Abstract
Sensory systems use stochastic mechanisms to diversify neuronal subtypes. In the Drosophila eye, stochastic expression of the PAS-bHLH transcription factor Spineless (Ss) determines a random binary subtype choice in R7 photoreceptors. Here, we show that a stochastic, cell-autonomous decision to express ss is made intrinsically by each ss locus. Stochastic on or off expression of each ss allele is determined by combinatorial inputs from one enhancer and two silencers acting at long range. However, the two ss alleles also average their frequency of expression through upregulatory and downregulatory interallelic cross-talk. This inter- or intra-chromosomal long-range regulation does not require endogenous ss chromosomal positioning or pairing. Therefore, although individual ss alleles make independent stochastic choices, interchromosomal communication coordinates expression state between alleles, ensuring that they are both expressed in the same random subset of R7s.
Developmental programs generally induce uniform or regionalized gene expression patterns to yield highly reproducible body plan outcomes. However, stochastic mechanisms are sometimes incorporated to diversify cell types in nervous systems. Non-autonomous stochastic mechanisms utilizing lateral inhibition strategies have been well-described, whereas cell autonomous stochastic mechanisms involved in color opsin and olfactory receptor selection in mammals are only partially understood 1,2.
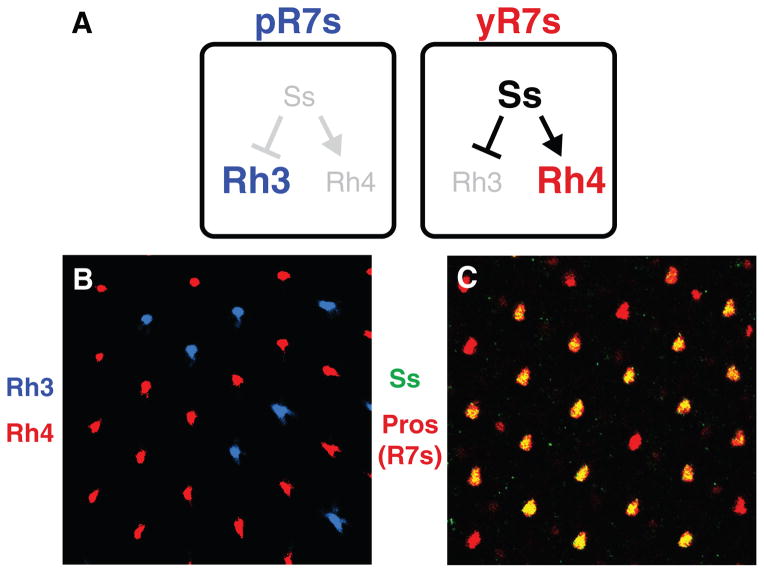
The fly eye is composed of two stochastically distributed subtypes of ommatidia (unit eyes) defined by expression of specific light-detecting Rhodopsin proteins in R7 photoreceptors (PRs). The random distribution is controlled by the stochastic expression of the PAS-bHLH transcription factor Spineless (Ss). Ss expression in ~65% of randomly distributed R7s induces ‘yellow’ (yR7) fate and expression of Rhodopsin4 (Rh4), whereas the absence of Ss in the remaining ~35% of R7s allows for ‘pale’ (pR7) fate and Rhodopsin3 (Rh3) expression (Fig. 1A–B). Loss of ss function leads to the transformation of all R7s to pR7-fate and Rh3 expression (Fig. S1A) whereas ectopic Ss causes all R7s to acquire yR7 fate and express Rh4 (Fig. S1B)1–5.
Figure 1. The stochastic decision to express ss is made early and maintained.
A. Ss is absent from pR7s allowing for Rh3 expression. Ss is expressed in yR7s activating Rh4 and repressing Rh3.
B. Stochastic distribution of Rh3- and Rh4-expressing R7s.
C. Ss is expressed in a random subset of R7s throughout development. Pros marks all R7s.
Ss was observed in 65% of randomly distributed R7s throughout development (Fig. 1C, S1D–G). Ss expression in adults perfectly correlated with Rh4 expression (Fig. S1C). We never observed switching of Rh expression6. Therefore, Ss expression is established and stably maintained throughout the lifetime of yR7 cells.
