Abstract
Background:
Src is a non-receptor tyrosine kinase involved in signalling and crosstalk between growth-promoting pathways. We aim to investigate the relationship of active Src in response to trastuzumab of HER2-positive breast carcinomas.
Methods:
We selected 278 HER2-positive breast cancer patients with (n=154) and without (n=124) trastuzumab treatment. We performed immunohistochemistry on paraffin-embedded tissue microarrays of active Src and several proteins involved in the PI3K/Akt/mTOR pathway, PIK3CA mutational analysis and in vitro studies (SKBR3 and BT474 cancer cells). The results were correlated with clinicopathological factors and patients' outcome.
Results:
Increased pSrc-Y416 was demonstrated in trastuzumab-resistant cells and in 37.8% of tumours that correlated positively with tumour size, necrosis, mitosis, metastasis to the central nervous system, p53 overexpression and MAPK activation but inversely with EGFR and p27. Univariate analyses showed an association of increased active Src with shorter survival in patients at early stage with HER2/hormone receptor-negative tumours treated with trastuzumab.
Conclusions:
Src activation participates in trastuzumab mechanisms of resistance and indicates poor prognosis, mainly in HER2/hormone receptor-negative breast cancer. Therefore, blocking this axis may be beneficial in those patients.
Keywords: Src, HER2 breast carcinoma, PTEN/PI3K/Akt, MAPK, trastuzumab resistance, prognosis
In patients with HER2-positive breast carcinoma, the relationship between poor clinical response to trastuzumab and alterations of the PI3K/Akt pathway has been recently demonstrated. This pathway is activated by several growth factor receptors such as IGF1R, HER1 (EGFR) (Lu et al, 2001; Nahta et al, 2005), HER3 (Lipton et al, 2013), low PTEN levels or PIK3CA-activating mutations (Nagata et al, 2004; Lerma et al, 2008; Serra et al, 2011; Gallardo et al, 2012). Nevertheless, combinations of these alterations can justify the development of resistance only in a subset of tumours.
The v-Src (Rous sarcoma virus) tyrosine kinase was the first oncogenic gene discovered by Stehelin et al (1977). c-Src is the corresponding cellular gene that codifies Src, a non-receptor (intracellular) tyrosine kinase that belongs to the Src family kinases (Thomas and Brugge, 1997). The 60-kDa human c-Src tyrosine kinase contains two phosphorylation sites regulating its enzymatic activity. Phosphorylation at Tyr527 leads to a reduced activity, whereas autophosphorylation at Tyr416 is associated with full kinase activity (Roskoski, 2005). Src can be activated by extracellular signals such as integrins and growth factors, including PDGFR, HER family members (HER1; HER2 and HER3), IGF1R, hepatocyte growth factor/scatter factor receptor, mitogen-activated protein kinases – MAPK – and STAT3 pathways, leading to cellular survival and proliferation (Olayioye et al, 2001). Once activated, Src acts as a common upstream regulator of the Ras/MAPK (Mitra and Schlaepfer, 2006) and PI3K pathways (Ingley, 2008) inducing malignant transformation. In fact, activation of Src has been observed in about 50% of tumours from the colon, liver, lung, breast and pancreas (Dehm and Bonham, 2004).
Previous experimental results strongly indicate an important role of Src in the development and progression of breast cancer (Zou et al, 2009) as well as a viable therapeutic option (Gnoni et al, 2011). Moreover, novel studies performed in animal models, breast cancer cells (Lu et al, 2003; Wang et al, 2009; Boyer et al, 2012) and human tumours (Nagata et al, 2004; Liang et al, 2010; Zhang et al, 2011) have suggested Src as a key modulator of trastuzumab response and a common node downstream of multiple trastuzumab-resistance pathways. Although the available experimental data are in fact very promising, the results in clinical series have shortcomings due to the small number of patients included. Furthermore, the underlying molecular mechanisms of Src-mediated trastuzumab resistance remain unclear.
To address this issue, we analysed the Src activation (phosphorylated at Tyr416) status together with several other Src-dependent biomarkers and patients' outcome in a large series of HER2-positive breast carcinomas. Here we show that Src activation is associated with MAPK and PI3K/Akt oncogenic pathways and imply worse patients' outcome, indicating resistance. In addition, we confirmed the activity of phosphorylated-Src in two resistant HER2-positive breast cancer cell lines (SKBR3 and BT474).
Materials and methods
Tumour samples and patients' follow-up
The current retrospective study is based on a cohort of 278 unselected consecutive HER2-positive breast carcinomas. Patients were classified into three groups depending on the modality of treatment as follows: group A (n=124) included those who received chemotherapy (CT) and no trastuzumab; group B (n=76) included patients who received trastuzumab for the treatment of metastatic disease (first-line therapy) after failure of the conventional CT with anthracyclines and/or taxanes, and group C (n=65) included those who received trastuzumab combined with CT with anthracyclines and/or taxanes for early stages in the adjuvant/neoadjuvant setting. The characteristics of trastuzumab-treated group have been described in detail previously (Gallardo et al, 2012). Treatment was given in a neoadjuvant setting in 4 patients in group A, 22 patients in group B and 21 in group C. In 13 patients, the type of treatment was unknown. Moreover, patients with ER and/or PR-positive tumours received tamoxifen or aromatase inhibitors for 2–5 years. Of note, trastuzumab untreated patients (group A) were diagnosed and treated before trastuzumab therapy was approved at both Institutions.
Tumours were collected from the Departments of Pathology of the University General Hospital of Alicante (n=196) and Hospital de la Santa Creu i Sant Pau (n=82), Barcelona, Spain. The study was conducted according to the Declaration of Helsinki principles. The Institutional Review Board at both Institutions approved the study and waived the requirement for informed consent in patients with the diagnosis before 2007.
Clinical–pathological staging was performed according to the WHO system and tumours were graded according to the Elston and Ellis method. The most representative areas of the infiltrating tumour were selected for the tissue microarray (TMA) construction (Beecher Instruments; Silver Spring, MD, USA) with three 1-mm cores.
The follow-up of the trastuzumab-treated cohort was updated (Gallardo et al, 2012). Median patients' follow-up in group A was 8.4 years (n=110; range 0.26–17.9 years), in group B it was 8.28 years (n= 46; range 1.34–20.9 years) and in group C it was 3.6 years (n=36; range 1.13–8.12 years).
We considered response or non-resistance to trastuzumab treatment when no progression o stable disease occurred. Progression-free survival was defined as the length of time after treatment during which a patient survived with no signs of the disease, and overall survival (OS) as the time to the patients' death or last follow-up.
