2004) encompassing mouse pre-implantation development and found that, while those studies correctly concluded that very few transcripts are induced between the metaphase II oocyte stage and the zygote stage, all Affymetrix control probesets against 18S rRNA show a distinctive up-regulation in the zygote in both studies (Figure S4A and S4B, see also RNA FISH in Figure 4B). Ribosomal RNA is transcribed by RNA Pol I, which is not inhibited by α-Amanitin (Bensaude, 2011), and our experiments using EU (Figure 3B) do not distinguish between RNA polymerases. We therefore set out to revisit the roles of DNA replication and RNA transcription during mid-to-late zygote development using specific small molecule inhibitors added at 5 hpi: Aphidicolin, for DNA replication, α-Amanitin for RNA Pol II-mediated transcription and CX-5461 (Bywater et al., 2012; Drygin et al., 2011; Haddach et al., 2012) for RNA Pol I-mediated transcription. Of note, activity of both RNA Pol I and II has been detected indirectly in zygotes using specific promoter-driven reporter plasmids (Nothias et al., 1996).
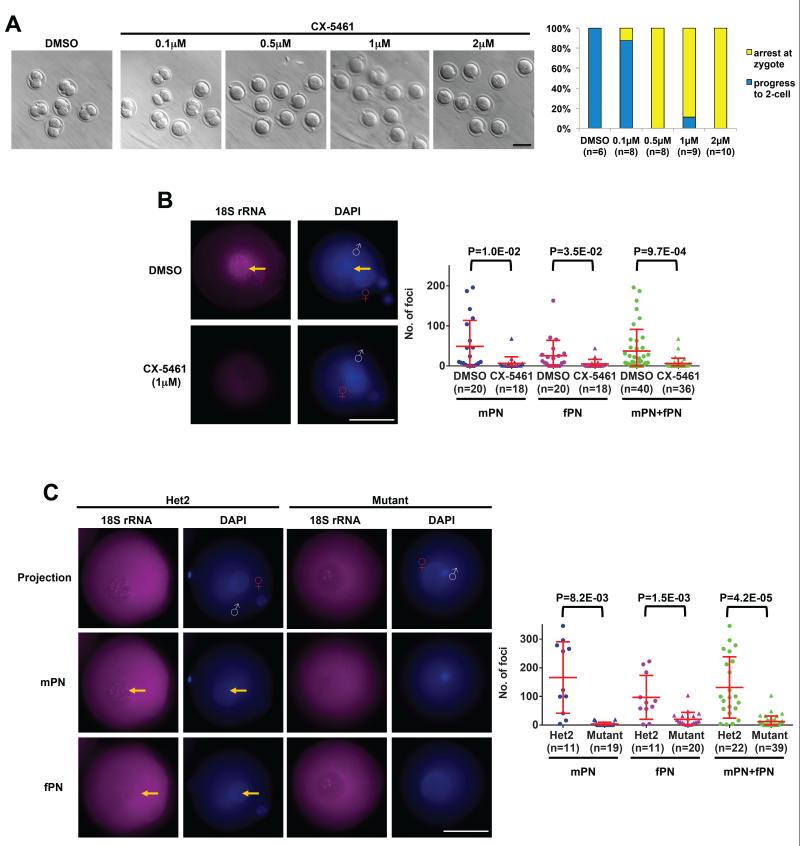
Figure 4. Hira-dependent RNA Pol I transcription is required for zygote development.
(A) Dose-response of wild-type zygotes to the RNA Pol I inhibitor CX-5461. 0.5μM is sufficient to completely block zygote cleavage. Embryos were imaged and quantified after overnight treatment with the indicated doses of CX-5461. Scale bar: 100 m.
(B) Detection of 18S ribosomal RNA by RNA FISH in wild-type zygotes shows a strong signal in DMSO-treated control zygotes. The signal is completely ablated by the addition of CX-5461. Shown are representative RNA FISH images (left panel) and quantification of RNA FISH-positive foci (right panel). Yellow arrows indicate foci inside DNA-dense regions that correspond to the nucleolar precursor bodies. Scale bar: 25 m.
(C) Ribosomal RNA transcription is abolished in Hira mutant zygotes. Representative 18S rRNA FISH images at different focal planes show distinct foci in both male and female pronuclei (left panel) in control but not mutant zygotes. Quantification of foci is shown (right panel). Scale bar: 25 m. White “♂” symbol indicates male genome; Red “♀” symbol indicates female pronucleus (fPN). Two-tailed t-test with unequal variance was used and error bars indicate s.d..
See also Figure S4.
As expected, we found that all control groups (untreated and DMSO, a solvent control) and α-Amanitin treated embryos develop to the 2-cell stage (Braude et al., 1979; Johnson, 1981; Schultz, 2002), whereas Aphidicolin treatment inhibits DNA replication and results in an arrest at the zygote stage (Howlett, 1986) (Figure S4C). Surprisingly, inhibition of RNA Pol I by CX-5461 also leads to a strict zygote stage arrest similar to that observed in Hira mutant zygotes and parthenotes (Figure 4A). CX-5461 has exquisite specificity for RNA Pol I and does not inhibit RNA Pol II (RNA Pol I IC50~0.1μM and IC75~1μM versus RNA Pol II IC50>25μM). A careful titration of CX-5461 revealed that a concentration of 0.5μM is sufficient to completely block zygote cleavage (Figure 4A). Our data are in contradiction with the idea (Zatsepina et al., 2003) that RNA Pol I transcription appears to be absent in the zygote based on the lack of BrUTP incorporation in the nucleolar precursor bodies (NPBs). These results were obtained using immunofluoresence detection of BrUTP after paraformaldehyde (PFA) fixation, but it has been reported that PFA fixation specifically hinders antibody penetration in the nucleolus and that methanol fixation should be used instead (Haukenes and Kalland, 1998). We sought to clarify this point and directly detect rRNA by using methanol fixation and a recently described high sensitivity RNA FISH protocol that involves the use of 48 singly labeled oligonucleotide probes (Raj et al., 2008). We took advantage of the fact that 18S rRNA probes can be used to label pre-rRNA inside the nucleus before the rRNA is exported to the cytoplasm (Kent et al., 2009) to design probes against 18S rRNA and test them in wild-type late PN stage zygotes at 10 hpi. We observed a robust pattern of 18S rRNA-positive foci inside both the mPN and the fPN at DNA-dense regions that correspond to the NPBs, and diffuse signal throughout the rest of the oocyte (Figure 4B). To validate the 18S rRNA FISH, we treated 5 hpi zygotes with CX-5461 for 5 h, and observed that the signal was essentially abolished in both mPN and fPN (Figure 4B). In contrast to the complete inhibition of DNA replication by Aphidicolin, zygotes treated with CX-5461 are able to replicate DNA normally, as assessed by EdU labeling (Figure S4D). This result indicates that the developmental arrest induced by inhibition of RNA Pol I transcription is independent of effects on DNA replication. Taken together, these data reveal that de novo RNA Pol I transcription of rRNA occurs during mid-to-late zygote stages and is required for progression to the 2-cell stage.