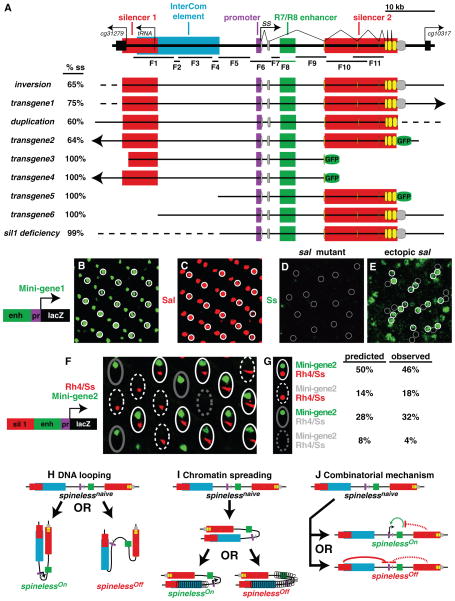
We evaluated reporter lines containing fragments of the ss gene 7. Frag 8 (R7/R8 enhancer) in mini-gene1 induced lacZ expression in all R7s and R8s (Fig. 2A–B), which closely resembled expression of the Salm zinc finger transcription factor (with Salr, collectively referred to as Sal) (Fig. 2C) that specifies R7 and R8 fate8. Ss expression was completely lost in sal mutants (Fig. 2D), whereas ectopic expression of Salm in all PRs led to the activation of Ss in a random subset of outer PRs (Fig. 2E, S2A) and expression of Mini-gene1 in outer PRs (Fig. S2B). Thus, Sal is necessary and sufficient to activate stochastic expression of Ss in PRs. The choice to express Ss is cell-autonomous since R7s and outer PRs within the same ommatidium made their decisions to express Ss independently of one another (Fig. 2E, S2A).
Figure 2. The cis-regulatory logic controlling intrinsically stochastic ss expression.
A. ss locus schematic. F = Fragment; red boxes = silencers; blue box = InterCom element; purple box = minimal promoter; green box = R7/R8 enhancer; gray circles = untranslated exons; yellow circles = translated exons; arrows = transcriptional starts.
For B–E, white circles indicate expression and gray circle indicate no expression.
B. Mini-gene1 is expressed in all R7s and R8s.
C. Sal is expressed in all R7s and R8s.
D. Ss expression is completely lost in sal mutants.
E. Ectopic Sal expression in svp mutants causes Ss expression in a random subset of PRs.
F. Mini-gene2 induces expression in a subset of R7s independently of endogenous Ss/Rh4 expression. Mini-gene2 localizes to the nucleus whereas Rh4/Ss localizes to membranous rhabdomere structures. The four possible combinations of expression are observed: 1. white solid ovals = Mini-gene2 and Ss/Rh4; 2. white dashed ovals = Ss/Rh4 only; 3. gray solid ovals = Transgene only; and 4. gray dotted ovals = no expression.
G. Four expression combinations in F.
H–J. Models for random expression decisions
H. The ss locus randomly assumes one of two (i.e. active and repressed) DNA looping configurations.
I. One silencer facilitates the nucleation of closed chromatin state spreading from the other silencer.
J. One silencer lowers expression in all R7s whereas the other specifically provides the stochastic input (through looping or spreading).
To identify DNA silencer elements required for stochastic Ss expression, we first defined the minimal ss DNA sequence required for stochastic ss expression. We used GFP from transgenes or Rh4 expression as a readout of Ss expression (“Ss/Rh4”), since Rh4 is always a perfect indication of Ss expression in R7s (Fig. S1C)5. An inversion 9 and transgene1 exhibited stochastic Ss expression and therefore defined the 5′ endpoint (Fig. 2A, S2C). A duplication with a breakpoint in the ss 3′ UTR 10 and transgene2 similarly exhibited stochastic Ss expression, defining the 3′ endpoint (Fig. 2A, S2D). These data determine a 55.5 kb minimal DNA sequence required for stochastic ss expression (Fig. 2A).
We identified two DNA elements that are critical for stochastic ss expression. transgene3 and transgene4 displayed expression in all R7s suggesting that an intragenic silencer (“silencer2”) is required for stochastic ss expression (Fig. 2A, S2E–F). transgene5 and transgene6 also displayed expression in all R7s, suggesting that a 5′ upstream silencer (“silencer1”) is also required for stochastic ss expression (Fig. 2A, S2G–H). A 36 kb deficiency that removed silencer1 (sil1 deficiency) and an inversion allele in which the ss coding region was moved 12 Mb away from silencer1 (ss high freq) showed expression of Ss/Rh4 in all R7s (Fig. 2A, S2I, S3A, E), validating the requirement for silencer1. Therefore, stochastic Ss expression requires an enhancer and two silencer elements.
When a ~3kb fragment of silencer1 was placed with the R7/R8 enh+prom element driving reporter expression (mini-gene2), we observed expression in a random subset of R7s (Fig. 2F), showing that silencer1 is sufficient to repress expression when present close to the enhancer and promoter. If the stochastic expression decision occurred intrinsically at each ss locus, mini-gene2 should induce reporter expression independently of expression from the endogenous ss loci. We compared expression of mini-gene2 to endogenous Ss/Rh4 expression and found all four possible expression combinations (Fig. 2F–G), suggesting that each ss locus makes an independent, stochastic expression decision.