Immunohistochemistry (IHC)
It was carried out on serial TMAs sections using the EnVision Flex detection system (Dako/Agilent Technologies; Carpinteria, CA, USA). Antibodies, suppliers, dilutions, conditions and cutoffs are listed in Table 1. Staining results were semiquantitatively scored according to the percentage of positive cells and intensity (0 to 3+) (Histo-score 0–300). Src activation (Src phosphorylated at Tyr416) status was studied in all tumours and correlated with several Src-dependent biomarkers and patients' outcome. Tumours with pSrc-Y416 membrane+/−cytoplasm staining of at least 5% of cells with moderate/strong (2–3+) intensity were considered positive. As negative controls, staining was carried out in the absence of the primary antibodies.
Table 1. Panel of antibodies for the immunohistochemical analysis.
| Antibody | Clone | Dilution | Supplier | Pretreatment | Incubation | Staining/cutoff |
|---|---|---|---|---|---|---|
| HER2 (HercepTest) |
Rabb Pol |
1 : 1 |
Dako |
Citrate buffer pH 9 |
30 min, RT |
Membrane, ⩾10% 3+ |
| ER-α |
6F11 |
1 : 40 |
Dako |
Citrate buffer pH 6 |
20 min, RT |
Nuclear, ⩾1% |
| PR |
PgR636 |
1 : 200 |
Dako |
Citrate buffer pH 6 |
20 min, RT |
Nuclear, ⩾1% |
| Ki67 |
MIB-1 |
1 : 1 |
Dako |
Citrate buffer pH 9 |
20 min, RT |
Nuclear, ⩾14% |
| pSrc (Tyr416) |
Rabbit Mn |
1 : 50 |
Cell Sig |
Citrate buffer pH 9 |
4 °C overnigth |
Membr/cytopl, HS >5% 2–3+ |
| EGFR (pharmaDx) |
H11 |
1 : 1 |
Dako |
Citrate buffer pH 9 |
30 min, RT |
Membrane, ⩾10% 3+ |
| α-IGF1R |
24-31 |
1 : 200 |
NeoM |
Citrate buffer pH 9 |
30 min, RT |
Membr/cytopl, HS ⩾220 |
| PTEN |
6H2.1 |
1 : 50 |
Dako |
Citrate buffer pH 6 |
20 min, RT |
Membr/cytopl, HS ⩾75 |
| p110α |
Rabbit Pol |
1 : 80 |
Cell Sig |
Citrate buffer pH 9 |
4 °C overnigth |
Membr/cytopl, HS ⩾150 |
| pAkt (Ser473) |
14-5 |
1 : 10 |
Dako |
Citrate buffer pH 9 |
20 min, RT |
Membr/cytopl, HS ⩾150 |
| pmTOR (Ser2448) |
Rabbit Pol |
1 : 50 |
Cell Sig |
Citrate buffer pH 9 |
20 min, RT |
Membr/cytopl, HS ⩾30 |
| pMAPK (Thr202/Tyr204) |
20G11 |
1 : 100 |
Cell Sig |
Citrate buffer pH 9 |
30 min, RT |
Membr/cytopl, HS ⩾150 |
| p53 |
DO-7 |
1 : 1 |
Dako |
Citrate buffer pH 9 |
20 min, RT |
Nuclear, 20% |
| p27 | SX53G8 | 1 : 50 | Dako | Citrate buffer pH 9 | 30 min, RT | Nuclear, 75% |
Abbreviations: EGFR=epidermal growth factor receptor; ER=oestrogen receptor; HER2=human epidermal growth factor receptor; HS=Histo-score; IGF1R=insulin-like growth factor 1-receptor; Membr/cytopl= membrane/cytoplasm; Mn=monoclonal; Pol=polyclonal; PR=progesterone receptor.
Dako, Dako Diagnostics (Glostrup, Denmark).
NeoMarkers Inc (Fremont, CA, USA).
Cell Signaling Technology (Danvers, MA, USA).
In situ hybridisation (ISH) analysis
HER2 gene status was confirmed by chromogenic ISH (Spot light; Zymed, San Francisco, CA, USA) or fluorescence ISH (Dako pharmaDx) in non-definitive cases (2+ and <10% 3+ cells) (Peiro et al, 2007).
Mutational analysis of PIK3CA
Genomic DNA was extracted from frozen or paraffin-embedded tumours and mutational analysis of PIK3CA was performed by PCR and direct sequencing using primers for exons 9 and 20, as previously described (Gallardo et al, 2012).
Cell culture
BT474 (HER2/ER-positive) and the SKBR3 (HER-positive/ER-negative) human breast cancer cells were cultured in DMEM/Ham's F-12 (1 : 1) (PAA, Cölbe, Germany), supplemented with 10% fetal bovine serum (PAA), penicillin (100 U ml−1) and streptomycin (100 μg/ml−1) and maintained at 37 °C in a humidified atmosphere of 5% CO2. Trastuzumab-resistant BT474 and SKBR3 cells were developed by adding freshly prepared trastuzumab (4 μg/ml−1 or 8 μg/ml−1) twice a week for 4 months; cells were passaged when 70–80% confluence was reached. The resistance to trastuzumab was tested by dose–response studies in parental and resistant cells and western blotting analysis, as described below.
Dose–response studies
SKBR3 and BT474 parental and trastuzumab-resistant cells were seeded in 96-well plates at a density of 1 or 2 × 103 cells per well, respectively. After 24 h, cell media was replaced and newly added with trastuzumab at different concentrations or DMSO (vehicle), using four wells per concentration. After 5 days, MTT reagent was added and incubated for 3 h at 37 °C in a humidified atmosphere of 5% CO2. Cell media was then replaced with 200 μl of DMSO, and after 30 min, optical density was measured at 570 nm in a microplate reader. The experiment was performed in triplicate at different times.
Western blotting analysis
Protein lysates were obtained from parental and trastuzumab-resistant cells using a lysis buffer composition of 20 mM Tris at pH 7.0, 1% Triton-X 100, 0.5% NP-40, 250 mM NaCl, 3 mM EDTA, 2 mM DTT and protease inhibitor cocktail. Cell lysate was collected after centrifugation at 14 000 r.p.m., 10 min at 4 °C, separated on SDS–PAGE gel at 8% polyacrilamyde and blotted onto a nitrocellulose membrane (Bio-Rad Laboratories, Hercules, CA, USA). The membrane was blocked in 5% milk in PBS-Tween for 1 h and then probed with primary antibodies Monoclonal Mouse Anti-Human HER2-pY1248 (Dako, Glostrup, Denmark); Src (36D10) #2146 and Src-Y416 #2101 (Cell Signaling Technology, Danvers, MA, USA) overnight at 4 °C. After washing the membrane in PBS-Tween, it was incubated with secondary antibodies according to the primary antibody used, for 1 h at room temperature, washed three times in PBS-Tween and then visualised with enhanced chemiluminescence reagent, following the manufacturer's instructions (Amersham ECL Western Blotting Analysis System, GE Healthcare UK Limited, Little Chalfont, UK).