Maternal Hira is Essential for RNA Pol I Transcription in the Zygote
The results above prompted us to ask the question of whether Hira mediates H3.3 incorporation into NPBs and regulates RNA Pol I transcription. Using methanol fixation, we found that H3.3 accumulates at high levels in the NPBs of both pronuclei and colocalizes with nucleolin, a major component of the nucleolus (Figure S4E). To remove the variable of fixation method altogether, we injected in vitro transcribed H3.3-GFP mRNA into zygotes, and found that it is incorporated at high levels into both mPN and fPN NPBs in unfixed, live embryos (Figure S4F). In support of these findings, H3.3 has also been reported to incorporate into rDNA in Drosophila somatic cells and mouse embryonic fibroblasts (3T3) (Ahmad and Henikoff, 2002; Kraushaar et al., 2013; Schneiderman et al., 2012). Moreover, we found that nascent 18S rRNA, assessed as above by quantification of FISH signal in the PNs, is drastically reduced in both mPN and fPN in Hira mutant embryos at 10 hpi (Figure 4C). Thus, fixation procedures that are suboptimal for studying the nucleolus (Haukenes and Kalland, 1998), lower sensitivity RNA FISH protocols and the lack of specific RNA Pol I inhibitors until recently (Bywater et al., 2012; Drygin et al., 2011; Haddach et al., 2012) may all have contributed to the idea that rRNA transcription is either absent or irrelevant in the zygote.
Depletion of H3.3 in the Oocyte Impairs Male Pronucleus Formation, DNA Replication and rRNA Transcription
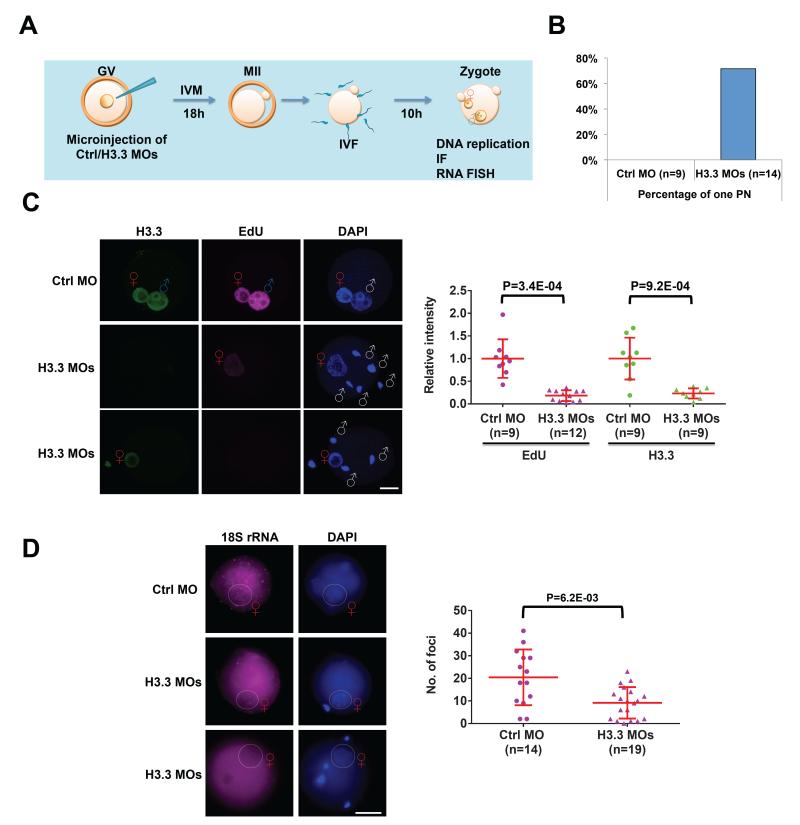
We next sought to determine whether the defects observed in Hira mutant oocytes can be attributed to a lack of H3.3 incorporation at fertilization. We took advantage of our previous approach of co-injecting two distinct and specific morpholino antisense oligonucleotides (MOs) into fully-grown oocytes to knockdown (KD) both genes that encode H3.3 (Lin et al., 2013) (Figure 5A). After in vitro maturation for 18 hours post-injection, we did not observe differences in oocyte maturation rates between Ctrl MO- and H3.3 MOs-injected oocytes, based on polar body extrusion rates (74.3% in Ctrl MO versus 71.2% in H3.3 MOs, P=0.34). We next removed the zona pellucida of MO-injected and in vitro matured MII oocytes and performed IVF. Interestingly, H3.3 KD zygotes, but not controls, display the same abnormal 1PN phenotype seen in Hira mutants (Figure 5B). Moreover, zygotes in the Ctrl MO group labeled with EdU for 5 hours show strong EdU signal in both pronuclei, but those treated with H3.3 MOs show no EdU signal in the mPN and a dramatic reduction in the fPN (18.3% compared with Ctrl MO, P=9.2E-04, Figure 5C). Finally, the number of foci of 18S rRNA FISH is significantly reduced in the fPNs of H3.3 MOs-treated embryos at 10 hpi (Figure 5D). Note that the number of foci that can be detected in this experimental paradigm is lower than in naturally fertilized zygotes (Figure 4B, C), most likely due to the additional manipulations required (GV stage injection, IVM, removal of zona pellucida, IVF). In summary, knocking down H3.3 in fully-grown GV oocytes recapitulates key phenotypes observed in maternal Hira mutants, namely abnormal male pronucleus formation and deficiency of DNA replication and rRNA transcription. These results strongly support the notion that the defects seen in Hira mutants are a direct consequence of the loss of H3.3 incorporation in zygotes, rather than being due to potential effects of Hira loss during oogenesis.