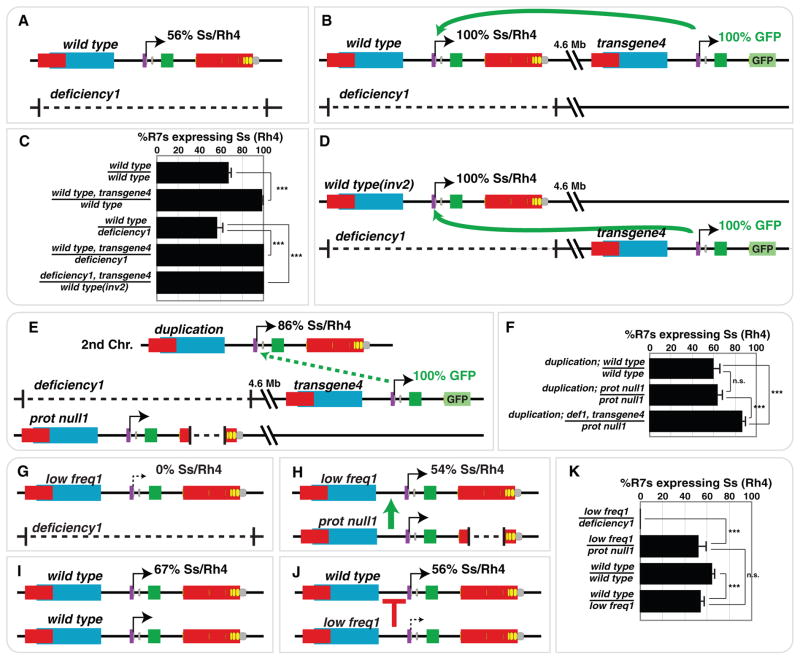
transgene4, which was inserted 4.6 Mb away from the ss locus, drove GFP expression in all R7s (Fig. 2A). Although transgene4 should not affect endogenous Ss expression, we observed a dramatic increase in the frequency of Ss/Rh4 expression in animals carrying transgene4, suggesting that transgene4 upregulated the frequency of Ss expression from the endogenous ss loci (Fig. 3C). transgene4 upregulated expression from ss loci in cis, or in trans (Fig. 3A–D), suggesting that it contains DNA elements that are sufficient to drive regulatory interactions in the absence of chromosomal pairing. transgene4 also upregulated expression, though less efficiently, from the ss locus translocated on a different chromosome (Fig. 3E–F), suggesting that ss alleles can interact at a distance, but chromosomal position plays a role in this process. These observations strongly implicate direct interactions between DNA elements in the transgene and endogenous loci, but do not exclude possible indirect mechanisms such as non-coding RNAs. transgene4 must contain a DNA element (“InterCom element”) between 25 kb to 8 kb upstream of the ss TSS, which is missing in mini-gene2 (Fig. 2F–G, Fig. S3).
Figure 3. ss regulatory regions upregulate and downregulate expression frequency through interchromosomal communication.
A. Wild type ss locus over deficiency1.
B. transgene4 upregulates expression frequency from the endogenous ss gene in cis.
C. Quantification of A, B, and D
D. transgene4 upregulates expression frequency from the endogenous ss gene in trans.
E. transgene4 upregulates expression, though less efficiently, from the ss locus on the non-homologous 2nd chromosome.
F. Quantification of E.
G. Ss/Rh4 is not expressed in sslow freq1 hemizygous mutants.
H. The normal regulatory regions of the ssprot null1 allele upregulate Ss/Rh4 expression from the sslow freq1 allele.
I. Wild type ss homozygous loci.
J. The regulatory regions of the sslow freq1 allele downregulate Ss/Rh4 expression from the wild type ss allele.
K. Quantification of G–J.
We next found that one ss allele could upregulate the frequency of expression from the other allele. sslow freq1, an allele affecting non-coding regions, was expressed at very low frequency when placed over ssdeficiency alleles (Fig 3G, K, Fig. S4A). When sslow freq1 was placed over ssprot null1, a protein coding null allele with normal cis-regulatory regions, the frequency of Ss/Rh4 expression dramatically increased (Fig. 3H, K), suggesting that the cis-regulatory elements from ssprot null1 upregulated expression frequency from sslow freq1. We verified our observations with additional allelic combinations (Fig. S4A–B).
