Abstract
Membrane bound vesicles, including microvesicles and exosomes, are secreted by both normal and cancerous cells into the extracellular space and in blood circulation. These circulating extracellular vesicles (cirEVs) and exosomes in particular are recognized as a potential source of disease biomarkers. However, to exploit the use of circulatory exosomes as a biomarker, a rapid, high-throughput and reproducible method is required for their isolation and molecular analysis. We have developed a simple, low cost microfluidic-based platform to isolate cirEVs enriched in exosomes directly from blood serum allowing simultaneous capture and quantification of exosomes in a single device. To capture specific exosomes, we employed “ExoChip”, a microfluidic device fabricated in polydimethylsiloxane (PDMS) and functionalized with antibodies against CD63, an antigen commonly overexpressed in exosomes. Subsequent staining with a fluorescent carbocyanine dye (DiO) that specifically labels the exosomes, we quantitated exosomes using a standard plate-reader. Ten independent ExoChip experiments performed using serum obtained from five pancreatic cancer patients and five healthy individuals revealed a statistically significant increase (2.34±0.31 fold, p <0.001) in exosomes captured in cancer patients when compared to healthy individuals. Exosomal origins of ExoChip immobilized vesicles were further confirmed using immuno-electron-microscopy and Western blotting. In addition, we demonstrate the ability of ExoChip to recover exosomes with intact RNA enabling profiling of exosomal-microRNAs through openarray analysis, which has potential applications in biomarker discovery. Based on our findings, ExoChip is a well suited platform to be used as an exosome-based diagnostic and research tool for molecular screening of human cancers.
INTRODUCTION
Diagnosing a cancer via cross sectional imaging (CT scan) and biopsy is an expensive and often uncomfortable approach for patients to undergo, yielding substantial false-negative rates and a limited potential for early diagnosis of disease1. One of the recent technological advancements in the detection of cancer is the use of microfluidic-based approaches to identify circulating tumor cells (CTCs) from the peripheral blood as a non-invasive procedure2–4. However, in early stage cancers few CTCs are present in the circulation; therefore it remains unclear if the measurement of CTCs with current technology will be an effective approach for early cancer detection4–9.
Studies have shown that circulatory extracellular vesicles (cirEVs), primarily originating from tumors, are a potential source of cancer biomarkers in cancer patients10–13. Furthermore, cancer patients exhibit a significantly higher quantity of total exosomes than healthy individuals as exosomes are secreted in large amounts during carcinogenesis. Additionally, it has been reported that certain proteins and nucleic acids including microRNAs (miRNAs), exclusively carried by exosomes, are associated with malignant tumors14–16. Interestingly, the functional relationships between tumor derived exosomes and cancer progression is not well understood yet, however, recognizing the diagnostic and prognostic potential of exosomes, certain exosome-based bio-assays have already been proposed for minimally invasive cancer detection10, 17.
The current methods commonly used for isolation and quantification of exosomes involve a series of differential centrifugations and filtration steps followed by a high-speed ultracentrifugation to pellet the membrane bound vesicles containing mainly exosomes (~30–300nm diameter), microvesicles of heterogeneous size (~ 0.5–5μm diameter) and protein aggregates. Exosomes are purified from these vesicles using a sucrose density gradient ultracentrifugation and finally are processed for morphological and molecular characterization using electron microscopy and immunophenotyping techniques18. These procedures for exosome isolation are lengthy (6–8 h), require an ultracentrifuge and yield a relatively low recovery of exosomes especially from blood samples making it difficult for clinical applications. Therefore, a rapid and reproducible isolation method for exosomes is essential to exploit them as a new diagnostic and therapeutic tool, as well as to carry out basic molecular analysis of exosome functions.
Recent advances in microfluidic based technologies have made it possible to extract EVs from the blood in an easily reproducible, convenient manner. The foremost among these approaches include use of a microfluidic device for isolating exosomes using an immuno-affinity approach, use of a porous silicon nanowire-on-micropillar structure, or isolating exosomes from whole blood using in situ prepared nanoporous membranes19–21. These new techniques provide faster separation than the standard approaches; however optimization of these microfluidic platforms is needed for application to the clinical settings. Furthermore, currently, microfluidic platforms have not been integrated with standard bio-analytical systems for molecular profiling and quantification of the exosomes. Taking into account the need for a significantly improved approach for exosome isolation, we designed the “ExoChip” which enables the isolation, on-chip quantification, and molecular characterization of exosomes.
PRINCIPLE
Engineering design of the ExoChip to integrate with existing read-out instruments for rapid quantification
Current techniques fail to accurately determine the concentration of vesicles, in particular the exosomes in blood samples, and rely on procedures of protein concentration or nanoparticle tracking analysis, which are semi-quantitative22, 23. The focus of our study is to present a microfluidic based methodology that is sensitive and readily can translate into clinical applications for the use of exosome based biomarker analysis in cancer research. The microfluidic approach as described by Chen et al19 captured serum vesicles using anti-CD63 capture antibody whereas Davies et al 21 reported the use of a PMMA-based membrane filter and Wang et al20 recently reported the use of ciliated micropillars (or nanowires) to separate the vesicles. The latter two prototypes are inherently vulnerable to clogging and none of these techniques allow direct measurement of immobilized exosomes, but rather require additional steps to extract exosomes for quantification. Hence, we selected the immuno-affinity-based approach of Chen et al,19 which offers selective capture of exosomes, and improvise the design such that, device is suitable for carrying out standardized studies using clinical samples. We have designed ExoChip, to perform isolation, detection and quantification of exosomes directly on-chip is schematically illustrated in Fig. 1. The series of alternatively placed circular chambers interconnected through straight narrow-cannels in the micro fluidic device allows for increased retention time (having reduced velocity, Fig S1) for the exosomes to interact with the functionalized surface, while straight channels present in between these circular areas allow intermittent mixing of the exosomes because of the changes in the fluid velocities, as determined by the simulation studies (Fig S1). Furthermore, the ExoChip was geometrically designed such that the standard plate reader can read them as standard wells, enabling the quantitative imaging. Additionally, the design aspects of ExoChip are fully scalable in throughput as the platform can be constructed as comprising single channel (Fig 1A) to multi-channels (Fig 1B,C). Microtiter-plate footprint where multichannel ExoChip allows the convenience of rapid analysis of multiple samples.