Figure 5. H3.3-depleted oocytes exhibit defects in male pronucleus formation, DNA replication and rRNA transcription.
(A) Schematic of experimental design. Denuded fully-grown oocytes were microinjected with control morpholino (Ctrl MO) or H3.3 MOs and subsequently in vitro matured for 18 hours. IVF was performed after removal of zona pellucida from mature MII oocytes. Immunofluorescence studies and analyses of DNA synthesis and RNA FISH were carried out in zygotes.
(B) Quantification of abnormal 1PN zygotes seen in H3.3 MOs-treated zygotes but not in the Ctrl MO-treated zygotes.
(C) DNA replication as detected by EdU staining is consistently absent in the paternal genome, while it is reduced in the maternal genome in H3.3 MOs zygotes. Representative immunofluorescence images of H3.3 and EdU labeling images in Ctrl MO-treated and in H3.3 MOs-treated zygotes are shown (left panel). Quantification of H3.3 and EdU intensity of female pronucleus (fPN) in Ctrl MO and H3.3 MOs zygotes (right panel). Scale bar: 20 m. White “♂” symbol indicates male genome; Red “♀” symbol indicates female pronucleus (fPN).
(D) Ribosomal RNA transcription is significantly reduced in H3.3 MOs-treated zygotes. Representative 18S rRNA FISH images show distinct foci in female pronuclei (left panel) in Ctrl MO zygotes, but the numbers of these foci are greatly reduced in H3.3 MOs zygotes.
Quantification of foci is shown (right panel). Note that the number of foci that can be detected in this experimental paradigm is lower than in naturally fertilized zygotes (Figure 4B, C), most likely due to the additional manipulations required (GV stage injection, IVM, removal of zona pellucida, IVF). Scale bar: 100 m. Red “♀” symbol indicates female pronucleus (fPN). Two-tailed t-test with unequal variance was used and error bars indicate s.d..
Ribosomal Protein Content and Localization are Impaired in Hira Mutant Zygotes
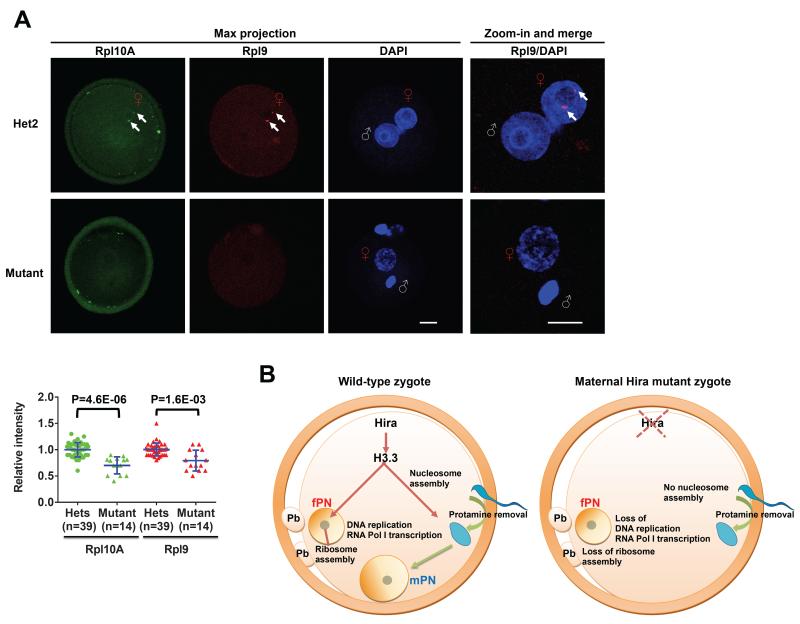
Ribosomal proteins are translated in the cytoplasm and imported back into the nucleolus for ribosome biogenesis (Kressler et al., 2010; Woolford and Baserga, 2013). Detection of ribosomal proteins in the nucleolus is therefore used as indicative of ribosome biogenesis (Bursac et al., 2012; Lam et al., 2007). The attenuation of rRNA synthesis is known to affect proper assembly of rRNAs and ribosomal proteins into ribosomes at the nucleolus, and their subsequent export to the cytoplasm (Laferte et al., 2006). We therefore examined the global levels of two ribosomal proteins (Rpl9 and Rpl10A) using immunofluorescence, as a measure of overall abundance of ribosomes. We found that both ribosomal proteins are significantly reduced in Hira mutant zygotes, indicative of a defect in ribosome biogenesis (Figure 6A). Remarkably, we observed that in controls both Rpl9 and Rpl10A accumulate in 1-2 distinct foci around the borders of the NPB in the female, but not the male PN (Figure 6A and Figure S5). Of note, this pattern is entirely lost in Hira mutants (Figure 6A and Figure S5). Taken together, the data suggest that ribosome assembly may occur specifically in the fPN in the zygote, and that the deficits in rRNA output in Hira mutants disrupt this assembly.