The upregulation of expression from one allele with impaired regulatory regions but normal protein function by another allele with normal regulatory regions but impaired protein function resembles transvection, initially described by Ed Lewis 11. Transvection is defined as the complementation of mutant alleles requiring position-dependent chromosomal pairing. Since the interallelic control of ss does not require position-dependent chromosomal pairing (Fig. 3B, D, E, S3B, S4B, C) and does not appear to require regulation by known mediators of transvection (Fig. S5), we conclude that this phenomenon is not a canonical case of transvection.
We also found that one ss allele could mediate the downregulation of expression frequency from the other allele. The sslow freq1 allele downregulated expression frequency from the sswild type alleles since the proportion of R7s expressing Ss/Rh4 was lower in sslow freq1/sswild type animals compared to sswild type homozygotes(Fig. 3I–K). Downregulation did not require endogenous ss chromosomal position since it also occurred for a wild type ss locus on an inversion(Fig. S4C). We confirmed downregulation with additional allelic combinations (Fig. S3A, B, E, S4C). Thus, ss alleles regulate one another through long-range interchromosomal activating and repressing mechanisms to determine the frequency of ss expression.
If each ss allele makes its own expression decision, expression states will sometimes agree (both alleles On or Off) and other times disagree (one allele On and the other Off). We tested whether interchromosomal communication functioned to coordinate the expression state from the two ss alleles.
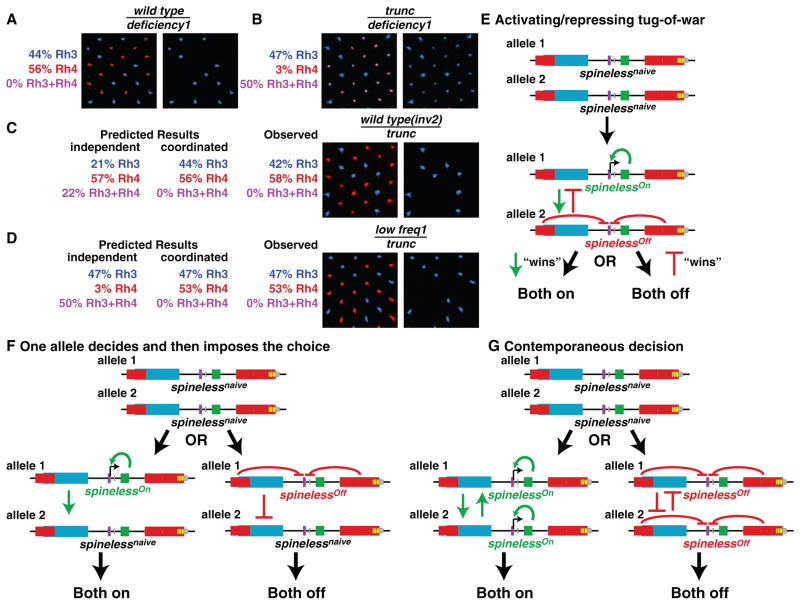
The sstrunc allele has normal regulatory regions but contains a mutation that truncates the Ss protein activation domain 5,10. This truncation weakens Ss protein function such that Ss activates Rh4 normally, but fails to repress Rh3, leading to co-expression of Rh3 and Rh4 in nearly all yR7s, and normal Rh3 expression in pR7s (Fig. 4B).
Figure 4. Interchromosomal communication coordinates expression from ss alleles.
A. Rh3 is expressed in pR7s and Rh4 is expressed in yR7s in wild type hemizygous animals.
B. Rh3 is expressed in pR7s and Rh4 and Rh3 are expressed in yR7s in sstrunc hemizygous animals.
C. In sswild type(inv2)/sstrunc animals, Rh3 and Rh4 are always expressed exclusively.
D. In sslow freq1/sstrunc animals, the normal regulatory regions of the sstrunc allele upregulate expression from sslow freq1 into the same subset of R7s since Rh3 and Rh4 are expressed exclusively.
E–G. Models for the coordination of expression state through interchromosomal communication
E. A temporally distinct two-step mechanism involving both alleles making independent expression decisions followed by an activating and repressing tug of war.
F. A temporally distinct two-step mechanism in which one allele makes the decision and then imposes the decision onto the other naïve allele.