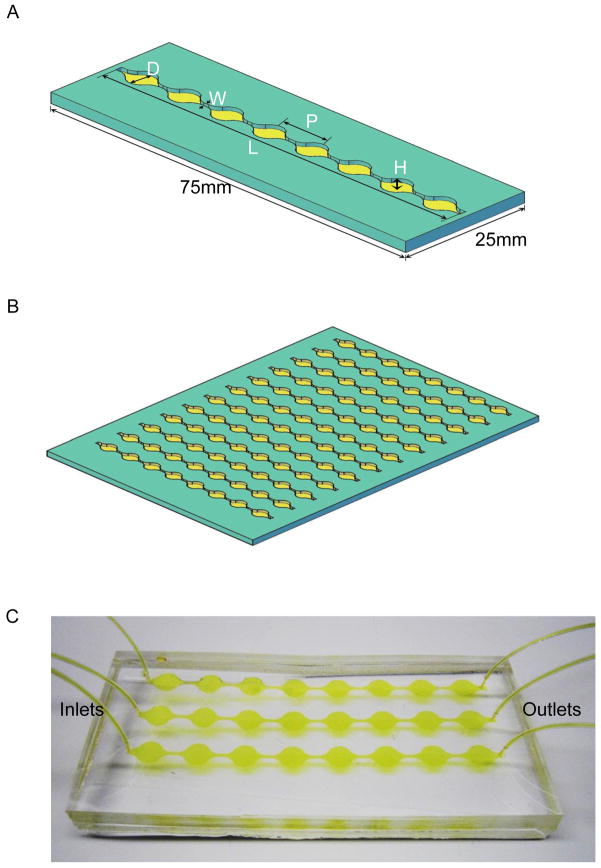
Fig. 1. ExoChip design and working prototype.
A: The ExoChip features a single channel of 73mm in length (L) and 100μm in height (H) consisting of eight circular chambers, with a diameter of 5mm (D) each, equally spaced at a distance 9mm (P) from each other connected through a shorter 0.75mm (W) wide channels. A single channel ExoChip design follows the overall dimension of a standard glass-side having 75mm×25mm size dimension whereas prototyped configuration of a multichannel ExoChip (B) follows a microtiter-plate footprint containing up to 12 channels laterally placed at 9mm distance, allowed its use for analyzing multiple samples simultaneously. C: A working prototype model of PDMS based ExoChip (three channel) depicting the flow of serum for exosomes capture in a typical experimental setup.
On-Chip Isolation, Detection and Quantification of Exosomes
To enable on-chip quantification of exosomes, we introduce a method for labeling exosomes bounded to the ExoChip with a fluorescent dye which allows accurate quantitation of exosomes using a plate reader. Fig. 2a represents the experimental strategy for isolation, quantification and characterization of exosomes using the ExoChip. The presented approach is a simple three-step process which involves the following procedure: (i) First, a serum sample obtained from healthy and/or diseased individual is infused through the inlet of ExoChip which is pre-coated with exosome-specific capture antibodies (anti-CD63). It should be noted that the CD63 protein, a member of a tetraspanin superfamily or transmembrane 4 superfamily (TM4SF), is one of the most abundant protein found in exosomes, thereby provide selectivity to the isolation procedure19,24. (ii) Next, the captured exosomes in the ExoChip are stained using fluorescence dye (DiO) which specifically stains only the membrane vesicles immobilized in the ExoChip. The added advantage of staining the exosomes with fluorescent dye is that it provides an easy visualization of otherwise microscopically invisible small sized vesicles (30–300nm) for imaging purposes. (iii) Finally, the total levels of exosomes are determined on-chip. Fluorescently stained vesicles can be measured using standard analytical tools such as a plate reader. Calibration of fluorescent intensity against the exosomes captured on chip and the ability of ExoChip to quantify relative abundance of exosomes were validated through serum samples containing increasing exosomes concentrations (volume/volume), viz., exosome-free serum (serum with 0% exosomes), 50% exosome-serum and 100% exosome-serum. A representative plot of fluorescence intensities against exosomes levels is shown in Fig S2 A. This data confirmed that ExoChip is capable to quantify the fluorescent intensities as obtained from a standard read-out instrument in multiple samples and also validate in principle our method to compare differentially secreted exosomes levels across healthy and diseased individuals.
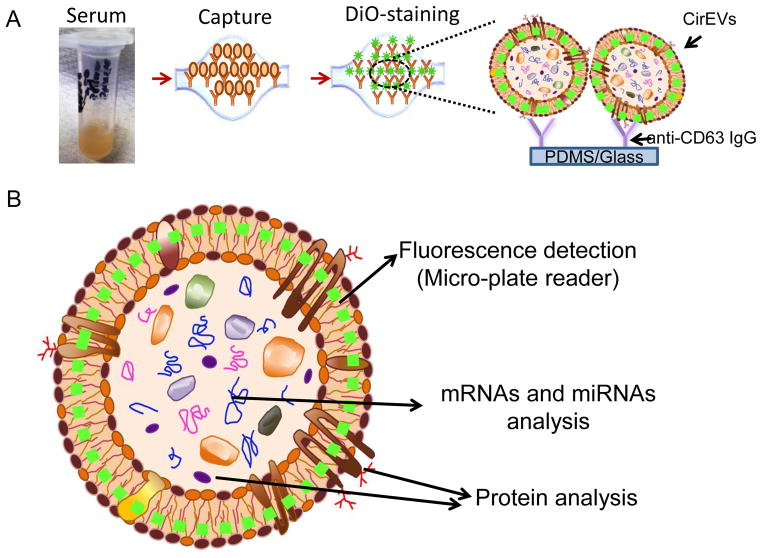
Fig. 2. Experimental strategy for exosomes immobilization and characterization using ExoChip.
A: Illustration depicting the scheme of exosomes capture and analysis procedure using ExoChip. The blood is collected for serum extraction from healthy or diseased individuals and then exosomes are captured by flowing serum through a CD63-antibody coated ExoChip. To visualize the captured exosomes, ExoChip is processed for membrane specific dye (DiO) staining. B: The ExoChip is designed to measure the levels of fluorescently stained exosomes through fluorescence intensity measurements using micro-plate readers and allows molecular characterization of Exosomes contents through variety of standard assays including protein analysis (western blots) and mRNA/miRNA analysis (RT-PCR/miRNA openarray).
The design of ExoChip, coupled with the method of exosome quantification as described above, therefore enables to perform the minimally invasive exosomes-based quick diagnostic test for cancer.
Material and methods
ExoChip fabrication and functionalization
The device is made of polydimethylsiloxane (PDMS), using standard soft lithography techniques. A SU-8 mold is fabricated with UV light shining through a transparent mask. The PDMS elastomer and curing agent are poured onto the mold and cured in an oven overnight. The PDMS chamber is then bonded to a glass slide (single channel ExoChip) or PDMS block (multichannel-ExoChip) after plasma treatments. Fig. 1 represents the prototyped fabrication design of ExoChip. The completed device is immediately treated with surface functionalization chemicals which consist of 3-mercaptopropyltrimethoxysilane (Gelest), GMBS (Pierce), NeutrAvidin (Invitrogen) and biotinylated anti-CD63 (Ancell). The control devices lacked biotinylated anti-CD63. We designed the ExoChip, either single channel or multichannel (for analysis of multiple samples), to be compatible for use with standard micro-well plate readers.