Statistical analyses
The chi-square or Fisher' tests were used to determine the distribution of the clinical–pathological, IHC and molecular characteristics. A receiver operating characteristic and area under the curve (AUC) were generated to determine a cutoff value of the protein expression and the potential clinical utility to predict trastuzumab response (recurrence vs death). Therefore, the cutoff with the largest AUC was chosen. Kaplan–Meier survival plots and log-rank tests were performed for the comparison of the survival curves. Multivariate analysis (Cox proportional hazards regression model) was carried out based on the results of the univariate analysis, to identify those factors that were independently associated with cancer-specific recurrence or death. Statistical analyses were performed with the SPSS/win 19.0 statistical software package (SPSS, Chicago, IL, USA). P-values <0.05 were considered statistically significant.
Results
The clinical–pathological data are summarised in Supplementary Table S1. Patients were classified into three groups as previously detailed. Median age was 56 years (range 30–92 years), and median tumour size was 23 mm (range 5–200 mm). Tumours were predominantly of grade 3 (171 out of 276; 61.5%) with necrosis (90 out of 171; 52.6%), no vascular invasion (203 out of 265; 76.6%), with positive lymph node status (146 out of 264; 55.3%) and presenting at early stage (IIA 28%). Thirteen patients were lost in the follow-up (5%), 141 (51%) were alive with no evidence of disease, 33 (12%) were alive with disease, 85 (31%) died of the disease and 6 (2%) were dead of other causes.
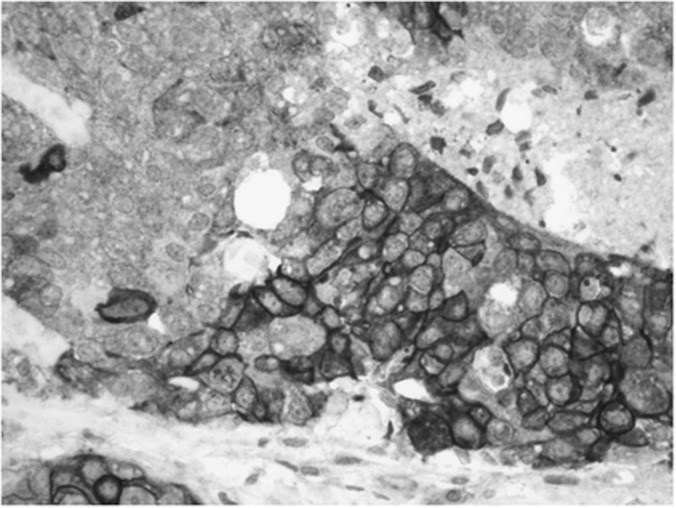
Table 2 includes the relationship between pSrc-Y416 and clinicopathological data. Src activation (Figure 1) was seen in 37.8% (105 out of 278) of the tumours, in relation with larger tumour size (P=0.049), necrosis (P=0.043), high mitotic index (P=0.021) or metastasis to the central nervous system (CNS; P=0.009). Table 3 shows the correlations between Src activation and other molecular biomarkers. An inverse correlation was found between Src and EGFR expression (8.4%, P=0.006). Significant positive correlations were observed with pMAPK activation (25.6% P<0.000), as well as p53 overexpression (43.4%, P=0.009) and p27 nuclear expression (74% P=0.028), and as a trend with α-IGF1R (33% P=0.16), p110α (22.9% P=0.11) or pAkt (28.4% P=0.2). However, no association was seen either with PTEN protein loss or with p-mTOR.
Table 2. Clinico-pathological features in relation to pSrc-Y416.
| Variables | All cases, n (%) | pSrcY416 (+), n (%) | pSrcY416 (−), n (%) | *P-value |
|---|---|---|---|---|
| Number |
278 (100) |
105 (37.8) |
173 (62.2) |
|
|
Age (years) | ||||
| <50 | 88 (31.7) | 34 (36.2) | 54 (32) | |
| ⩾50 | 175 (62.9) | 60 (63.8) | 115 (68) | 0.487 |
| Unknown |
15 (5.4) |
|
|
|
|
Size | ||||
| 5–20 mm | 103 (37.1) | 45 (48.9) | 58 (36.3) | |
| >20 mm | 149 (53.6) | 47 (51.1) | 102 (63.8) | 0.049 |
| Unknown |
26 (9.4) |
|
|
|
|
Grade | ||||
| 1 | 12 (4.3) | 2 (1.9) | 10 (5.8) | |
| 2 | 93 (33.5) | 33 (31.4) | 61 (35.3) | |
| 3 |
171 (61.5) |
70 (66.7) |
102 (59) |
0.203 |
|
Necrosis | ||||
| Absent | 81 (29.1) | 19 (35.8) | 62 (52.5) | |
| Present | 90 (32.4) | 34 (64.2) | 56 (47.5) | 0.043 |
| Unknown |
107 (38.5) |
|
|
|
|
LVI | ||||
| Absent | 203 (73) | 70 (71.4) | 133 (79.6) | |
| Present | 62 (22.3) | 28 (28.6) | 34 (20.4) | 0.127 |
| Unknown |
13 (4.7) |
|
|
|
|
LN status | ||||
| Negative | 118 (42.4) | 46 (46.9) | 72 (43.4) | |
| Positive | 146 (52.5) | 52 (53.1) | 94 (56.6) | 0.573 |
| Unknown |
14 (5) |
|
|
|
|
Stage | ||||
| I | 61 (21.9) | 23 (22.8) | 38 (22.6) | |
| II | 109 (39.2) | 39 (38.6) | 70 (41.7) | |
| III | 89 (32) | 33 (32.7) | 56 (33.3) | |
| IV | 10 (3.6) | 6 (5.9) | 4 (2.4) | 0.512 |
| Unknown | 9 (3.2) | |||
Abbreviations: LN=lymph nodes; LVI=lymph vascular invasion.
*All comparisons by Chi2 or Fisher tests.
Figure 1.