Figure 6. Ribosomal protein dosage and subcellular localization are impaired in Hira mutant zygotes.
(A) Rpl9 and Rpl10A proteins are both significantly reduced and fail to localize to the female pronucleus in Hira mutant zygotes. Representative immunofluorescence images of Rpl9 and Rpl10A in Hira mutant and heterozygous zygotes (top panel). Quantification of intensity of whole zygotes (lower panel). Arrows indicate the ribosomal protein foci detected at the border of nucleolar precursor body specifically in the fPN of heterozygotes. Scale bar: 20 m. White “♂” symbol indicates male genome; Red “♀” symbol indicates female pronucleus (fPN). Two-tailed t-test with unequal variance was used and error bars indicate s.d..
(B) Model for the dual role of maternal Hira during early zygotic development. In wild-type zygotes, Hira mediates H3.3 incorporation into decondensed sperm head and facilitates nucleosome assembly and male pronucleus formation. Hira also regulates DNA replication and RNA Pol I transcription in both pronuclei. In mutant zygotes lacking maternal Hira, H3.3 cannot be incorporated into the decondensed sperm head, no nucleosomes are assembled and the paternal pronucleus fails to form. In addition, Hira deficiency results in impaired DNA replication and the complete absence of RNA Pol I transcription from both genomes, coupled with the loss of ribosome assembly in the maternal pronucleus, culminating in the 1-cell arrest of Hira mutant embryos.
See also Figure S5.
DISCUSSION
This study establishes that maternal Hira is strictly required for development of the mouse zygote. Our data reveal that the role of Hira and H3.3 in protamine-to-histone reprogramming of the sperm genome is a conserved biological process from Drosophila (Loppin et al., 2005) to mouse. However, in Drosophila maternal Hira is neither involved in maternal genome reprogramming, nor in rRNA transcription, nor in zygote development since those mutants can complete embryogenesis (Loppin et al., 2005). This work therefore uncovers essential roles for Hira during mammalian embryogenesis (Figure 6B), discussed below.
Our data indicate that the male PN is largely devoid of histones in Hira mutants, rather than simply lacking H3.3. These observations likely are due to the fact that chromatinization of the sperm genome in the early zygote happens prior to DNA replication. H3.3 is the only H3 that can be incorporated outside of S phase via the replication-independent pathway, of which Hira is a core component (Maze et al., 2014). Thus, in the absence of Hira-mediated H3.3 incorporation, no H3/H4 tetramers can be incorporated, and no seeding of H2A/H2B is likely to occur.
Most studies of reprogramming in the zygote have focused on the sperm genome, and understandably so given the dramatic global chromatin changes that it undergoes. We provide here functional data to support the notion that the female genome is not a mere passenger at this stage but instead undergoes dynamic chromatin reprogramming that is critical for zygote development. In the maternal genome, Hira likely incorporates H3.3 both during oogenesis as well as in the zygote (Akiyama et al., 2011). While this incorporation does not appear to be essential for oogenesis, fertilization and completion of meiosis, we show here that H3.3 incorporation around the time of fertilization sets a chromatin state required for DNA replication, rRNA transcription and ribosomal assembly in the female PN at late zygote stages. Future studies should determine how the lack of H3.3-containing nucleosomes hinders access and/or processivity of the replication and transcription machineries in the parental genomes.
Our results also overturn an idea that has stood since the 70’s that transcription in the zygote is both minor and irrelevant for development, and that Zygotic Gene Activation (ZGA) only becomes functional at the 2-cell stage, when there is a major burst in mRNA synthesis (Braude et al., 1979; Johnson, 1981; Schultz, 2002; Zernicka-Goetz et al., 2009). We report a critical role for RNA Pol I transcription in the zygote, and show that this transcription is Hira-dependent. Therefore, functional ZGA can actually be considered to begin at the zygote stage, and the component of rRNA transcription is essential for progression to the 2-cell stage. Interestingly, both 18S rRNA (Paynton et al., 1988) and ribosomal protein mRNAs (Su et al., 2007) have been shown to decrease in abundance during oocyte maturation (similar data in Figure S6), suggesting that the MII oocyte may be in a “poised” state of low translation capacity until fertilization. Indeed, earlier studies have reported that nascent RNA synthesis, including at the nucleolus, ceases in fully-grown oocytes with condensed chromatin, prior to germinal vesicle breakdown (Bouniol-Baly et al., 1999; Wassarman and Letourneau, 1976), and this is accompanied by a global reduction in protein synthesis (Schultz and Wassarman, 1977). Relative to other model organisms, mouse oocytes are not as loaded with yolk and other maternal factors, and thus strictly depend on RNA Pol I transcription at the 1-cell stage (this study) and RNA Pol II transcription at the 2-cell stage (Braude et al., 1979; Johnson, 1981; Schultz, 2002). It is possible that mammalian oocytes adjust their pool of rRNA and ribosomes to match the mRNA pool and protein requirements, with high levels during oocyte growth, a relative quiescent MII stage, and a return to high levels upon fertilization and embryonic development. It will be of interest to determine the potential role of newly-synthesized rRNA in protein translation in the zygote, and how it may affect the first cell division. The peculiar fPN-specific pattern of new ribosome assembly suggested by our results also deserves further studies. More broadly, it will be important to consider from now on defects in rRNA synthesis and/or new ribosome assembly as one of the possible reasons for developmental failure in other genetic or pharmacological studies of fertilization and early embryogenesis.