Serum Preparation
Blood samples drawn in lavender EDTA tubes (BD Biosciences) were obtained from healthy individuals and pancreatic cancer patients in the Multidisciplinary Pancreatic Tumor Clinic at University of Michigan, Ann Arbor, USA using IRB approved protocols. Samples were centrifuged immediately for 10 min at 2000 RCF and the top layer of plasma was carefully collected for serum extraction. Serum was prepared by coagulating plasma fibrinogen and clotting factors using the Pacific Hemostasis Thromboplastin D reagent (Thermo). For this, an equal amount of plasma and thromboplastin D reagent were mixed rapidly and incubated at 37°C for 15 min to achieve coagulation of plasma proteins. Samples were then centrifuged at 10,000 rpm at room temperature for 10 min to collect the clear supernatant as serum and stored immediately at −80°C until use.
Exosome capture
Exosome isolation in ExoChip was based on the immuno-affinity approach originally reported by Chen et al in 2010 19. In a typical experiment, biotinylayted anti-CD-63 treated and control devices were first blocked with 3% BSA- PBS solution infused at a flow rate of 50 μl/min for 10 min and then incubated for 30 min. Serum samples (400 μl) derived from healthy volunteers or pancreatic cancer patients were infused through the device at a flow rate of 8μl/min followed by a PBS rinse at a flow rate of 50 μL/min for 10 min. Serum infused ExoChip channels were analyzed for the amount of exosomes captured by various on-chip-characterization assays including quantitation methods to determine total exosomes levels following the fluorescence labeling method as well as quantitating total protein and nucleic acid in ExoChip.
Visualization and On-chip Quantification of exosomes
The captured exosomes were labeled with fluorescent carbocyanine dye, Vybrant™ DiO (Molecular Probes) at a flow rate of 10μl/min for 10 min followed by 10 min incubation at 37°C required for DiO to react with exosomal membrane vesicles in the device. After a PBS rinse (at a flow rate of 10μl/min for 10 min), initial visualization of immobilized vesicles was done through fluorescence (Nikon Eclipse Ti equipped with NIS Elements software) and confocal microscopy examinations. For quantification of exosomes levels, the relative fluorescence intensities were measured at excitation and emission wavelengths of 485 and 510nm respectively, using a BioTek-Synergy Neo multi-purpose plate reader. The relative fluorescent intensity (RFI) measured for healthy and disease samples were normalized with the fluorescence intensity of a control. The relative fold-change in fluorescence intensities was computed by comparing the normalized RFI values for healthy and diseased samples.
Electron Microscope (EM) analysis of ExoChip captured exosomes
Exosomes captured in the ExoChip were fixed in 2.5% glutaraldehyde-Sorensen’s buffer for 30 minutes and then rinsed for 3 × 5 minutes with Sorensen’s buffer. The samples were post-fix for 15 minutes in 1% osmium tetroxide and rinsed (3 × 5 minutes) with Sorensen’s buffer. The samples were dehydrated in a graded series of ethanols (30%, 50%, 70%, 95% and 100%) for 2 × 10 min at each step. The samples were then coated with gold using a high resolution ion bean coater and examined using a FEI Nova 200 Nanolab Dualbeam FIB scanning electron microscope at the Electron Microscopy Analysis Lab (EMAL) at University of Michigan.
Immunogold Labeling of ExoChip- Exosomes for EM Analysis
Exosomes attached to the PDMS surface of ExoChip were initially fixed in 4% PFA solution for 1 h and then incubated for 15 min in 0.05M glycine solution to inactivate residual aldehyde groups present after the aldehyde fixation. Blocking was done for 15 min using 5% BSA-PBS + 5% normal serum with the same species as that of secondary antibodies, followed by 3 × 5 min wash in incubation buffer (0.1–0.2% Aurion BSA-c™ containing 10 mM NaN3). Subsequently, samples were incubated overnight at 4°C in primary antibodies (20 μg/ml) prepared in incubation buffer followed by a 6 × 5 min wash in incubation buffer. Samples were incubated overnight at 4°C in corresponding secondary antibodies using the AURION Conventional Immunogold Reagent (containing gold particles of 25 nm diameter) at a 1:20 dilution, which was followed by 6 × 5 min wash in incubation buffer. Samples were rinsed 3 × 5 min in PBS and processed for EM analysis.
Western Blotting
ExoChip-immobilized exosomes were lysed in RIPA buffer (Sigma). Equal amount of protein samples prepared in Laemmli sample buffer were boiled for 7 minutes, and subjected to SDS-PAGE. The proteins were transferred onto nitrocellulose membranes (Novex, Life technologies) using a dry Transfer-Blot kit (Novex, Life technologies). Blots were first incubated in PBS blocking buffer containing 5% milk for 1 hour at room temperature and then with the respective primary antibodies (anti-CD63 and anti-Rab5, Santa Cruz Biotechnology) diluted in PBST (containing 0.1% Tween 20) overnight at 4°C. Subsequently, blots were washed and incubated with appropriate secondary antibodies (Millipore) in PBST and detected using SuperSignal West Pico Chemiluminescent Substrate (Thermo Scientific).
RNA extraction and Openarray miRNAs analysis
Total RNA was extracted from the ExoChip using RNeasy Micro kit (QIAGEN) and the samples were analyzed for miRNAs expression at the DNA Sequencing Core, University of Michigan. RNA integrity and quantity was analyzed using the Bioanalyzer. 100 ng of RNA was used for making cDNA using Megaplex™ RT Primers and each 2.5uL of RT product was preamplified to increase the quantity of desired cDNA prior to PCR on the TaqMan® OpenArray® MiRNAs plates for miRNAs expression analysis following the instructions provided by the manufacturer (Applied Biosystems, Life technologies). The DCT values were calculated as the difference of CT values of each miRNAs to an endogenous reference gene U6. The fold changes were calculated as 2−DDCT with the DDCT being the difference between the cancer patients and healthy samples being compared.
Statistical Analyses
Statistical differences for indicated assays were determined using Student’s t-test. A P value less than 0.05 was considered statistically significant.