Representative example of the immunohistochemical expression of pSrc-Y416 (membrane±cytoplasm) ( × 400).
Table 3. Relationship between pSrc-Y416 and molecular biomarkers.
| Variables | All cases, n (%) | pSrcY416 (+), n (%) | pSrcY416 (−), n (%) | *P-value |
|---|---|---|---|---|
| Number |
278 (100) |
105 (37.8) |
173 (62.2) |
|
|
HR status | ||||
| Positive | 150 (54) | 51 (53.1 | 99 (57.6) | |
| Negative | 118 (42.4) | 45 (46.9) | 73 (42.4) | 0.483 |
| Unknown |
10 (3.6) |
|
|
|
|
p53 | ||||
| <20% | 174 (62.6) | 56 (56.6) | 118 (72.4) | |
| ⩾20% | 88 (31.7) | 43 (43.4) | 45 (27.6) | 0.009 |
| Unknown |
16 (5.8) |
|
|
|
|
Ki67 | ||||
| <14% | 95 (34.2) | 32 (33) | 63 (40.1) | |
| ⩾14% | 159 (57.2) | 65 (67) | 94 (59.9) | 0.253 |
| Unknown |
24 (8.6) |
|
|
|
|
PTEN | ||||
| Loss | 47 (16.9) | 15 (14.9) | 32 (18.9) | |
| Present | 224 (80.6) | 87 (85.3) | 137 (81.1) | 0.373 |
| Unknown |
7 (2.5) |
|
|
|
|
α-IGF1R | ||||
| No overexpression | 191 (68.7) | 69 (67) | 122 (74.8) | |
| Overexpression | 75 (27) | 34 (33) | 41 (25.2) | 0.165 |
| Unknown |
12 (4.3) |
|
|
|
|
EGFR | ||||
| Negative | 221 (79.5) | 93 (92.1) | 128 (79.5) | |
| Positive | 41 (14.7) | 8 (7.9) | 33 (20.5) | 0.006 |
| Unknown |
16 (5.8) |
|
|
|
|
PIK3CA
mut | ||||
| Negative | 201 (72.3) | 78 (86.7) | 123 (80.4) | |
| Positive | 42 (15.1) | 12 (13.3) | 30 (19.6) | 0.212 |
| Unknown |
35 (12.6) |
|
|
|
|
p110α | ||||
| No overexpression | 186 (66.9) | 64 (77.1) | 122 (85.3) | |
| Overexpression | 40 (14.4) | 19 (22.9) | 21 (14.7) | 0.119 |
| Unknown |
52 (18.7) |
|
|
|
|
pAkt | ||||
| No overexpression | 186 (66.9) | 63 (71.6) | 123 (77.8) | |
| Overexpression | 60 (21.6) | 25 (28.4) | 35 (22.2) | 0.273 |
| UNKNOWN |
32 (11.5) |
|
|
|
|
p-mTOR | ||||
| No overexpression | 161 (57.9) | 58 (56.9) | 103 (62.4) | |
| Overexpression | 106 (38.1) | 44 (43.1) | 62 (37.6) | 0.367 |
| Unknown |
11 (4) |
|
|
|
|
pMAPK | ||||
| No overexpression | 203 (73) | 61 (74.4) | 142 (92.8) | |
| Overexpression | 32 (11.5) | 21 (25.6) | 11 (7.2) | <0.000 |
| Unknown |
43 (15.5) |
|
|
|
|
p27 | ||||
| Loss | 89 (32) | 25 (26) | 64 (39.5) | |
| Present | 169 (60.8) | 71 (74) | 98 (60.5) | 0.028 |
| Unknown | 20 (7.2) | |||
Abbreviations: EGFR=epidermal growth factor receptor; HR=hormone receptor; IGF1R=insulin-like growth factor 1-receptor.
*All comparisons by Chi2 or Fisher tests.
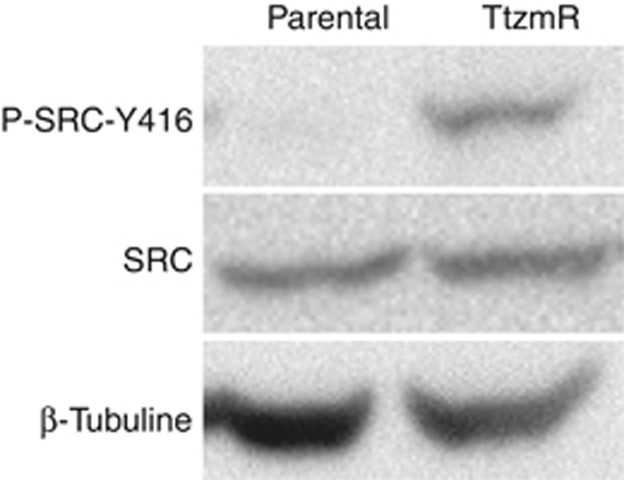
The expression level of inactive/non-phosphorylated Src did not change in BT474- or SKBR3-resistant cells when compared with the parental cell line. In contrast, it was higher for pSrc-Y416 in trastuzumab-resistant cells (Figure 2).
Figure 2.
Western blot results of Src and pSrc-Y416 (active) expression in BT474 parental and resistant cells. The expression of total Src did not change in resistant cells when compared with the parental cell line. In contrast, increased pSrc-Y416 was demonstrated in trastuzumab-resistant cells. Similar results were seen in SKBR3 cells (data not shown).
At last follow-up, 44% of patients had distant metastases, which were located in the liver (41%), bone (38%), lung (28%), lymph nodes (21%), pleura (18%) or CNS (18%). Increased pSrc-Y416 expression was found in tumours that metastasised to CNS (P=0.009). Inverse associations were found between liver metastases and EGFR (P=0.042) and bone metastases with α-IGF1R (P=0.010), p-mTOR (P=0.008) or Ki67 (P=0.012).