The findings reported here may also be of relevance for human assisted reproduction technologies, because an abnormal 1PN phenotype similar to that found in maternal Hira mutants is often observed in cases of ICSI that fail to develop past the zygote stage (Flaherty et al., 1995). It may prove useful to determine whether or not in such cases the sperm genome has incorporated nucleosomes adequately. In addition, it will be of interest to assess if human zygotes also express and require nascent rRNA. Finally, our results suggest that a role in rRNA synthesis should be considered in other settings where Hira or H3.3 have been implicated, such as development of the blastocyst (Lin et al., 2013), mesoderm (Szenker et al., 2012) and neural crest (Cox et al., 2012), primordial germ cell reprogramming (Hajkova et al., 2008) and tumorigenesis (Lewis et al., 2013).
EXPERIMENTAL PROCEDURES
Mice
Mice carrying a “knockout-first” conditional-ready allele for Hira (Hiratm1a(EUCOMM)Wtsi) were obtained from the European Conditional Mouse Mutagenesis Program (EUCOMM) at the Wellcome Trust Sanger Institute (White et al., 2013). The mice were crossed to ROSA26-Flpe mice (Gt(ROSA)26Sortm1(FLP1)Dym) (Farley et al., 2000) that express Flp recombinase ubiquitously to delete the promoterless LacZ cassette, while leaving behind exon 4 flanked by two loxP sites. The resulting Hiraflox allele (Hiratm1c(EUCOMM)Wtsi) was crossed to Zp3-Cre mice (Tg(Zp3-cre)93Knw) (de Vries et al., 2000) that express Cre recombinase in the female germline. All of the studies were performed on a C57BL/6 genetic background. The care and use of mice were in accordance with the guidelines of the University of California, San Francisco (UCSF) Institutional Animal Care and Use Committee (IACUC).
H3.3 Knockdown in Oocytes
Microinjection platform and sequences of control and H3.3 morpholinos were as previously described (Lin et al., 2013). Wildtype denuded fully-grown oocytes were isolated from 5-week-old mice. Injected oocytes were in vitro matured in M16 medium (Millipore) for 18 hours. Only matured MII oocytes (based on polar body extrusion) were selected and cultured shortly in acidic Tyrode’s Solution (Millipore) to removed the zona pellucida prior to IVF.
In Vitro Fertilization and Parthenogenesis
IVF was performed as described (Sztein et al., 2000) with minor modifications. Briefly, sperm was collected from vasa deferentia from 8-12 weeks old male B6 mice and then capacitated for 1.5 h in human tubal fluid (HTF) medium (Millipore) at 37°C in 5% CO 2 incubator. Sperm cells were incubated with intact cumulus-oocyte complexes or zona-free oocytes for 5-6 h. After removing excess sperm and cumulus lysate, presumptive fertilized eggs were transferred to equilibrated KSOM + AA medium (Millipore) for further culture and analysis. MII stage oocytes were parthenogenetically activated in calcium-free CZB medium containing 10mM strontium and supplement with 5μg/ml cytochalasin B for 6 h. Parthenogenic embryos and then transferred into KSOM + AA medium for further culture.
RNA Isolation and Quantitative RT-PCR
Total RNA was isolated from embryos using PicoPure RNA extraction kit (Arcturus). cDNA was generated using iScript kit (BioRad) and qRT-PCR reactions were performed in duplicate using the SYBR Green quantitative RT-PCR Master Mix (Applied BioSystems) and ran on an Applied BioSystems 7900HT Sequence Detection System. Relative abundance of mRNAs was calculated by normalization to Hprt mRNA levels. Primer sequences used in this study are listed in Supplementary Experimental Procedures.
Inhibitor Treatment
Aphidicolin (3μg/ml, Sigma)(Hamatani et al., 2004), α-Amanitin (0.1mg/ml, Sigma), and CX-5461(Drygin et al., 2011) (0.1μM to 2μM, Xcess Biosciences) were added to the culture medium at the concentrations indicated.
Detection of DNA and RNA Synthesis
Zygotes at 5 hpi were incubated with 50μM EdU for 5 h, fixed in PFA and stained using the Click-iT EdU imaging kit (Invitrogen) for DNA replication analysis. Alternatively, 5 hpi zygotes were incubated with 2mM EU for 5 h, fixed in 100% methanol for 20 mins in -20°C and stained using the Click-iT RNA imaging kit (Invitrogen) for nascent RNA analysis.
Immunofluorescence
With the exception of extra decondensed treatment(Hammoud et al., 2009) for protamine antibody and methanol fixation (100% methanol for 20 mins in -20°C) for H3.3 and nucleolin co-localization, standard IF was conducted as previously described (Lin et al., 2013). Antibodies used for this study are listed in Supplementary Experimental Procedures. Embryos were imaged on a laser-scanning confocal microscope (CTR 6500, Leica). IF signal intensity was quantified using ImageJ software (NIH).
RNA FISH
With the exception of using methanol fixation, FISH was performed according to the manufacturer’s instructions (Biosearch Technologies). Fixed zygotes were permeabilized using 70% ethanol and hybridized at 37°C overnight with 1 :250 of 18S rRNA Stellaris probes in RNA FISH Hybridization beta Buffer (Biosearch Technologies). Embryos were imaged using a 100x oil-immersion objective, every 0.25μM using the Z-stack function of the MetaMorph software (Molecular Devices). Only foci located inside each pronucleus (defined based on DAPI staining) were quantified. The StarSearch software (Raj Laboratory, University of Pennsylvania) was used to identify and count foci, applying an identical threshold for foci counting in all embryos.
Statistical Analyses
Two-tailed t-test with unequal variance was used for analyses of number of pups, IF intensity and number of FISH foci. All error bars indicate s.d..