RESULTS AND DISCUSSION
On-chip Visualization, Quantification and Characterization of ExoChip-exosomes
The current methods of isolation and quantification of exosomes do not account for the relative secretion of exosomes in a cancer state as compared to the native levels of exosomes in a healthy state. Hence, the quantification is limited to the identification of exosomes, and cannot be used directly for diagnostics. Previously, various fluorescent lipophilic tracers have been used in the fluorescent exosomes localization technique to analyze the purity of exosomes obtained from cell culture supernatant through differential centrifugation techniques25–27. To improve upon this approach, we selected to use Molecular Probes’ Vybrant™ DiO (3,3′-dihexadecyloxacarbocyanine) as it offers several advantages over the previously used structurally related PKH dyes including: (1) the DiO dye labeling protocol requires only one step whereas PKH labeling includes multiple steps, including an application of iso-osmotic mannitol solution and a longer incubation time, thereby increasing the risk of losing captured exosomes, and (2) use of DiO improves the successful use of exosomes for downstream assays to be performed on ExoChip27. Therefore, to visualize the immobilized exosomes captured in ExoChip we stained the exosomes with DiO. Figure 3 presents several images of the ExoChip and isolated, fluorescently labeled exosomes. Our results show that only those channels that were coated with anti-CD63 captured the vesicles (Fig. 3A). In the control channels, although bleak fluorescence was noticed on the surface, no immobilization of vesicles was observed (Fig. 3B). Interestingly, ExoChip bounded native exosomes were observed to be present in clusters as well as single vesicles. Per the limits of light microscopy only those exosomes that were in clusters were visualized (Fig. 3C) however, confocal microscopy results indicated that clusters are formed from smaller vesicles (Fig. 3D). The results of EM analyses further confirmed these observations, as vesicles were present only in the anti-CD63 coated devices as single and in clusters that were comparable to our confocal microscopy results (Fig. 3E–G). Of particular importance is the fact that the traditional methods of isolating exosomes are known to interfere with the structural morphology of the exosomes and therefore, fails to present the true picture of native exosomes23. It is currently not known whether exosomes function as single entities or if exosomes function as small clusters or aggregates, based on traditional methods of exosome isolation previous reports do suggest that single exosome vesicles size may vary from 30–100 nm28. Our findings as revealed through EM, clearly showed that most of the exosomes captured by ExoChip were in the size range from 30–300nm diameter (Fig. 3F) where 300nm exosomes were seen as comprised of smaller sized vesicles clustered together (Fig. 3G). These findings provide direct evidence that circulating exosomes of ~100nm size may tend to form aggregates. Further, an overall morphology and size of captured vesicles were found to closely resemble to those reported for exosomes in previously characterized studies using EM29, 30.
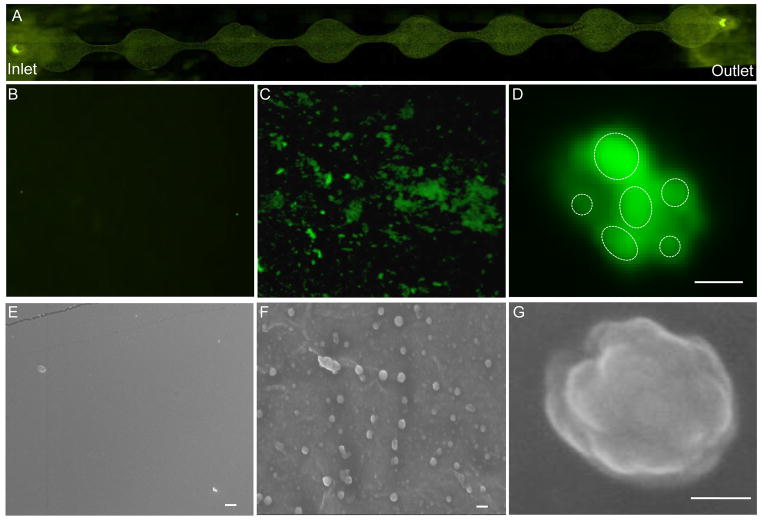
Fig. 3. Characterization of Exosomes captured using ExoChip.
A: Fluorescence microscopy image of ExoChip channel depicting immobilization of exosomes in native forms after DiO staining. High magnification (400X) images of control one of the control chamber (B) and anti-CD63 coated chambers depicting exosomes capture (C). Per the light-microscopy resolution limits the exosomes as visualized were found to be in clusters which were also confirmed through confocal microscopy. D: A confocal microscopic image showing a group of smaller exosomes (depicted as dotted-circles) forming a cluster (bar = 2μm). The native morphology of the exosomes vesicles were revealed following EM analysis. Electron micrograph images showed the absence of any vesicle immobilizations in control ExoChip (E), whereas the anti-CD63 coated ExoChip (F) showed the presence of exosomes both in clusters as well as single vesicles (bar = 500nm). G: A magnified view of a single exosome vesicles as examined under EM depicting a cluster of exosomes (bar = 100nm).
Immuno-characterization of ExoChip-exosomes: EM Studies
The biogenesis of exosomes and their biological role in disease pathogenesis is an emerging field of research. Studies suggest that Rab5 protein, which belongs to a family of GTPases localized to distinct membrane bound compartments, play a crucial role in the regulation of membrane traffic during biogenesis of exosomes31. Rab5 is specifically expressed during early stage endosome formation and later carried as membrane-bound during exosomes formation31. To confirm the exosomal origin of the vesicles captured in the ExoChip, we performed on-chip immuno-detection of Rab5 using immuno-labeled nanogold particles in the exosomes and analyzed through EM. The immuno-electronmicroscopy technique using gold-nanoparticles is a validated method for ultrastructure studies of cellular antigens32. We have utilized the commonly used “two step” method, in which primary antibodies are applied, followed by detection using immunogold conjugate reagents, for localizing Rab5 on the exosomes captured in the ExoChip. Our EM analysis clearly revealed the localization of gold-nanoparticles bound to Rab5 on the membrane of the exosomes which were visualized as a typical cup-shaped vesicles ranging from 30–300 nm diameter (Figure 4A, B)23. Once again, our ExoChip-EM results showed the presence of both small sized vesicles of ~ 30 nm diameter (Figure 4A, shown encircled) and larger vesicles of up to 300 nm diameter (Fig. 4A, B). Our EM results on the size and morphology of exosomes presents a striking resemblance to previously reported EM analysis performed using classical methods of exosomes isolation thereby validate the specificity of our exosomes isolation 33, 34.
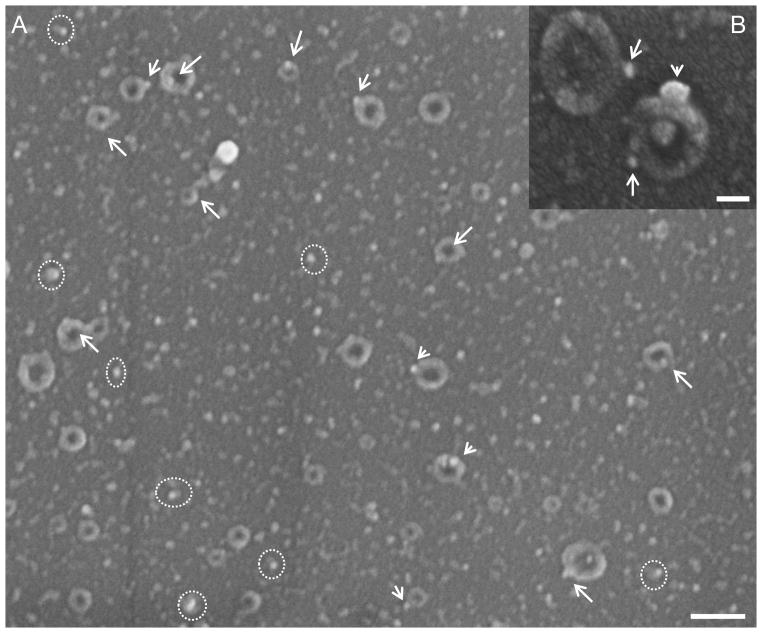
Fig. 4. Immuno-characterization of ExoChip bounded-exosomes.