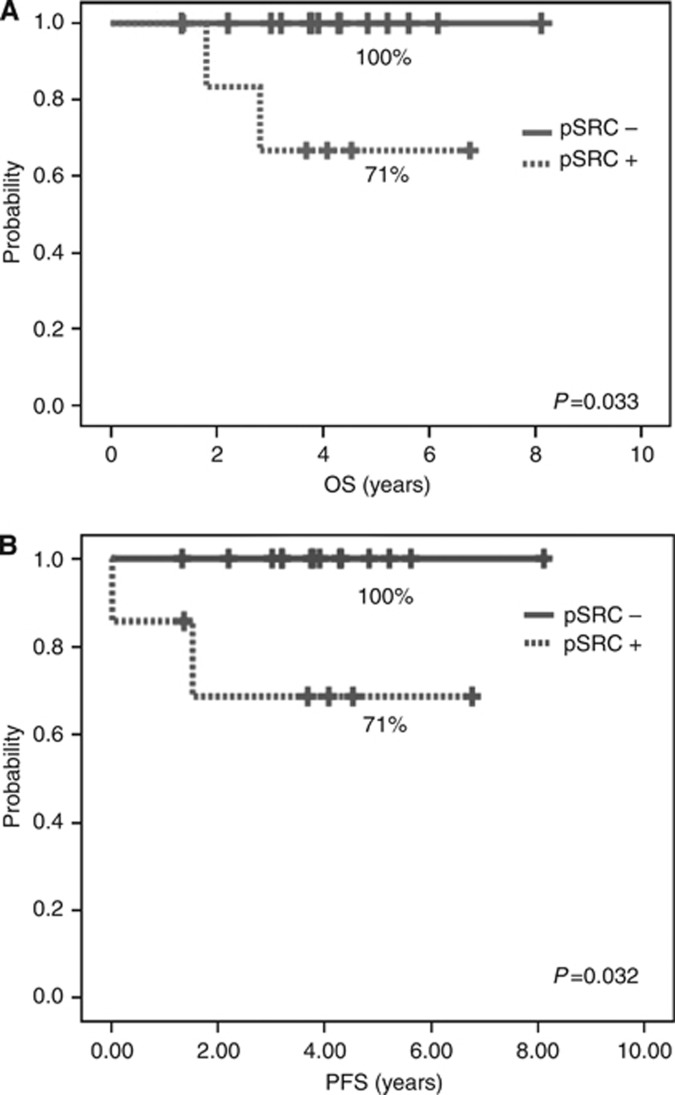
Table 4 shows the results of the univariate analysis (Kaplan–Meier; log-rank test) for the group without trastuzumab. No differences in survival were observed for the activated status of pSrc-Y416. In contrast, in patients treated with trastuzumab-based therapy, increased Src activation implied poorer survival in patients at early stage (OS P=0.034) and in those with metastatic disease, as a trend (OS P=0.148). Interestingly, further analysis showed that specifically in patients under adjuvant trastuzumab with HR-negative tumours, increased pSrc-Y416 correlated with worse DFS (71% vs 100% P=0.032) and OS (71% vs 100% P=0.033) (Figures 3A and B).
Table 4. Clinico-pathological, immunohistochemical and molecular features of trastuzumab untreated breast cancer patients (Kaplan–Meier; log-rank test).
| DFS | P | OS | P | |
|---|---|---|---|---|
| Age <50 vs ⩾50 years |
86% vs 73% |
0.061 |
89% vs 82% |
0.148 |
| Size <2 vs >2 cm |
85% vs 71% |
0.049 |
90% vs 79% |
0.095 |
| Grade (1+2 vs 3) |
84% vs 72% |
NS |
89.5% vs 80.6% |
NS |
| LVI (−) vs (+) |
76% vs 82% |
NS |
83% vs 91% |
NS |
| LN (−) vs (+) |
83% vs 69% |
0.021 |
88% vs 79% |
0.102 |
| Stage (I+II vs III) |
81% vs 61% |
0.004 |
88% vs 67% |
0.001 |
| CNS mets |
0% vs 0% |
NS |
40% vs 0% |
NS |
| Liver mets |
0% vs 0% |
NS |
33% vs 33% |
NS |
| Lung mets |
0% vs 0% |
NS |
50% vs 0% |
0.039 |
| ER (−) vs (+) |
67% vs 83% |
0.067 |
79% vs 87% |
NS |
|
α-IGF1R |
76% vs 82% |
NS |
83% vs 89% |
NS |
| EGFR |
80% vs 57% |
0.003 |
87% vs 71% |
0.016 |
| PTEN loss |
72% vs 96% |
NS |
83% vs 100% |
NS |
|
PI3KCA mutations |
77% vs 77% |
NS |
83.5% vs 90.5% |
NS |
| p110α |
100% vs 75% |
NS |
100% vs 83% |
NS |
| pAkt |
74% vs 79% |
NS |
74% vs 88% |
NS |
| p-mTOR |
73% vs 82% |
NS |
82% vs 89% |
NS |
| pMAPK |
78% vs 78% |
NS |
78% vs 86% |
NS |
| p27 |
75% vs 79.5% |
NS |
85% vs 82% |
NS |
| pSrc-Y416 | 76% vs 80% | NS | 84% vs 85% | NS |
Abbreviations: CNS=central nervous system; EGFR=epidermal growth factor receptor; ER=oestrogen receptor; IGF1R=insulin-like growth factor 1-receptor; LN=lymph nodes; LVI=lymph vascular invasion; mets=metastasis.
Figure 3.
Kaplan-Meier plots for pSrc-Y416 results in patients under adjuvant trastuzumab therapy with HR-negative tumours: (A) overall survival, (B) disease-free survival.
Discussion
Current research has increased substantially the understanding of the abnormalities involved in the mechanisms of trastuzumab resistance. However, there are no validated biomarkers of resistance to this therapy. Our studies focussed on acquired trastuzumab-resistant cells, and a clinical series of HER2 breast cancer patients showed activated Src in breast cancer cells and in a significant proportion of human tumours whose patients had poorer prognosis. This is in line with previous in vitro and in vivo preclinical resistance model studies (Lu et al, 2003; Nagata et al, 2004; Wang et al, 2009; Liang et al, 2010; Boyer et al, 2012; Rexer et al, 2012), indicating that activation status of pSrc-Y416 is involved in the mechanisms of resistance to trastuzumab.