G. A mechanism involving contemporaneous decisions that average the activating and repressing inputs from each allele.
We evaluated sswild type/sstrunc animals to determine whether these alleles were expressed in an independent or coordinated manner. If the two ss alleles were expressed independently, sswild type/sstrunc would produce three Rh expression outcomes: (i) Rh3 alone (neither sswild type nor sstrunc are expressed), (ii) Rh4 alone (sswild type alone or both sswild type and sstrunc are expressed), and (iii) Rh3 and Rh4 co-expression (sstrunc alone is expressed) (Fig. S6C). Alternatively, coordinated expression from the two alleles would yield two Rh expression outcomes: (i) Rh3 alone (neither sswild type nor sstrunc are expressed) and (ii) Rh4 alone (both sswild type and sstrunc are expressed) (Fig. S6D). For these experiments, the wild type ss allele was on an inverted chromosome (sswild type(inv2)) to prevent pairing of homologous chromosomes. For sswild type(inv2)/sstrunc flies, we observed expression of Rh4 alone and Rh3 alone, but never co-expression of Rh3 and Rh4, (Fig. 4C). The wild type ss locus on a different inverted chromosome (sswild type(inv3)) over sstrunc displayed similar expression coordination (Supplemental materials). Together, these data suggest that expression from the two ss alleles is coordinated and that endogenous ss position on homologous chromosomes is not critical.
We next tested whether interchromosomal communication was able to coordinate expression from two ss alleles with widely different expression frequencies. sslow freq1 expressed fully functional Ss protein but at a low frequency (Fig. 3G). We predicted that sslow freq1/sstrunc animals should display upregulation of Ss expression from sslow freq1 due to interchromosomal communication from the normal cis-regulatory elements of sstrunc. sslow freq1/sstrunc flies displayed nearly perfect coordination of expression from the two alleles with almost no co-expression of Rh3 and Rh4 (Fig. 4D), verifying that interchromosomal communication coordinates expression from the two ss alleles.
Since stochastic Ss expression requires an enhancer and two silencer elements, we propose three possible mechanistic models controlling the decision: (i) the ss locus randomly assumes one of two (i.e. active and repressed) DNA looping configurations, (ii) one silencer facilitates the nucleation of closed chromatin state spreading from the other silencer, and (iii) one silencer generally lowers expression in all R7s whereas the other specifically provides the stochastic input (through looping or spreading) (Fig. 2H–J).
Similarly, we envision three models for how interchromsomal communication coordinates expression: (i) a temporally distinct two-step mechanism involving both alleles making independent expression decisions followed by an activating and repressing tug of war, (ii) a temporally distinct two-step mechanism in which one allele makes the decision and then imposes the decision onto the other naïve allele, and (iii) a mechanism involving contemporaneous decisions that average the activating and repressing inputs from each allele (Fig. 4E–G).
Interchromosomal communication is reminiscent of transvection. In contrast to transvection-like processes that allow allelic complementation between null alleles whose biological meaning is unclear, interchromosomal communication regulating ss appears to have dedicated biological functions to average the frequency and coordinate expression state between stochastically-expressed alleles.
The color vision systems of flies and humans present an interesting case of convergent evolution. In both species, the apparent goal is the same: use stochastic mechanisms to diversify cell fates and distribute color sensory capacities across the eye. The fly eye requires an enhancer and two silencer elements to achieve stochastic expression of ss whereas the human eye uses random LCR-mediated activation of M or L opsins. To avoid disagreement in allelic expression states, interchromosomal communication coordinates expression in flies, whereas X-inactivation completely turns off expression from one allele in females and there is only one copy of the locus in males, creating a mono-allelic expression decision in both cases1,2.
Stochastic gene expression mechanisms may be a “cost-effective” way to diversify the repertoire of cell fates within a tissue. Though these phenomena involve stochastic processes, this randomness is very often well-controlled, incorporating multiple steps apparently to ensure robustness. Evolution has yielded many different mechanisms to determine stochastic cell fate specification in bacteria, flies, and vertebrates2. As our understanding of stochastic phenomena increases, it will be interesting to see whether common, ancestral strategies become apparent or if novel stochastic gene expression mechanisms arise in individual species.
Supplementary Material
A. ss mutants express Rh3 and lose Rh4 in all R7s.
B. Expression of Ss in all R7s induces Rh4 and represses Rh3 in all R7s.
C. Ss (green) is expressed in the nucleus in the same R7s that express Rh4 in the rhabdomere (white ovals). R7s that lack Ss also lack Rh4 (gray ovals). Pros (red) marks all R7 cell nuclei.
D–F. Ss (green) is expressed in a random subset of R7s throughout development including 0%, 25%, 50%, 75% and 100% pupation (adult). Pros (red) marks all R7 cells. Upper panel: Ss and Pros; lower panel: Ss alone. * indicates Ss expression in bristle cells that are in the same focal plane as R7s at 25% pupation.
G. Quantification of A–E.
A. Ectopic Sal induces Ss (green) in a random subset of R1 and R6 PRs. Each cell in an ommatidium makes its own stochastic decision showing that the ss expression decision is cell-autonomous. Three cells are marked with white (Ss+) or gray (Ss−) circles indicating R1, R7, or R6.