A: EM analysis of the ExoChip chambers after Rab5-immunogold labeling showing typical features of cup-shaped exosomes with nano-gold particles (arrows). Also Rab-5 immunogold particles are seen bounded to smaller sized exosomes vesicles (dotted-circle), bar = 300nm. Inset B: magnified view of the exosome vesicles having diameter <300 nm labeled with Rab5-immunogold particles (bar = 100nm).
Quantification of ExoChip-Exosomes derived from Clinical samples
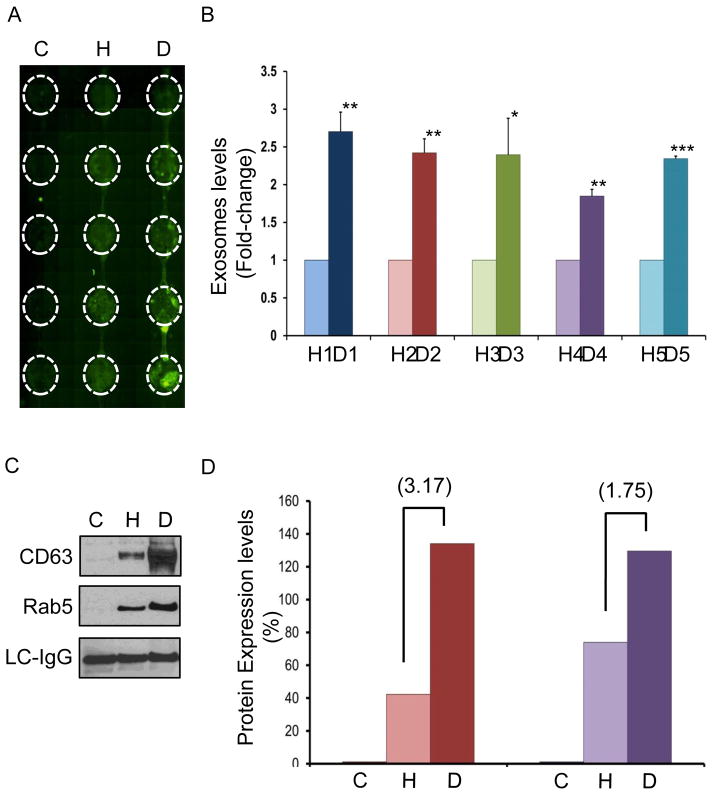
To establish clinical utility of ExoChip in determining the levels of exosomes, we quantified the exosomes in serum from pancreatic cancer patients and healthy individuals. Ten independent experiments using serum samples from five healthy individuals (healthy) and five pancreatic cancer patients (disease) were carried out to analyze the vesicles captured through ExoChip. A representative fluorescent image as captured through scanning the device using a fluorescence microscope is shown in Fig. 5A. On-chip quantification results showed an overall 2.34±0.31 fold increased (p <0.001) presence of circulating-exosomes in the cancer patients’ samples as compared to healthy controls (Fig. 5B, and Fig S2B). The data corroborates previous reports of increased secretion of exosomes in cancer patients and further, promises its utility in identifying cancer biomarkers23, 35, 36. This experiment demonstrates that a rapid-quantification of exosomes can be achieved in an easy way using ExoChip because of its compatibility with the standard plate reader. In addition, the design of ExoChip also offers an advantage to have multi-channels for simultaneous processing of multiple samples.
Fig. 5. On-chip Quantification and protein characterization of exosomes using clinical samples.
A: A typical image of a 3-channel ExoChip obtained through fluorescence microscope scanning following DiO staining of the immobilized exosomes, depicting the areas (encircled) of fluorescent intensity measurement for exosomes quantification. B: Exosomes levels in the serum obtained from pancreatic cancer patients (n=5) and healthy individuals (n=5) as calculated from fluorescence intensity measurements. The mean fluorescent intensity values obtained from ExoChip-chambers corresponding to healthy and diseased were normalized against the mean value of the control and used to calculate the fold-change in the levels of exosomes by comparing the means of disease (D) and healthy (H). The data represent the means of duplicate experiments carried out using ExoChip for each subject (Supplementary Information, Fig. S1). C: A representative western blot analysis of the proteins isolated from ExoChip and characterized for CD63 and Rab5 expression levels in diseased (D) and healthy (H) serum sample. A non-specific light chains immunoglobulins (LC-IgG) were detected to confirm equal loading of the proteins. D: Densitometry analysis of the protein expression levels were performed using image J and relative density of control (C) healthy (H) and pancreatic cancer patient (D) samples were plotted. Fold change in the expression levels for the diseased (D) when compared with healthy (H) are shown in brackets.
Protein Characterization of ExoChip-exosomes
Further, we observed the expression of two different classes of proteins CD63 and Rab5 using Western blot analysis on the exosomes isolated using ExoChip. The presence of these proteins, as derived from cytoplasmic organelles such as lysosomes (CD63) and endosomes (Rab5), is also used to exclude the possible presence of membrane vesicles derived from membranes shedding (ectosomes) and disruption (necrosis and apoptosis)37, 38. We observed that compared to healthy individuals, exosomes isolated from pancreatic cancer patients have an overall increase in the levels of both CD63 (3.17 fold) and Rab5 (1.75 fold) proteins (Fig. 5C,D). Although few reports are available on the roles of CD63 and Rab5 in pancreatic cancer derived exosomes, at the cellular level CD-63 family of proteins typically regulate tumor cell movement by altering cell–cell and cell–matrix interactions31. The high levels of CD63 expressing exosomes have been reported in the plasma of melanoma patients whereas, over-expression of the RAB5 gene in tumors has been correlated to a high metastatic potential in human lung, pancreatic and stomach cancers38–40. Recognizing the fact that clinicopathologic or prognostic implications of CD63 and Rab5 proteins in exosomes derived from pancreatic tumors are still unclear we plan to conduct further studies to unravel the potential of these molecules as diagnostic or prognostic biomarkers for pancreatic cancer using ExoChip. Nevertheless, from these findings we can conclude that the vesicles being immobilized in ExoChip represent true exosomes, as defined by their shape and size (determined using standard EM techniques) and also confirmed through various immunophenotyping assays.