Src has a role in signalling and crosstalk between growth-promoting pathways (Yeatman, 2004). Activated Src expression has been reported in 18–39% of breast carcinomas (Chu et al, 2007; Schmitz et al, 2005). In agreement, we found that 37.8% of our tumours showed active Src, especially in those of larger size, with high proliferation index, necrosis, mitosis, metastasis to the CNS and p53 overexpression, all related with a more aggressive phenotype. Moreover, several molecular alterations were also frequent, such as overexpression of IGF1R, p110α/pAkt, MAPK and p27. In contrast, there were no differences in PTEN or mTOR status. Collectively, our results support that Src activation is associated with an IGF1R-dependent mechanism involving activation of the MAPK and PI3K/Akt pathways, as reported in other types of neoplasia (Michels et al, 2013). However, an inverted correlation of Src with EGFR and p27 loss and not association with mTOR makes probable that other than IGF1R-dependent pathways may mediate Src activation in breast carcinoma as well (Chu et al, 2007; Ishizawar et al, 2007). In fact, experimental data indicates that Src and PTEN may regulate each other to promote trastuzumab resistance. On the one hand, PTEN inactivation occurs by Src through phosphorylation on tyrosine (Lu et al, 2003) and on serine/threonine at the carboxyl terminal (S380/T382/T383) of PTEN. The latter phosphorylation leads to increased PTEN stability but loss of its function (Vazquez et al, 2000). On the other hand, a novel mechanism suggests that PTEN directly and specifically dephosphorylates Src-Y416 by its protein phosphatase activity (Zhang et al, 2011). In the current study, we found no correlation between PTEN and Src activation status. Of note, among our tumours with active Src, 85.3% had PTEN-preserved expression; therefore, it is plausible that in some cases PTEN was non-functional. However, this issue was out of the scope of our study as we analysed the expression levels but not the functional status or subcellular location (cytoplasm vs membrane).
Deregulation of the PI3K/Akt pathway has been associated with resistance to the HER2 inhibitors (Nagata et al, 2004; Esteva et al, 2010), and adverse outcome has been observed in patients with neoadjuvant- or adjuvant-trastuzumab treatment (Jensen et al, 2012; Cizkova et al, 2013). In the current study, the fact that p110α (PI3K catalytic subunit) and pAkt were overexpressed supports the influence of Src in this pathway in a subset of tumours.
Despite the observed changes in mTOR levels in 39.7% of our cases, a significant association neither with Src activation status nor with prognosis was found, in line with in vitro studies in trastuzumab-resistant and -sensitive cells (Liu et al, 2011). Therefore, this action appears to be independent of mTOR signalling.
Other factors have also been recently associated with the mechanisms of trastuzumab resistance involving Src and PI3K signalling, such as EpoR/Jak2 (Liang et al, 2010), phosphorylation of beta1 integrin subunit upregulation and GDF15-mediated activation of TGF beta receptor-Src-HER2 signalling crosstalk (Joshi et al, 2011), but their clinical relevance has not been confirmed.
p27, a key mediator of cell cycle arrest, is present in normal quiescent epithelial tissues. Loss of nuclear p27 was observed in 34.5% of our cases in correlation with HR-negative status, PTEN loss, high Ki67 and overexpression of p53 and EGFR but without impact or survival. Nevertheless, either p27 downregulation (Lane et al, 2001; Nahta et al, 2004) or upregulation (Huober et al, 2012) has been involved in the mechanisms of resistance of trastuzumab.
Recently, Zhang et al (2011) demonstrated in experimental models that inhibition of Src enhanced trastuzumab-mediated growth inhibition by promoting apoptosis. Further, in a small series of 57 breast cancer patients treated with trastuzumab-based therapies, high amounts of pSrc-Y416 in tumours correlated with lower clinical response, a higher progressive disease and shorter overall survival rates than patients having low active Src tumours. In line, our experimental studies in BT474 and SKBR3 cell lines showed increased pSrc-Y416 levels in trastuzumab-resistant cells. Furthermore, in our large cohort of patients, Src correlated with poorer outcome in patients under trastuzumab treatment. Comparing our three groups of patients, adjuvant trastuzumab vs first line in metastatic disease vs no trastuzumab, activated Src correlated with shorter overall survival in patients under trastuzumab treatment, especially in earlier stages. More interesting, however, was the fact that in subgroup analysis, those patients with HR-negative and increased pSrc-Y416 tumours had even more recurrences or died of the disease.
Preliminary preclinical as well as pharmacodynamic data suggest that Src inhibition is a viable therapeutic option in patients with Src-dependent neoplasms (Gnoni et al, 2011). Furthermore, it may represent a novel therapeutic strategy, with the potential to delay or prevent the acquisition of subsequent resistance to anti-growth factor therapies (Chen et al, 2011; Rexer et al, 2012).
In summary, our data in acquired trastuzumab-resistant breast cancer cells and a large clinical series of patients with HER2 breast carcinoma reinforces that activation of Src in coexistence with alterations in the MAPK and PI3K/Akt pathways are associated with a lower response to trastuzumab. These findings argue in favour of Src as a potential therapeutic target in those patients. Moreover, considering that different resistance mechanisms may coexist in the same tumour, combination with other targeted agents with a potential for synergistic activity might be recommendable to restore sensitivity to trastuzumab. Nevertheless, further understanding of the mechanistic basis for progression of HER2-overexpressing breast cancer will allow more effective targeted treatment options to be developed.
Acknowledgments
We are grateful to Dr. MA Japón (Hospital Virgen del Rocío, Sevilla, Spain) and Dr. F Rojo (Fundación Jiménez Díaz, Madrid, Spain) for providing the cell lines used in this study. We also thank Cristina Pomares and Daniel Fernández for their excellent technical assistance.
This work was supported by grants from Fondo de Investigaciones Sanitarias (FIS PI06/0709 and PI06/1495), Mutua Madrileña Foundation (448/2008), ACOMP/2009-195, FCVI-HGUA PC-03/2010; Instituto Carlos III RTICCCFIS RD06/0020/0015, Pfizer Laboratories, and ROCHE Diagnostics-Sociedad Española de Anatomía Patológica (SEAP).
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on British Journal of Cancer website (http://www.nature.com/bjc)
This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution-NonCommercial-Share Alike 3.0 Unported License.
This paper was presented in part at the 102nd USCAP Meeting in Baltimore, 2–8 March 2013.