B. Ectopic Sal induces Mini-gene1 (green) expression in outer PRs. Elav (blue) marks all PRs.
C. Ss/Rh4 (red) is expressed in a random subset of R7s in flies with a chromosomal inversion with a breakpoint in the gene 5′ to ss.
D. Ss/Rh4 (red) is expressed in a random subset of R7s in flies with a duplication of ss on the 2nd chromosome with a breakpoint in the ss 3′ UTR.
E. Ss>GFP (green) is expressed in all R7s from transgene3 that lacks silencer2.
F. Ss>GFP (green) is expressed in all R7s from transgene4 that lacks silencer2.
G. Ss>GFP (green) is expressed in all R7s from transgene5 that lacks silencer1.
H. Ss/Rh4 (red) is expressed in all R7s from transgene6 that lacks silencer1.
I. Ss/Rh4 (red) is expressed in all R7s in flies with the sil1 deficiency allele that removes silencer1.
A. Ss/Rh4 is expressed in 100% of R7s in sshigh freq hemizygous mutants. For the sshigh freq allele, the ss locus is moved 12 Mb away from silencer1.
B. The normal regulatory regions of the ssprot null1 allele mediates downregulation of Ss/Rh4 expression frequency from the sshigh freq allele that retains silencer1 and the InterCom element. The inversion breakpoint defines the proximal boundary of the InterCom element at 8 kb upstream of the ss TSS.
C. The normal regulatory regions of the ssprot null1 allele are unable to mediate downregulation of Ss/Rh4 expression frequency from the sssil1 deficiency allele that has a similar breakpoint as sshigh freq but completely lacks silencer1 and the InterCom element.
D. Ss/Rh4 is expressed in 100% of R7s sssil1 deficiency homozygous mutants.
E. Quantification of data for the sshigh freq and sssil1 deficiency alleles including A–D.
A. ssprot null alleles with normal regulatory regions but impaired protein-coding regions upregulate sslow freq alleles with normal protein-coding regions but impaired regulatory regions.
B. We generated the ssprot null2 allele using homologous recombination10 to replace exons 5–8 (removing partially the first PAS domain and completely the second PAS domain, PAC domain and transcriptional activation region) and 2 kb of the 3′ region with a marker gene. ssprot null2 upregulated expression frequency significantly more than ssprot null1 or sswild type (sslow freq1/ ssprot null1 = 54.1% +/− 7.6; sslow freq2/ ssprot null1 = 56.2% +/− 5.6; sslow freq1/ sswild type = 56.1% +/− 3.3; sslow freq2/ sswild type(inv2) = 54.5 +/− 3.6)(Fig. S3A), suggesting that a critical repressive cis-element lies in exons 5–8 or their intervening introns. ssprot null2 also upregulated expression from a ss locus on an inverted chromosome (sswild type(inv2)/ ssprot null2 = 75.5% +/− 3.3) and a ss locus duplicated on chromosome 2 (ssduplication) (from 59.5% +/− 6.3 in ssduplication; sswild type animals to 79.4% +/− 5.9 in ssduplication; ssprot null2 animals), consistent with the ability of interchromosomal communication to upregulate expression from inverted and non-homologous chromosomes.
C. sslow freq alleles downregulate expression from ss in its normal (sswild type) or heterologous position (sswild type(inv2)). sslow freq1 downregulates expression frequency from sslow freq2.
A. The frequency of Rh3 and Rh4 expression is normal in Condensin II/Cap-H2 mutants.
B. The frequency of Rh3 and Rh4 expression is normal in zeste/za mutants.
C. The frequency of Rh3 and Rh4 expression is normal in zeste/z1 mutants.
D. Expression frequency and coordination between sswild type and sstrunc alleles (i.e. complete exclusive expression of Rh3 and Rh4) is normal in flies with ectopic expression of Condensin II/Cap-H2.
E. Blue channel (Rh3) only of Fig. S1D.
F. transgene4 upregulates Ss/Rh4 expression normally in flies with ectopic expression of Condensin II/Cap-H2.
G. transgene4 upregulates Ss/Rh4 expression normally in zeste/za mutants.
Schematics for ss locus as in Figure 2. For A–B, E–F, Rh3 is in blue and Rh4 is in red.
A. Rh3 is expressed in pR7s and Rh4 is expressed in yR7s in wild type hemizygous animals.
B. Rh3 is expressed in pR7s and Rh4 and Rh3 are expressed in yR7s in sstrunc hemizygous animals.
C. If expression from ss alleles is independent, there are four combinations of expression from sswild type and sstrunc yielding 3 Rh expression states in R7s (Rh3 alone; Rh4 alone; Rh3 and Rh4).