MicroRNAs analysis of ExoChip-exosomes
Accumulating evidence suggests that exosomes act as mediators of tumor progression and metastasis22, 41, 42. Several studies in cancer patients have implicated exosomes as playing a key role in the activation of oncogenes and inactivation of tumor suppressor genes23, 42, whereas, other group of researchers have explored the potential of circulating exosomes to serve as biomarkers in cancer38, 43–45. MicroRNAs are known to be carried by exosomes and shown to play a key role in intercellular communications and have emerged as an important candidate for biomarker discovery22, 43, 46. In the present study, we tested our ExoChip for its applications in performing miRNA analysis of exosomes. Hence, we carried out the RNA isolation from the ExoChip captured exosomes for miRNA expression profiling in two cases of pancreatic cancer patients and compared this to a healthy control. We observed that about 90 miRNAs were differentially expressed in exosomes of pancreatic cancer patients in comparison to healthy control. In our analysis, we selected an upper threshold of >1.5 fold-change in expression for identifying upregulated miRNAs whereas to identify the down-regulated miRNAs, those showing < 0.67 fold-change were selected. Among these we observed that expression levels of certain miRNAs, specifically, hsa-miR-130a, hsa-miR-29b, hsa-miR-30b, hsa-miR-518d, hsa-miR-551b and hsa-miR-646 were up-regulated whereas, expression of hsa-miR-601, hsa-miR-106b, hsa-miR-92a, hsa-miR-1275, and hsa-miR-302c* were down-regulated in both the cases (Table 1). The complete list of individual miRNAs that were upregulated and down-regulated in both of the cases are presented in Supplementary Table 1. Interestingly, among those miRNAs that are upregulated in the two cases, hsa-miR-130a has previously been found to be upregulated in plasma of pancreatic cancer patients, while hsa-miR-29b has been reported to be upregulated in pancreatic cancer tumors 47, 48. Our preliminary analysis showed that most of the miRNAS detected in ExoChip derived exosomes are known to be associated with various types of cancers. Although, our findings encourage conducting a detailed prospective trial with a large sample population and performing detailed molecular analysis nonetheless, present work clearly demonstrates the potential application of ExoChip in transcriptome- based predictive biomarker exploration research.
Table 1. miRNA expression profile analysis on ExoChip bound exosomes.
List of miRNAs identified to be commonly expressed (upregulated or downregulated) in the two cases of pancreatic cancer patients.
| Upregulated miRNAs (>1.5-fold) | Downregulated miRNAs (0–0.67 fold) | |
|---|---|---|
| Case#1 | 48 | 42 |
| Case#2 | 31 | 25 |
| Common | hsa-miR-130a hsa-miR-29b hsa-miR-30b hsa-miR-518d hsa-miR-551b hsa-miR-646 |
hsa-miR-601 hsa-miR-106b hsa-miR-92a hsa-miR-1275 hsa-miR-302c* |
follows the convention of nomenclature for assigning names to miRNA subtypes
ExoChip design and on-chip quantification: bridging the gap between technology and clinical application
In this paper, we demonstrated a platform technology for isolation and quantification of exosomes using the newly designed ExoChip and introduced its application for diagnostics using clinical samples. This approach is superior to other described microfluidic approaches, as we not only capture, but also quantitate and study exosomes with sensitivity and specificity. Compared to previously reported work for assaying blood-based exosomes our study has the following foremost distinctions: (i) Exochip is very different in engineering design than the earlier presented device. In fact, it’s the design of the channels that provides the unique advantage in the application of microfluidic based exosomes capture for clinical applications. The earlier work of Chen etc. al. used 20um height channels with herringbone structures on the top to facilitate mixing of vesicles, which enabled the smaller vesicles binding to the surface. In our work, to avoid the structures on the top, which can hinder imaging and quick quantification using a standard plate reader, we have designed channels incorporating multiple chambers connected by narrowed channel yielding an expanding and contracting channel structure. The hydrodynamics of the fluid in ExoChip was analyzed by performing fluid flow simulation studies using COMSOL Multiphysics 4.3 software (COMSOL, Inc., Burlington, MA, USA). As was expected, the simulation results showed that as the fluid goes through the expanded sections, the velocity is low and hence captures the vesicles and when the fluids pass through the contracting portions, the velocity is faster and enables mixing of vesicles (Fig S1). By repeating these patterns of capture and mixing, along with immuno-selective antibody-coating we were able to achieve the sensitive capture of vesicles. (ii) Earlier work used SEM as a way to verify the presence of vesicles. SEM prep method is laborious, and also needs expensive equipment and does not result in quantitative assessment of vesicles. This is fine for a proof of principle work. On the other hand, we want to go beyond this, and demonstrate a quick diagnostic test that is based on the quantitative yield differences between healthy and diseased states. To enable this, we developed a fluorescent intensity measurement method for analyzing the exosome yield, the advantage of which is manifold. One of the major challenges with the works involving these nanometer vesicles is their visualization. Using a lipophilic dye (DIO, presented in this work), we can get a higher level of fluorescence signals which can be readily detected from these nanometer scale entities. The detection limit or the sensitivity of the fluorescence measurement is up to 0.5 pM which is typical for the type of instrument used in present studies. (iii) Yet another significant difference is the characterization of these vesicles. Using immuno-gold nanoparticles as secondary imaging agents we have demonstrated that these vesicles do present the tumor-specific exosomal proteins. Additionally, the exosome yield of the ExoChip was found to be in the range of 15–18 μg of total proteins and 10–15 ng of total nucleic acids. The amount of exosomal yield for both protein and nucleic acids is high enough to allow downstream analysis through current molecular assay technologies that are sensitive enough to be performed at pico-and femto-levels. As for example, we also demonstrated the successful isolation of total RNA and specifically look for microRNAs that were relevant to pancreatic cancers.
Clearly, ease of simultaneous on-chip capture and quantification of exosomes along with robust validation are important prerequisites for the acceptance of a new technology to be successfully implemented in any research or clinical diagnosis setting. Furthermore, unlike existing ELISA and antibody-based detection methods such as Exotest and ExoELISA as well as polymer-based isolation methods, ExoChip design has functional advantages of simultaneous isolation, detection and characterization of exosomes38, 49, 50. In addition, ExoChip allows the convenience of performing customized assays to meet a variety of research and clinical objectives. As such, ExoChip is suitable for rapid screening of the large number of clinical samples, to carry out highly sensitive on-chip in-situ hybridization assays for nucleosome analysis and automated microscopy/imaging for biomarker exploration. As one of the promising applications of ExoChip in performing exosome-based molecular assays, we demonstrated its use for biomarker discovery through miRNA analysis using openarray advanced technology.