Supplementary Material
References
- Boyer AP, Collier TS, Vidavsky I, Bose R. Quantitative proteomics with siRNA screening identifies novel mechanisms of trastuzumab resistance in HER2 amplified breast cancers. Mol Cell Proteomics. 2012;12 (1:180–193. doi: 10.1074/mcp.M112.020115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cizkova M, Dujaric ME, Lehmann-Che J, Scott V, Tembo O, Asselain B, Pierga JY, Marty M, de Cremoux P, Spyratos F, Bieche I. Outcome impact of PIK3CA mutations in HER2-positive breast cancer patients treated with trastuzumab. Br J Cancer. 2013;108:1807–1809. doi: 10.1038/bjc.2013.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Alvarez EA, Azzam D, Wander SA, Guggisberg N, Jorda M, Ju Z, Hennessy BT, Slingerland JM. Combined Src and ER blockade impairs human breast cancer proliferation in vitro and in vivo. Breast Cancer Res Treat. 2011;128:69–78. doi: 10.1007/s10549-010-1024-7. [DOI] [PubMed] [Google Scholar]
- Chu I, Sun J, Arnaout A, Kahn H, Hanna W, Narod S, Sun P, Tan CK, Hengst L, Slingerland J. p27 phosphorylation by Src regulates inhibition of cyclin E-Cdk2. Cell. 2007;128:281–294. doi: 10.1016/j.cell.2006.11.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehm SM, Bonham K. SRC gene expression in human cancer: the role of transcriptional activation. Biochem Cell Biol. 2004;82:263–274. doi: 10.1139/o03-077. [DOI] [PubMed] [Google Scholar]
- Esteva FJ, Guo H, Zhang S, Santa-Maria C, Stone S, Lanchbury JS, Sahin AA, Hortobagyi GN, Yu D. PTEN, PIK3CA, p-AKT, and p-p70S6K status: association with trastuzumab response and survival in patients with HER2-positive metastatic breast cancer. Am J Pathol. 2010;177:1647–1656. doi: 10.2353/ajpath.2010.090885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallardo A, Lerma E, Escuin D, Tibau A, Munoz J, Ojeda B, Barnadas A, Adrover E, Sanchez-Tejada L, Giner D, Ortiz-Martinez F, Peiro G. Increased signalling of EGFR and IGF1R, and deregulation of PTEN/PI3K/Akt pathway are related with trastuzumab resistance in HER2 breast carcinomas. Br J Cancer. 2012;106:1367–1373. doi: 10.1038/bjc.2012.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gnoni A, Marech I, Silvestris N, Vacca A, Lorusso V. Dasatinib: an anti-tumour agent via Src inhibition. Curr Drug Targets. 2011;12:563–578. doi: 10.2174/138945011794751591. [DOI] [PubMed] [Google Scholar]
- Huober J, Fasching PA, Barsoum M, Petruzelka L, Wallwiener D, Thomssen C, Reimer T, Paepke S, Azim HA, Ragosch V, Kubista E, Baumgartner AK, Beckmann MW, May C, Nimmrich I, Harbeck N. Higher efficacy of letrozole in combination with trastuzumab compared to letrozole monotherapy as first-line treatment in patients with HER2-positive, hormone-receptor-positive metastatic breast cancer - results of the eLEcTRA trial. Breast. 2012;21:27–33. doi: 10.1016/j.breast.2011.07.006. [DOI] [PubMed] [Google Scholar]
- Ingley E. Src family kinases: regulation of their activities, levels and identification of new pathways. Biochim Biophys Acta. 2008;1784:56–65. doi: 10.1016/j.bbapap.2007.08.012. [DOI] [PubMed] [Google Scholar]
- Ishizawar RC, Miyake T, Parsons SJ. c-Src modulates ErbB2 and ErbB3 heterocomplex formation and function. Oncogene. 2007;26:3503–3510. doi: 10.1038/sj.onc.1210138. [DOI] [PubMed] [Google Scholar]
- Jensen JD, Knoop A, Laenkholm AV, Grauslund M, Jensen MB, Santoni-Rugiu E, Andersson M, Ewertz M. PIK3CA mutations, PTEN, and pHER2 expression and impact on outcome in HER2-positive early-stage breast cancer patients treated with adjuvant chemotherapy and trastuzumab. Ann Oncol. 2012;23:2034–2042. doi: 10.1093/annonc/mdr546. [DOI] [PubMed] [Google Scholar]
- Joshi JP, Brown NE, Griner SE, Nahta R. Growth differentiation factor 15 (GDF15)-mediated HER2 phosphorylation reduces trastuzumab sensitivity of HER2-overexpressing breast cancer cells. Biochem Pharmacol. 2011;82:1090–1099. doi: 10.1016/j.bcp.2011.07.082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane HA, Motoyama AB, Beuvink I, Hynes NE. Modulation of p27/Cdk2 complex formation through 4D5-mediated inhibition of HER2 receptor signaling. Ann Oncol. 2001;12 (Suppl 1:S21–S22. doi: 10.1093/annonc/12.suppl_1.s21. [DOI] [PubMed] [Google Scholar]
- Lerma E, Catasus L, Gallardo A, Peiro G, Alonso C, Aranda I, Barnadas A, Prat J. Exon 20 PIK3CA mutations decreases survival in aggressive (HER-2 positive) breast carcinomas. Virchows Arch. 2008;453:133–139. doi: 10.1007/s00428-008-0643-4. [DOI] [PubMed] [Google Scholar]
- Liang K, Esteva FJ, Albarracin C, Stemke-Hale K, Lu Y, Bianchini G, Yang CY, Li Y, Li X, Chen CT, Mills GB, Hortobagyi GN, Mendelsohn J, Hung MC, Fan Z. Recombinant human erythropoietin antagonizes trastuzumab treatment of breast cancer cells via Jak2-mediated Src activation and PTEN inactivation. Cancer Cell. 2010;18:423–435. doi: 10.1016/j.ccr.2010.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipton A, Goodman L, Leitzel K, Cook J, Sperinde J, Haddad M, Kostler WJ, Huang W, Weidler JM, Ali S, Newton A, Fuchs EM, Paquet A, Singer CF, Horvat R, Jin X, Banerjee J, Mukherjee A, Tan Y, Shi Y, Chenna A, Larson J, Lie Y, Sherwood T, Petropoulos CJ, Williams S, Winslow J, Parry G, Bates M. HER3, p95HER2, and HER2 protein expression levels define multiple subtypes of HER2-positive metastatic breast cancer. Breast Cancer Res Treat. 2013;141:43–53. doi: 10.1007/s10549-013-2665-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Fan Z, Edgerton SM, Yang X, Lind SE, Thor AD. Potent anti-proliferative effects of metformin on trastuzumab-resistant breast cancer cells via inhibition of erbB2/IGF-1 receptor interactions. Cell Cycle. 2011;10:2959–2966. doi: 10.4161/cc.10.17.16359. [DOI] [PubMed] [Google Scholar]