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to Qun-Tian Wang and Magdalena Zernicka-Goetz for sharing raw microarray data; to Andrej Susor, Pankaj Sahai and Arturo Orjalo for technical support for RNA FISH; and to Michael McManus for sharing the ROSA26-Flpe mice. We thank Robert Blelloch and Diana Laird for input during the course of the work, and members of the Santos lab for critical reading of the manuscript. F.M.K. was supported by a National Science Scholarship from the Agency for Science, Technology and Research (Singapore). This project was supported in part by Eunice Kennedy Shriver National Institute of Child Health and Human Development/National Institutes of Health cooperative agreement 1U54HD055764, as part of the Specialized Cooperative Centers Program in Reproduction and Infertility Research. This work was funded by R01 GM097165 to M.C. and an NIH New Innovator Award DP2OD004698 to M.R.-S.
Footnotes
Author Contributions: M.R.-S. directed the project. F.M.K. initially conceived of using a conditional allele of Hira and procured the mice. C.-J.L., M.C. and M.R.-S. designed experiments. C.-J.L. performed all experiments with the following exceptions. F.M.K. performed bioinformatics analyses. F.M.K. and P.W. managed the mouse colony and P.W. performed genotyping. C.-J.L., F.M.K., M.C. and M.R.-S. interpreted the data and wrote the manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Adenot PG, Mercier Y, Renard JP, Thompson EM. Differential H4 acetylation of paternal and maternal chromatin precedes DNA replication and differential transcriptional activity in pronuclei of 1-cell mouse embryos. Development. 1997;124:4615–4625. doi: 10.1242/dev.124.22.4615. [DOI] [PubMed] [Google Scholar]
- Ahmad K, Henikoff S. The histone variant H3.3 marks active chromatin by replication-independent nucleosome assembly. Molecular Cell. 2002;9:1191–1200. doi: 10.1016/s1097-2765(02)00542-7. [DOI] [PubMed] [Google Scholar]
- Akiyama T, Suzuki O, Matsuda J, Aoki F. Dynamic replacement of histone H3 variants reprograms epigenetic marks in early mouse embryos. PLoS Genet. 2011;7:e1002279. doi: 10.1371/journal.pgen.1002279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bensaude O. Inhibiting eukaryotic transcription: Which compound to choose? How to evaluate its activity? Transcription. 2011;2:103–108. doi: 10.4161/trns.2.3.16172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouniol-Baly C, Hamraoui L, Guibert J, Beaujean N, Szollosi MS, Debey P. Differential transcriptional activity associated with chromatin configuration in fully grown mouse germinal vesicle oocytes. Biol Reprod. 1999;60:580–587. doi: 10.1095/biolreprod60.3.580. [DOI] [PubMed] [Google Scholar]
- Bouniol C, Nguyen E, Debey P. Endogenous Transcription Occurs at the 1-Cell Stage in the Mouse Embryo. Experimental Cell Research. 1995;218:57–62. doi: 10.1006/excr.1995.1130. [DOI] [PubMed] [Google Scholar]
- Braude P, Pelham H, Flach G, Lobatto R. Post-transcriptional control in the early mouse embryo. Nature. 1979;282:102–105. doi: 10.1038/282102a0. [DOI] [PubMed] [Google Scholar]
- Bursac S, Brdovcak MC, Pfannkuchen M, Orsolic I, Golomb L, Zhu Y, Katz C, Daftuar L, Grabusic K, Vukelic I, et al. Mutual protection of ribosomal proteins L5 and L11 from degradation is essential for p53 activation upon ribosomal biogenesis stress. Proc Natl Acad Sci U S A. 2012;109:20467–20472. doi: 10.1073/pnas.1218535109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bywater MJ, Poortinga G, Sanij E, Hein N, Peck A, Cullinane C, Wall M, Cluse L, Drygin D, Anderes K, et al. Inhibition of RNA Polymerase I as a Therapeutic Strategy to Promote Cancer-Specific Activation of p53. Cancer Cell. 2012;22:51–65. doi: 10.1016/j.ccr.2012.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clift D, Schuh M. Restarting life: fertilization and the transition from meiosis to mitosis. Nat Rev Mol Cell Bio. 2013;14:549–562. doi: 10.1038/nrm3643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox SG, Kim H, Garnett AT, Medeiros DM, An W, Crump JG. An Essential Role of Variant Histone H3.3 for Ectomesenchyme Potential of the Cranial Neural Crest. Plos Genetics. 2012;8 doi: 10.1371/journal.pgen.1002938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vries WN, Binns LT, Fancher KS, Dean J, Moore R, Kemler R, Knowles BB. Expression of Cre recombinase in mouse oocytes: A means to study maternal effect genes. Genesis. 2000;26:110–112. [PubMed] [Google Scholar]
- Dorn ES, Cook JG. Nucleosomes in the neighborhood: new roles for chromatin modifications in replication origin control. Epigenetics. 2011;6:552–559. doi: 10.4161/epi.6.5.15082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drygin D, Lin A, Bliesath J, Ho CB, O’Brien SE, Proffitt C, Omori M, Haddach M, Schwaebe MK, Siddiqui-Jain A, et al. Targeting RNA polymerase I with an oral small molecule CX-5461 inhibits ribosomal RNA synthesis and solid tumor growth. Cancer Res. 2011;71:1418–1430. doi: 10.1158/0008-5472.CAN-10-1728. [DOI] [PubMed] [Google Scholar]
- Farley FW, Soriano P, Steffen LS, Dymecki SM. Widespread recombinase expression using FLPeR (flipper) mice. Genesis. 2000;28:106–110. [PubMed] [Google Scholar]