Conclusion
We introduce a microfluidic “ExoChip” device as a platform for on-chip isolation of exosomes and developed a novel on-chip fluorescent assay for rapid quantification of exosomes using a standard read-out plate reader. The ability of ExoChip to measure the levels of exosomes establishes its significance as a rapid-screening tool for monitoring exosome levels in patient serum. As a proof of concept, we demonstrated the use of ExoChip as a platform technology for the characterization of exosomes through conducting a comparative study using serum from healthy persons and pancreatic cancer patients. Further, Western blot and miRNA openarray analysis are shown to detect a broader range of suitable markers that can be used to validate unique diagnostic biomarkers for pancreatic cancer. The ExoChip is a promising platform that should advance our ability to use circulating exosomes as potential cancer biomarkers and allow the study of their contribution to human cancer biology.
Supplementary Material
Acknowledgments
We acknowledge Vasudha Murlidhar for her assistance in carrying out simulation studies on COMSOL Multiphysics 4.3 Software, Dr. Meggie M. G. Grafton for the use of fluorescence microscope scanning; Dorothy Sorenson from Microcopy and Image-Analysis Laboratory and Dr. John Mansfield from North Campus Electron Microbeam Analysis Laboratory, University of Michigan for their assistance in EM studies; Kirk Herman and Missy Tuck for collecting patients’ blood samples. This work was supported by NIH Director’s New Innovator Award (1DP2OD006672-01), a Career Development Grant from the University of Michigan Gastrointestinal Specialized Program of Research Excellence (GI SPORE) award (CA130810) to SN. Resources and support from Lurie Nanofabrication Facility and DNA Sequencing Core at University of Michigan are greatly appreciated.
References
- 1.Gress F, Gottlieb K, Sherman S, Lehman G. Annals of Internal Medicine. 2001;134:459–464. doi: 10.7326/0003-4819-134-6-200103200-00010. [DOI] [PubMed] [Google Scholar]
- 2.Nagrath S, Sequist LV, Maheswaran S, Bell DW, Irimia D, Ulkus L, Smith MR, Kwak EL, Digumarthy S, Muzikansky A, Ryan P, Balis UJ, Tompkins RG, Haber DA, Toner M. Nature. 2007;450:1235–1239. doi: 10.1038/nature06385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tan S, Yobas L, Lee G, Ong C, Lim C. Biomedical Microdevices. 2009;11:883–892. doi: 10.1007/s10544-009-9305-9. [DOI] [PubMed] [Google Scholar]
- 4.Cima I, Yee CW, Iliescu FS, Phyo WM, Lim KH, Iliescu C, Tan MH. Biomicrofluidics. 2013;7:011810–011816. doi: 10.1063/1.4780062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yoon HJ, Kim TH, Zhang Z, Azizi E, Pham TM, Paoletti C, Lin J, Ramnath N, Wicha MS, Hayes DF, Simeone DM, Nagrath S. Nature nanotechnology. 2013;8:735–741. doi: 10.1038/nnano.2013.194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li P, Stratton ZS, Dao M, Ritz J, Huang TJ. Lab on a chip. 2013;13:602–609. doi: 10.1039/c2lc90148j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hou HW, Warkiani ME, Khoo BL, Li ZR, Soo RA, Tan DS-W, Lim W-T, Han J, Bhagat AAS, Lim CT. Sci Rep. 2013:3. doi: 10.1038/srep01259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kanwar SS, Grafton MMG, Nagrath S. Microfluidics, Nanotechnology and Disease Biomarkers for Personalized Medicine Applications. In: Muhammad EJHW, Shiddiky JA, Rauf Sakandar, Trau Matt, editors. Bio-MEMS (Microfluidics) for CTC Detection in Cancer Patients. Nova Science Publishers, Inc; United States of America: 2013. pp. 7–10. (NBC-C) [Google Scholar]
- 9.Ozkumur E, Shah AM, Ciciliano JC, Emmink BL, Miyamoto DT, Brachtel E, Yu M, Chen PI, Morgan B, Trautwein J, Kimura A, Sengupta S, Stott SL, Karabacak NM, Barber TA, Walsh JR, Smith K, Spuhler PS, Sullivan JP, Lee RJ, Ting DT, Luo X, Shaw AT, Bardia A, Sequist LV, Louis DN, Maheswaran S, Kapur R, Haber DA, Toner M. Sci Transl Med. 2013;5:179ra147. doi: 10.1126/scitranslmed.3005616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Taylor DD, Gercel-Taylor C. Gynecologic Oncology. 2008;110:13–21. doi: 10.1016/j.ygyno.2008.04.033. [DOI] [PubMed] [Google Scholar]
- 11.Nilsson J, Skog J, Nordstrand A, Baranov V, Mincheva-Nilsson L, Breakefield XO, Widmark A. Br J Cancer. 2009;100:1603–1607. doi: 10.1038/sj.bjc.6605058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lau C, Kim Y, Chia D, Spielmann N, Eibl G, Elashoff D, Wei F, Lin Y-L, Moro A, Grogan T, Chiang S, Feinstein E, Schafer C, Farrell J, Wong DTW. Journal of Biological Chemistry. 2013 doi: 10.1074/jbc.M113.452458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Khan S, Jutzy JMS, Valenzuela MMA, Turay D, Aspe JR, Ashok A, Mirshahidi S, Mercola D, Lilly MB, Wall NR. PLoS ONE. 2012;7:e46737. doi: 10.1371/journal.pone.0046737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Peinado H, Aleckovic M, Lavotshkin S, Matei I, Costa-Silva B, Moreno-Bueno G, Hergueta-Redondo M, Williams C, Garcia-Santos G, Ghajar CM, Nitadori-Hoshino A, Hoffman C, Badal K, Garcia BA, Callahan MK, Yuan J, Martins VR, Skog J, Kaplan RN, Brady MS, Wolchok JD, Chapman PB, Kang Y, Bromberg J, Lyden D. Nat Med. 2012;18:883–891. doi: 10.1038/nm.2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee T, D’Asti E, Magnus N, Al-Nedawi K, Meehan B, Rak J. Seminars in immunopathology. 2011;33:455–467. doi: 10.1007/s00281-011-0250-3. [DOI] [PubMed] [Google Scholar]
- 16.Kogure T, Lin WL, Yan IK, Braconi C, Patel T. Hepatology. 2011;54:1237–1248. doi: 10.1002/hep.24504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li J, Sherman-Baust C, Tsai-Turton M, Bristow R, Roden R, Morin P. BMC Cancer. 2009;9:244. doi: 10.1186/1471-2407-9-244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cantin R, Diou J, Bélanger D, Tremblay AM, Gilbert C. Journal of Immunological Methods. 2008;338:21–30. doi: 10.1016/j.jim.2008.07.007. [DOI] [PubMed] [Google Scholar]