- Lu Y, Yu Q, Liu JH, Zhang J, Wang H, Koul D, McMurray JS, Fang X, Yung WK, Siminovitch KA, Mills GB. Src family protein-tyrosine kinases alter the function of PTEN to regulate phosphatidylinositol 3-kinase/AKT cascades. J Biol Chem. 2003;278:40057–40066. doi: 10.1074/jbc.M303621200. [DOI] [PubMed] [Google Scholar]
- Lu Y, Zi X, Zhao Y, Mascarenhas D, Pollak M. Insulin-like growth factor-I receptor signaling and resistance to trastuzumab (Herceptin) J Natl Cancer Inst. 2001;93:1852–1857. doi: 10.1093/jnci/93.24.1852. [DOI] [PubMed] [Google Scholar]
- Michels S, Trautmann M, Sievers E, Kindler D, Huss S, Renner M, Friedrichs N, Kirfel J, Steiner S, Endl E, Wurst P, Heukamp L, Penzel R, Larsson O, Kawai A, Tanaka S, Sonobe H, Schirmacher P, Mechtersheimer G, Wardelmann E, Buttner R, Hartmann W. SRC signaling is crucial in the growth of synovial sarcoma cells. Cancer Res. 2013;73:2518–2528. doi: 10.1158/0008-5472.CAN-12-3023. [DOI] [PubMed] [Google Scholar]
- Mitra SK, Schlaepfer DD. Integrin-regulated FAK-Src signaling in normal and cancer cells. Curr Opin Cell Biol. 2006;18:516–523. doi: 10.1016/j.ceb.2006.08.011. [DOI] [PubMed] [Google Scholar]
- Nagata Y, Lan KH, Zhou X, Tan M, Esteva FJ, Sahin AA, Klos KS, Li P, Monia BP, Nguyen NT, Hortobagyi GN, Hung MC, Yu D. PTEN activation contributes to tumour inhibition by trastuzumab, and loss of PTEN predicts trastuzumab resistance in patients. Cancer Cell. 2004;6:117–127. doi: 10.1016/j.ccr.2004.06.022. [DOI] [PubMed] [Google Scholar]
- Nahta R, Takahashi T, Ueno NT, Hung MC, Esteva FJ. P27(kip1) down-regulation is associated with trastuzumab resistance in breast cancer cells. Cancer Res. 2004;64:3981–3986. doi: 10.1158/0008-5472.CAN-03-3900. [DOI] [PubMed] [Google Scholar]
- Nahta R, Yuan LX, Zhang B, Kobayashi R, Esteva FJ. Insulin-like growth factor-I receptor/human epidermal growth factor receptor 2 heterodimerization contributes to trastuzumab resistance of breast cancer cells. Cancer Res. 2005;65:11118–11128. doi: 10.1158/0008-5472.CAN-04-3841. [DOI] [PubMed] [Google Scholar]
- Olayioye MA, Badache A, Daly JM, Hynes NE. An essential role for Src kinase in ErbB receptor signaling through the MAPK pathway. Exp Cell Res. 2001;267:81–87. doi: 10.1006/excr.2001.5242. [DOI] [PubMed] [Google Scholar]
- Peiro G, Aranda FI, Adrover E, Niveiro M, Alenda C, Paya A, Segui J. Analysis of HER2 by chromogenic in situ hybridization and immunohistochemistry in lymph node-negative breast carcinoma: Prognostic relevance. Hum Pathol. 2007;38:26–34. doi: 10.1016/j.humpath.2006.07.013. [DOI] [PubMed] [Google Scholar]
- Rexer BN, Ham AJ, Rinehart C, Hill S, Granja-Ingram Nde M, Gonzalez-Angulo AM, Mills GB, Dave B, Chang JC, Liebler DC, Arteaga CL. Phosphoproteomic mass spectrometry profiling links Src family kinases to escape from HER2 tyrosine kinase inhibition. Oncogene. 2012;30:4163–4174. doi: 10.1038/onc.2011.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roskoski R., Jr. Src kinase regulation by phosphorylation and dephosphorylation. Biochem Biophys Res Commun. 2005;331:1–14. doi: 10.1016/j.bbrc.2005.03.012. [DOI] [PubMed] [Google Scholar]
- Schmitz KJ, Grabellus F, Callies R, Otterbach F, Wohlschlaeger J, Levkau B, Kimmig R, Schmid KW, Baba HA. High expression of focal adhesion kinase (p125FAK) in node-negative breast cancer is related to overexpression of HER-2/neu and activated Akt kinase but does not predict outcome. Breast Cancer Res. 2005;7:R194–R203. doi: 10.1186/bcr977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serra V, Scaltriti M, Prudkin L, Eichhorn PJ, Ibrahim YH, Chandarlapaty S, Markman B, Rodriguez O, Guzman M, Rodriguez S, Gili M, Russillo M, Parra JL, Singh S, Arribas J, Rosen N, Baselga J. PI3K inhibition results in enhanced HER signaling and acquired ERK dependency in HER2-overexpressing breast cancer. Oncogene. 2011;30:2547–2557. doi: 10.1038/onc.2010.626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stehelin D, Fujita DJ, Padgett T, Varmus HE, Bishop JM. Detection and enumeration of transformation-defective strains of avian sarcoma virus with molecular hybridization. Virology. 1977;76:675–684. doi: 10.1016/0042-6822(77)90250-1. [DOI] [PubMed] [Google Scholar]
- Thomas SM, Brugge JS. Cellular functions regulated by Src family kinases. Annu Rev Cell Dev Biol. 1997;13:513–609. doi: 10.1146/annurev.cellbio.13.1.513. [DOI] [PubMed] [Google Scholar]
- Vazquez F, Ramaswamy S, Nakamura N, Sellers WR. Phosphorylation of the PTEN tail regulates protein stability and function. Mol Cell Biol. 2000;20:5010–5018. doi: 10.1128/mcb.20.14.5010-5018.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang SE, Xiang B, Zent R, Quaranta V, Pozzi A, Arteaga CL. Transforming growth factor beta induces clustering of HER2 and integrins by activating Src-focal adhesion kinase and receptor association to the cytoskeleton. Cancer Res. 2009;69:475–482. doi: 10.1158/0008-5472.CAN-08-2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeatman TJ. A renaissance for SRC. Nat Rev Cancer. 2004;4:470–480. doi: 10.1038/nrc1366. [DOI] [PubMed] [Google Scholar]
- Zhang S, Huang WC, Li P, Guo H, Poh SB, Brady SW, Xiong Y, Tseng LM, Li SH, Ding Z, Sahin AA, Esteva FJ, Hortobagyi GN, Yu D. Combating trastuzumab resistance by targeting SRC, a common node downstream of multiple resistance pathways. Nat Med. 2011;17:461–469. doi: 10.1038/nm.2309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou D, Yoon HS, Anjomshoaa A, Perez D, Fukuzawa R, Guilford P, Humar B. Increased levels of active c-Src distinguish invasive from in situ lobular lesions. Breast Cancer Res. 2009;11:R45. doi: 10.1186/bcr2332. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.