Summary
Distal enhancers commonly contact target promoters via chromatin looping. In erythroid cells, the locus control region (LCR) contacts β-type globin genes in a developmental stage-specific manner to stimulate transcription. Previously, we induced LCR-promoter looping by tethering the self-association domain (SA) of Ldb1 to the β-globin promoter via artificial zinc fingers. Here, we show that targeting the SA to a developmentally silenced embryonic globin gene in adult murine erythroblasts triggered its transcriptional reactivation. This activity depended on the LCR, consistent with an LCR-promoter looping mechanism. Strikingly, targeting SA to the fetal γ-globin promoter in primary adult human erythroblasts increased γ-globin promoter-LCR contacts, stimulating transcription to approximately 85% of total β-globin synthesis with a reciprocal reduction in adult β-globin expression. Our findings demonstrate that forced chromatin looping can override a stringent developmental gene expression program and suggest a novel approach to control the balance of globin gene transcription for therapeutic applications.
Keywords: hemoglobin switching, gene looping, chromatin, transcription, Ldb1
Introduction
Long-range enhancers physically contact their target promoters to form chromatin loops. The β-globin locus has been at the forefront of studies on the dynamics and mechanisms of chromatin looping and gene regulation. In erythroid cells, a powerful distal enhancer called the locus control region (LCR), composed of multiple DNase I hypersensitive sites (HSs), is in physical proximity to the β-globin genes in a developmentally dynamic manner (Carter et al., 2002; Tolhuis et al., 2002). In primitive murine erythroid cells the LCR loops to the embryonic type β-globin gene promoters (εy and βh1), whereas in definitive erythroid cells the LCR exclusively contacts the adult type β-globin genes (β-major and β-minor) with the intervening embryonic globin genes looped out (Palstra et al., 2003). Humans additionally bear fetal stage specific β-like globin (γ-globin) genes that contact the LCR and are transcribed from the beginning of fetal liver erythropoiesis until birth when blood formation gradually shifts to the bone marrow. Several transcription factors have been implicated in loop formation at the β-globin locus, including Klf1, GATA1, and its co-regulators Ldb1 and FOG1 (Drissen, 2004; Song et al., 2007; Vakoc et al., 2005). In previous work we addressed whether any of these factors might be sufficient for chromatin looping, and whether looping is a causal event during transcription initiation or merely a reflection of it (Deng et al., 2012). Specifically, in immature murine erythroblasts in which the β-globin promoter is not in contact with the LCR, tethering of Ldb1 to the promoter via artificial zinc finger (ZF) proteins established an LCR-promoter loop similar to that observed in mature erythroid cells and stimulated transcription (Deng et al., 2012). The self-association (SA) domain of Ldb1 was sufficient for this activity. These results demonstrated that forced juxtaposition of an enhancer and promoter via chromatin looping can trigger gene activation, and identified a relatively small functional domain of Ldb1 as a powerful mediator of such long-range interactions. Therefore, it should be possible to activate any other similarly regulated nearby gene by producing an interaction between it and the active LCR.
The molecular mechanisms mediating the developmental switch in globin gene expression have been widely investigated. A variety of nuclear complexes contribute to the silencing of the embryonic and fetal globin genes in adult erythroid cells (Sankaran et al., 2010). Among these, arguably the most powerful one is nucleated by Bcl11a (Sankaran et al., 2008; 2009) which does not bind directly to promoters of the silenced human fetal globin genes but seems to repress them via a mechanism involving higher order chromatin looping (Kiefer et al., 2011; Xu et al., 2010). Hence, manipulating chromatin loops might be a viable strategy to reverse the globin switch. The overarching interest in these questions stems in part from the clinical importance of understanding hemoglobin switching. Patients affected by sickle cell anemia and β-thalassemia experience a milder course of the disease if they express elevated levels of fetal hemoglobin in adulthood (Platt et al., 1994; Weatherall, 2001). This observation in particular provided a major impetus for studies over the last decades aimed at unraveling the mechanisms by which the fetal globin genes are silenced with the ultimate goal of reactivating their expression.