- 19.Chen C, Skog J, Hsu CH, Lessard RT, Balaj L, Wurdinger T, Carter BS, Breakefield XO, Toner M, Irimia D. Lab on a chip. 2010;10:505–511. doi: 10.1039/b916199f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang Z, Wu H-j, Fine D, Schmulen J, Hu Y, Godin B, Zhang JXJ, Liu X. Lab on a chip. 2013;13:2879–2882. doi: 10.1039/c3lc41343h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Davies RT, Kim J, Jang SC, Choi EJ, Gho YS, Park J. Lab on a chip. 2012;12:5202–5210. doi: 10.1039/c2lc41006k. [DOI] [PubMed] [Google Scholar]
- 22.Gyorgy B, Szabo TG, Pasztoi M, Pal Z, Misjak P, Aradi B, Laszlo V, Pallinger E, Pap E, Kittel A, Nagy G, Falus A, Buzas EI. Cell Mol Life Sci. 2011;68:2667–2688. doi: 10.1007/s00018-011-0689-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Raposo G, Stoorvogel W. J Cell Biol. 2013;200:373–383. doi: 10.1083/jcb.201211138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pols MS, Klumperman J. Experimental cell research. 2009;315:1584–1592. doi: 10.1016/j.yexcr.2008.09.020. [DOI] [PubMed] [Google Scholar]
- 25.Hood JL, San RS, Wickline SA. Cancer Research. 2011;71:3792–3801. doi: 10.1158/0008-5472.CAN-10-4455. [DOI] [PubMed] [Google Scholar]
- 26.Tian T, Wang Y, Wang H, Zhu Z, Xiao Z. Journal of Cellular Biochemistry. 2010;111:488–496. doi: 10.1002/jcb.22733. [DOI] [PubMed] [Google Scholar]
- 27.Xiang X, Poliakov A, Liu C, Liu Y, Deng Z-b, Wang J, Cheng Z, Shah SV, Wang G-J, Zhang L, Grizzle WE, Mobley J, Zhang H-G. International Journal of Cancer. 2009;124:2621–2633. doi: 10.1002/ijc.24249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zomer A, Vendrig T, Hopmans ES, van Eijndhoven M, Middeldorp JM, Pegtel DM. Communicative & integrative biology. 2010;3:447–450. doi: 10.4161/cib.3.5.12339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Harding CV., III . PhD. Washington University; 1985. 8520798. [Google Scholar]
- 30.Harding C, Heuser J, Stahl P. European journal of cell biology. 1984;35:256–263. [PubMed] [Google Scholar]
- 31.Brennwald P. J Cell Biol. 2000;149:1–4. doi: 10.1083/jcb.149.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.de Harven E, Leung R, Christensen H. J Cell Biol. 1984;99:53–57. doi: 10.1083/jcb.99.1.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mathivanan S, Ji H, Simpson RJ. Journal of proteomics. 2010;73:1907–1920. doi: 10.1016/j.jprot.2010.06.006. [DOI] [PubMed] [Google Scholar]
- 34.Thery C, Ostrowski M, Segura E. Nat Rev Immunol. 2009;9:581–593. doi: 10.1038/nri2567. [DOI] [PubMed] [Google Scholar]
- 35.Filipazzi P, Bürdek M, Villa A, Rivoltini L, Huber V. Seminars in Cancer Biology. 2012;22:342–349. doi: 10.1016/j.semcancer.2012.02.005. [DOI] [PubMed] [Google Scholar]
- 36.Rak J, Guha A. BioEssays: news and reviews in molecular, cellular and developmental biology. 2012;34:489–497. doi: 10.1002/bies.201100169. [DOI] [PubMed] [Google Scholar]
- 37.Cocucci E, Racchetti G, Meldolesi J. Trends in Cell Biology. 2009;19:43–51. doi: 10.1016/j.tcb.2008.11.003. [DOI] [PubMed] [Google Scholar]
- 38.Logozzi M, De Milito A, Lugini L, Borghi M, Calabrò L, Spada M, Perdicchio M, Marino ML, Federici C, Iessi E, Brambilla D, Venturi G, Lozupone F, Santinami M, Huber V, Maio M, Rivoltini L, Fais S. PLoS ONE. 2009;4:e5219. doi: 10.1371/journal.pone.0005219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li Y, Meng X, Feng H, Zhang G, Liu C, Li P. Chinese medical sciences journal = Chung-kuo i hsueh k’o hsueh tsa chih/Chinese Academy of Medical Sciences. 1999;14:96–101. [PubMed] [Google Scholar]
- 40.Li Y, Feng H, Chen Y. Chinese journal of oncologyZhonghua zhong liu za zhi. 1999;21:178–181. [PubMed] [Google Scholar]
- 41.Skog J, Wurdinger T, van Rijn S, Meijer DH, Gainche L, Curry WT, Carter BS, Krichevsky AM, Breakefield XO. Nat Cell Biol. 2008;10:1470–1476. doi: 10.1038/ncb1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Al-Nedawi K, Meehan B, Micallef J, Lhotak V, May L, Guha A, Rak J. Nat Cell Biol. 2008;10:619–624. doi: 10.1038/ncb1725. [DOI] [PubMed] [Google Scholar]
- 43.Rosell R, Wei J, Taron M. Clinical Lung Cancer. 2009;10:8–9. doi: 10.3816/CLC.2009.n.001. [DOI] [PubMed] [Google Scholar]
- 44.Simpson RJ, Lim JWE, Moritz RL, Mathivanan S. Expert Review of Proteomics. 2009;6:267–283. doi: 10.1586/epr.09.17. [DOI] [PubMed] [Google Scholar]
- 45.Park JO, Choi DY, Choi DS, Kim HJ, Kang JW, Jung JH, Lee JH, Kim J, Freeman MR, Lee KY, Gho YS, Kim KP. Proteomics. 2013;13:2125–2134. doi: 10.1002/pmic.201200323. [DOI] [PubMed] [Google Scholar]
- 46.Valadi H, Ekstrom K, Bossios A, Sjostrand M, Lee JJ, Lotvall JO. Nat Cell Biol. 2007;9:654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 47.Ali S, Almhanna K, Chen W, Philip PA, Sarkar FH. Am J Transl Res. 2010;3:28–47. [PMC free article] [PubMed] [Google Scholar]
- 48.Yu J, Li A, Hong SM, Hruban RH, Goggins M. Clinical cancer research: an official journal of the American Association for Cancer Research. 2012;18:981–992. doi: 10.1158/1078-0432.CCR-11-2347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mitsuhashi M, Taub DD, Kapogiannis D, Eitan E, Zukley L, Mattson MP, Ferrucci L, Schwartz JB, Goetzl EJ. The FASEB Journal. 2013;27:5141–5150. doi: 10.1096/fj.13-238980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yeatts K, Millis M, Smith S, Duncan J, Moran B, Ortiz G, Little T, Conrad A, Spetzler DB, Pawlowksi T. A novel blood-based method for evaluating KRAS in circulating microvesicles from colorectal cancer patients. Chicago, IL. Philadelphia (PA): 2012. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.