D. If expression from ss allele is coordinated, there are two combinations of expression from sswild type and sstrunc yielding 2 Rh expression states in R7s (Rh3 alone; Rh4 alone).
E. In sswild type(inv2)/sstrunc animals, Rh3 and Rh4 are always expressed exclusively showing that expression from ss alleles is coordinated.
F. In sslow freq1/sstrunc animals, the normal regulatory regions of the sstrunc allele upregulate expression from sslow freq1 into the same subset of R7s since Rh3 and Rh4 are expressed exclusively.
References and Notes
- 1.Johnston RJ, Jr, Desplan C. Stochastic neuronal cell fate choices. Curr Opin Neurobiol. 2008;18:20–27. doi: 10.1016/j.conb.2008.04.004. S0959-4388(08)00028-7 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Johnston RJ, Jr, Desplan C. Stochastic mechanisms of cell fate specification that yield random or robust outcomes. Annu Rev Cell Dev Biol. 2010;26:689–719. doi: 10.1146/annurev-cellbio-100109-104113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wernet MF, et al. Stochastic spineless expression creates the retinal mosaic for colour vision. Nature. 2006;440:174–180. doi: 10.1038/nature04615. nature04615 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Johnston RJ, Jr, et al. Interlocked feedforward loops control cell-type-specific Rhodopsin expression in the Drosophila eye. Cell. 2011;145:956–968. doi: 10.1016/j.cell.2011.05.003. S0092-8674(11)00529-0 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Thanawala SU, et al. Regional modulation of a stochastically expressed factor determines photoreceptor subtypes in the Drosophila retina. Dev Cell. 2013;25:93–105. doi: 10.1016/j.devcel.2013.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vasiliauskas D, et al. Feedback from rhodopsin controls rhodopsin exclusion in Drosophila photoreceptors. Nature. 2011;479:108–112. doi: 10.1038/nature10451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Emmons RB, Duncan D, Duncan I. Regulation of the Drosophila distal antennal determinant spineless. Dev Biol. 2007;302:412–426. doi: 10.1016/j.ydbio.2006.09.044. S0012-1606(06)01261-9 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mollereau B, et al. Two-step process for photoreceptor formation in Drosophila. Nature. 2001;412:911–913. doi: 10.1038/35091076. 35091076[pii] [DOI] [PubMed] [Google Scholar]
- 9.Matzkin LM, Merritt TJ, Zhu CT, Eanes WF. The structure and population genetics of the breakpoints associated with the cosmopolitan chromosomal inversion In(3R)Payne in Drosophila melanogaster. Genetics. 2005;170:1143–1152. doi: 10.1534/genetics.104.038810. genetics.104.038810 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Duncan DM, Burgess EA, Duncan I. Control of distal antennal identity and tarsal development in Drosophila by spineless-aristapedia, a homolog of the mammalian dioxin receptor. Genes Dev. 1998;12:1290–1303. doi: 10.1101/gad.12.9.1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lewis E. The theory and application of a new method of detecting chromosomal rearrangements in Drosophila melanogaster. Am Nat. 1954;88:225–239. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A. ss mutants express Rh3 and lose Rh4 in all R7s.
B. Expression of Ss in all R7s induces Rh4 and represses Rh3 in all R7s.
C. Ss (green) is expressed in the nucleus in the same R7s that express Rh4 in the rhabdomere (white ovals). R7s that lack Ss also lack Rh4 (gray ovals). Pros (red) marks all R7 cell nuclei.
D–F. Ss (green) is expressed in a random subset of R7s throughout development including 0%, 25%, 50%, 75% and 100% pupation (adult). Pros (red) marks all R7 cells. Upper panel: Ss and Pros; lower panel: Ss alone. * indicates Ss expression in bristle cells that are in the same focal plane as R7s at 25% pupation.
G. Quantification of A–E.
A. Ectopic Sal induces Ss (green) in a random subset of R1 and R6 PRs. Each cell in an ommatidium makes its own stochastic decision showing that the ss expression decision is cell-autonomous. Three cells are marked with white (Ss+) or gray (Ss−) circles indicating R1, R7, or R6.
B. Ectopic Sal induces Mini-gene1 (green) expression in outer PRs. Elav (blue) marks all PRs.
C. Ss/Rh4 (red) is expressed in a random subset of R7s in flies with a chromosomal inversion with a breakpoint in the gene 5′ to ss.
D. Ss/Rh4 (red) is expressed in a random subset of R7s in flies with a duplication of ss on the 2nd chromosome with a breakpoint in the ss 3′ UTR.