- Flaherty SP, Payne D, Swann NJ, Matthews CD. Assessment of fertilization failure and abnormal fertilization after intracytoplasmic sperm injection (ICSI) Reprod Fertil Dev. 1995;7:197–210. doi: 10.1071/rd9950197. [DOI] [PubMed] [Google Scholar]
- Gu TP, Guo F, Yang H, Wu HP, Xu GF, Liu W, Xie ZG, Shi L, He X, Jin SG, et al. The role of Tet3 DNA dioxygenase in epigenetic reprogramming by oocytes. Nature. 2011;477:606–610. doi: 10.1038/nature10443. [DOI] [PubMed] [Google Scholar]
- Haddach M, Schwaebe MK, Michaux J, Nagasawa J, O’Brien SE, Whitten JP, Pierre F, Kerdoncuff P, Darjania L, Stansfield R, et al. Discovery of CX-5461, the First Direct and Selective Inhibitor of RNA Polymerase I, for Cancer Therapeutics. Acs Med Chem Lett. 2012;3:602–606. doi: 10.1021/ml300110s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajkova P, Ancelin K, Waldmann T, Lacoste N, Lange UC, Cesari F, Lee C, Almouzni G, Schneider R, Surani MA. Chromatin dynamics during epigenetic reprogramming in the mouse germ line. Nature. 2008;452:877–881. doi: 10.1038/nature06714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamatani T, Carter MG, Sharov AA, Ko MS. Dynamics of global gene expression changes during mouse preimplantation development. Dev Cell. 2004;6:117–131. doi: 10.1016/s1534-5807(03)00373-3. [DOI] [PubMed] [Google Scholar]
- Hammoud S, Liu L, Carrell DT. Protamine ratio and the level of histone retention in sperm selected from a density gradient preparation. Andrologia. 2009;41:88–94. doi: 10.1111/j.1439-0272.2008.00890.x. [DOI] [PubMed] [Google Scholar]
- Haukenes G, Kalland KH. Visualisation of ribosomal RNA (rRNA) synthesis in eukaryotic cells in culture. Methods in Cell Science. 1998;19:295–302. [Google Scholar]
- Howlett SK. The effect of inhibiting DNA replication in the one-cell mouse embryo. Roux’s archives of developmental biology. 1986;195:499–505. doi: 10.1007/BF00375890. [DOI] [PubMed] [Google Scholar]
- Johnson MH. The molecular and cellular basis of preimplantation mouse development. Biol Rev Camb Philos Soc. 1981;56:463–498. doi: 10.1111/j.1469-185x.1981.tb00356.x. [DOI] [PubMed] [Google Scholar]
- Kent T, Lapik YR, Pestov DG. The 5′ external transcribed spacer in mouse ribosomal RNA contains two cleavage sites. Rna. 2009;15:14–20. doi: 10.1261/rna.1384709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraushaar DC, Jin W, Maunakea A, Abraham B, Ha M, Zhao K. Genome-wide incorporation dynamics reveal distinct categories of turnover for the histone variant H3.3. Genome Biol. 2013;14:R121. doi: 10.1186/gb-2013-14-10-r121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kressler D, Hurt E, Bassler J. Driving ribosome assembly. Biochim Biophys Acta. 2010;1803:673–683. doi: 10.1016/j.bbamcr.2009.10.009. [DOI] [PubMed] [Google Scholar]
- Laferte A, Favry E, Sentenac A, Riva M, Carles C, Chedin S. The transcriptional activity of RNA polymerase I is a key determinant for the level of all ribosome components. Gene Dev. 2006;20:2030–2040. doi: 10.1101/gad.386106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam YW, Lamond AI, Mann M, Andersen JS. Analysis of nucleolar protein dynamics reveals the nuclear degradation of ribosomal proteins. Curr Biol. 2007;17:749–760. doi: 10.1016/j.cub.2007.03.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latham KE, Akutsu H, Patel B, Yanagimachi R. Comparison of gene expression during preimplantation development between diploid and haploid mouse embryos. Biol Reprod. 2002;67:386–392. doi: 10.1095/biolreprod67.2.386. [DOI] [PubMed] [Google Scholar]
- Lewis PW, Muller MM, Koletsky MS, Cordero F, Lin S, Banaszynski LA, Garcia BA, Muir TW, Becher OJ, Allis CD. Inhibition of PRC2 activity by a gain-of-function H3 mutation found in pediatric glioblastoma. Science. 2013;340:857–861. doi: 10.1126/science.1232245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin CJ, Conti M, Ramalho-Santos M. Histone variant H3.3 maintains a decondensed chromatin state essential for mouse preimplantation development. Development. 2013;140:3624–3634. doi: 10.1242/dev.095513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loppin B, Bonnefoy E, Anselme C, Laurencon A, Karr TL, Couble P. The histone H3.3 chaperone HIRA is essential for chromatin assembly in the male pronucleus. Nature. 2005;437:1386–1390. doi: 10.1038/nature04059. [DOI] [PubMed] [Google Scholar]
- Maze I, Noh KM, Soshnev AA, Allis CD. Every amino acid matters: essential contributions of histone variants to mammalian development and disease. Nat Rev Genet. 2014;15:259–271. doi: 10.1038/nrg3673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modlinski JA. Haploid Mouse Embryos Obtained by Microsurgical Removal of One Pronucleus. J Embryol Exp Morph. 1975;33:897–905. [PubMed] [Google Scholar]
- Nakamura T, Liu YJ, Nakashima H, Umehara H, Inoue K, Matoba S, Tachibana M, Ogura A, Shinkai Y, Nakano T. PGC7 binds histone H3K9me2 to protect against conversion of 5mC to 5hmC in early embryos. Nature. 2012;486:415–419. doi: 10.1038/nature11093. [DOI] [PubMed] [Google Scholar]
- Nothias JY, Miranda M, DePamphilis ML. Uncoupling of transcription and translation during zygotic gene activation in the mouse. EMBO J. 1996;15:5715–5725. [PMC free article] [PubMed] [Google Scholar]