E. Ss>GFP (green) is expressed in all R7s from transgene3 that lacks silencer2.
F. Ss>GFP (green) is expressed in all R7s from transgene4 that lacks silencer2.
G. Ss>GFP (green) is expressed in all R7s from transgene5 that lacks silencer1.
H. Ss/Rh4 (red) is expressed in all R7s from transgene6 that lacks silencer1.
I. Ss/Rh4 (red) is expressed in all R7s in flies with the sil1 deficiency allele that removes silencer1.
A. Ss/Rh4 is expressed in 100% of R7s in sshigh freq hemizygous mutants. For the sshigh freq allele, the ss locus is moved 12 Mb away from silencer1.
B. The normal regulatory regions of the ssprot null1 allele mediates downregulation of Ss/Rh4 expression frequency from the sshigh freq allele that retains silencer1 and the InterCom element. The inversion breakpoint defines the proximal boundary of the InterCom element at 8 kb upstream of the ss TSS.
C. The normal regulatory regions of the ssprot null1 allele are unable to mediate downregulation of Ss/Rh4 expression frequency from the sssil1 deficiency allele that has a similar breakpoint as sshigh freq but completely lacks silencer1 and the InterCom element.
D. Ss/Rh4 is expressed in 100% of R7s sssil1 deficiency homozygous mutants.
E. Quantification of data for the sshigh freq and sssil1 deficiency alleles including A–D.
A. ssprot null alleles with normal regulatory regions but impaired protein-coding regions upregulate sslow freq alleles with normal protein-coding regions but impaired regulatory regions.
B. We generated the ssprot null2 allele using homologous recombination10 to replace exons 5–8 (removing partially the first PAS domain and completely the second PAS domain, PAC domain and transcriptional activation region) and 2 kb of the 3′ region with a marker gene. ssprot null2 upregulated expression frequency significantly more than ssprot null1 or sswild type (sslow freq1/ ssprot null1 = 54.1% +/− 7.6; sslow freq2/ ssprot null1 = 56.2% +/− 5.6; sslow freq1/ sswild type = 56.1% +/− 3.3; sslow freq2/ sswild type(inv2) = 54.5 +/− 3.6)(Fig. S3A), suggesting that a critical repressive cis-element lies in exons 5–8 or their intervening introns. ssprot null2 also upregulated expression from a ss locus on an inverted chromosome (sswild type(inv2)/ ssprot null2 = 75.5% +/− 3.3) and a ss locus duplicated on chromosome 2 (ssduplication) (from 59.5% +/− 6.3 in ssduplication; sswild type animals to 79.4% +/− 5.9 in ssduplication; ssprot null2 animals), consistent with the ability of interchromosomal communication to upregulate expression from inverted and non-homologous chromosomes.
C. sslow freq alleles downregulate expression from ss in its normal (sswild type) or heterologous position (sswild type(inv2)). sslow freq1 downregulates expression frequency from sslow freq2.
A. The frequency of Rh3 and Rh4 expression is normal in Condensin II/Cap-H2 mutants.
B. The frequency of Rh3 and Rh4 expression is normal in zeste/za mutants.
C. The frequency of Rh3 and Rh4 expression is normal in zeste/z1 mutants.
D. Expression frequency and coordination between sswild type and sstrunc alleles (i.e. complete exclusive expression of Rh3 and Rh4) is normal in flies with ectopic expression of Condensin II/Cap-H2.
E. Blue channel (Rh3) only of Fig. S1D.
F. transgene4 upregulates Ss/Rh4 expression normally in flies with ectopic expression of Condensin II/Cap-H2.
G. transgene4 upregulates Ss/Rh4 expression normally in zeste/za mutants.
Schematics for ss locus as in Figure 2. For A–B, E–F, Rh3 is in blue and Rh4 is in red.
A. Rh3 is expressed in pR7s and Rh4 is expressed in yR7s in wild type hemizygous animals.
B. Rh3 is expressed in pR7s and Rh4 and Rh3 are expressed in yR7s in sstrunc hemizygous animals.
C. If expression from ss alleles is independent, there are four combinations of expression from sswild type and sstrunc yielding 3 Rh expression states in R7s (Rh3 alone; Rh4 alone; Rh3 and Rh4).
D. If expression from ss allele is coordinated, there are two combinations of expression from sswild type and sstrunc yielding 2 Rh expression states in R7s (Rh3 alone; Rh4 alone).
E. In sswild type(inv2)/sstrunc animals, Rh3 and Rh4 are always expressed exclusively showing that expression from ss alleles is coordinated.
F. In sslow freq1/sstrunc animals, the normal regulatory regions of the sstrunc allele upregulate expression from sslow freq1 into the same subset of R7s since Rh3 and Rh4 are expressed exclusively.