- Paynton BV, Rempel R, Bachvarova R. Changes in state of adenylation and time course of degradation of maternal mRNAs during oocyte maturation and early embryonic development in the mouse. Dev Biol. 1988;129:304–314. doi: 10.1016/0012-1606(88)90377-6. [DOI] [PubMed] [Google Scholar]
- Raj A, van den Bogaard P, Rifkin SA, van Oudenaarden A, Tyagi S. Imaging individual mRNA molecules using multiple singly labeled probes. Nat Methods. 2008;5:877–879. doi: 10.1038/nmeth.1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray-Gallet D, Quivy JP, Scamps C, Martini EMD, Lipinski M, Almouzni G. HIRA is critical for a nucleosome assembly pathway independent of DNA synthesis. Molecular Cell. 2002;9:1091–1100. doi: 10.1016/s1097-2765(02)00526-9. [DOI] [PubMed] [Google Scholar]
- Schneiderman JI, Orsi GA, Hughes KT, Loppin B, Ahmad K. Nucleosome-depleted chromatin gaps recruit assembly factors for the H3.3 histone variant. P Natl Acad Sci USA. 2012;109:19721–19726. doi: 10.1073/pnas.1206629109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz RM. The molecular foundations of the maternal to zygotic transition in the preimplantation embryo. Hum Reprod Update. 2002;8:323–331. doi: 10.1093/humupd/8.4.323. [DOI] [PubMed] [Google Scholar]
- Schultz RM, Wassarman PM. Specific changes in the pattern of protein synthesis during meiotic maturation of mammalian oocytes in vitro. Proc Natl Acad Sci U S A. 1977;74:538–541. doi: 10.1073/pnas.74.2.538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su YQ, Sugiura K, Woo Y, Wigglesworth K, Kamdar S, Affourtit J, Eppig JJ. Selective degradation of transcripts during meiotic maturation of mouse oocytes. Dev Biol. 2007;302:104–117. doi: 10.1016/j.ydbio.2006.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szenker E, Lacoste N, Almouzni G. A Developmental Requirement for HIRA-Dependent H3.3 Deposition Revealed at Gastrulation in Xenopus. Cell Reports. 2012;1:730–740. doi: 10.1016/j.celrep.2012.05.006. [DOI] [PubMed] [Google Scholar]
- Sztein JM, Farley JS, Mobraaten LE. In vitro fertilization with cryopreserved inbred mouse sperm. Biology of Reproduction. 2000;63:1774–1780. doi: 10.1095/biolreprod63.6.1774. [DOI] [PubMed] [Google Scholar]
- Tagami H, Ray-Gallet D, Almouzni G, Nakatani Y. Histone H3.1 and H3.3 complexes mediate nucleosome assembly pathways dependent or independent of DNA synthesis. Cell. 2004;116:51–61. doi: 10.1016/s0092-8674(03)01064-x. [DOI] [PubMed] [Google Scholar]
- Torres-Padilla ME, Bannister AJ, Hurd PJ, Kouzarides T, Zernicka-Goetz M. Dynamic distribution of the replacement histone variant H3.3 in the mouse oocyte and preimplantation embryos. Int J Dev Biol. 2006;50:455–461. doi: 10.1387/ijdb.052073mt. [DOI] [PubMed] [Google Scholar]
- van der Heijden GW, Dieker JW, Derijck AA, Muller S, Berden JH, Braat DD, van der Vlag J, de Boer P. Asymmetry in histone H3 variants and lysine methylation between paternal and maternal chromatin of the early mouse zygote. Mech Dev. 2005;122:1008–1022. doi: 10.1016/j.mod.2005.04.009. [DOI] [PubMed] [Google Scholar]
- Wang QT, Piotrowska K, Ciemerych MA, Milenkovic L, Scott MP, Davis RW, Zernicka-Goetz M. A genome-wide study of gene activity reveals developmental signaling pathways in the preimplantation mouse embryo. Developmental Cell. 2004;6:133–144. doi: 10.1016/s1534-5807(03)00404-0. [DOI] [PubMed] [Google Scholar]
- Wassarman PM, Letourneau GE. RNA synthesis in fully-grown mouse oocytes. Nature. 1976;261:73–74. doi: 10.1038/261073a0. [DOI] [PubMed] [Google Scholar]
- White JK, Gerdin AK, Karp NA, Ryder E, Buljan M, Bussell JN, Salisbury J, Clare S, Ingham NJ, Podrini C, et al. Genome-wide Generation and Systematic Phenotyping of Knockout Mice Reveals New Roles for Many Genes. Cell. 2013;154:452–464. doi: 10.1016/j.cell.2013.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolford JL, Jr., Baserga SJ. Ribosome biogenesis in the yeast Saccharomyces cerevisiae. Genetics. 2013;195:643–681. doi: 10.1534/genetics.113.153197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamanaka S, Blau HM. Nuclear reprogramming to a pluripotent state by three approaches. Nature. 2010;465:704–712. doi: 10.1038/nature09229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Shi L, Wang BA, Liang D, Zhong C, Liu W, Nie Y, Liu J, Zhao J, Gao X, et al. Generation of genetically modified mice by oocyte injection of androgenetic haploid embryonic stem cells. Cell. 2012;149:605–617. doi: 10.1016/j.cell.2012.04.002. [DOI] [PubMed] [Google Scholar]
- Zatsepina O, Baly C, Chebrout M, Debey P. The step-wise assembly of a functional nucleolus in preimplantation mouse embryos involves the cajal (coiled) body. Dev Biol. 2003;253:66–83. doi: 10.1006/dbio.2002.0865. [DOI] [PubMed] [Google Scholar]
- Zeng F, Baldwin DA, Schultz RM. Transcript profiling during preimplantation mouse development. Dev Biol. 2004;272:483–496. doi: 10.1016/j.ydbio.2004.05.018. [DOI] [PubMed] [Google Scholar]
- Zernicka-Goetz M, Morris SA, Bruce AW. Making a firm decision: multifaceted regulation of cell fate in the early mouse embryo. Nat Rev Genet. 2009;10:467–477. doi: 10.1038/nrg2564. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.