Here we tested whether developmentally silenced embryonic or fetal globin genes can be re-activated in adult erythroid cells by juxtaposing them with the LCR, and whether re-activation would lead to a concomitant reduction in the adult type globin genes. We found that zinc finger mediated tethering of the SA domain of Ldb1 to the murine embryonic βh1-globin promoter activated transcription in an adult type erythroblast cell line and primary definitive erythroid cells. This activity was abrogated in erythroid cells from mice in which the LCR had been deleted, demonstrating that embryonic globin gene activation was dependent on a long range LCR interaction. Primary human erythroid cells expressing the SA domain fused to a zinc finger moiety that targets the γ-globin promoters produced high quantities of γ-globin mRNA and protein in the vast majority of cells with an accompanying reduction in adult type globin expression. The reciprocal alteration in fetal versus adult globin gene transcription was reflected in corresponding changes in contact frequencies with the LCR.
Together, these studies demonstrate that stringent developmental regulation of a gene can be overcome through the manipulation of higher order chromatin structure, and that such manipulations have the potential for therapeutic applications.
Results
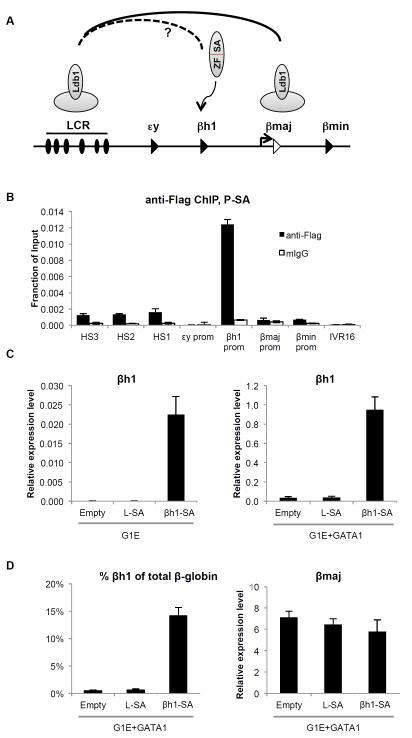
βh1-SA mediated activation of an embryonic globin gene in adult erythroblasts
To examine the potential of forced gene looping in reprogramming the β-globin locus, we began by using G1E erythroblasts. G1E is a murine adult type erythroid cell line that lacks the erythroid transcription factor GATA1 (Weiss et al., 1997). As a result, a productive interaction between the LCR and the adult β-globin (β-major) promoter fails to form and β-major mRNA production is low (Vakoc et al., 2005). GATA1 deficiency is associated with little to no Ldb1 recruitment to the promoter (Tripic et al., 2009). Tethering Ldb1 or its SA domain to the β-major promoter restored looping in a manner highly similar to that in normal erythroid cells or G1E cells upon GATA1 restoration (Deng et al., 2012). Here we asked whether a similar approach could be employed to achieve a distinct goal, which is to activate the embryonic βh1-globin gene by tethering the Ldb1 SA domain to its promoter to induce an interaction with the LCR (Figure 1A). We designed custom zinc finger proteins, and screened them for binding to βh1-globin promoter sequences in vitro (see Experimental Procedures). Functional proteins were fused to the SA domain of Ldb1 (βh1-SA) and introduced into G1E cells by retroviral infection. Infected cells were isolated by fluorescence-activated cell sorting (FACS). Chromatin immunoprecipitation (ChIP) revealed specific binding of βh1-SA to the βh1-globin promoter but not the promoters of the other β-like globin genes (Figure 1B). We observed a low but reproducible ChIP signal at the DNase 1 hypersensitive (HS) sites of the LCR, which is likely due to βh1-SA binding to endogenous Ldb1 containing complexes (Deng et al., 2012). We next examined the effects of βh1-SA on βh1-globin transcription by RT-qPCR. βh1-globin mRNA levels were increased 368-fold over non-expressing G1E cells or cells that express a zinc finger protein targeting HS2 of the LCR (L-SA) (Figure 1C). Restoration of GATA1 activity in G1E cells via expression of an estradiol-inducible form of GATA1 (GATA1-ER) leads to erythroid maturation and high-level induction of β-major transcription as well as low level induction of βh1-globin (Figure 1C,D) (Weiss et al., 1997; Welch et al., 2004). Hence we investigated whether βh1-SA expression was capable of raising βh1-globin transcription in differentiating β-major producing erythroid cells. Notably, βh1-SA induced βh1-globin levels 26-fold over control cells, amounting to ~15% of total β-globin expression compared to ~0.5% in vector only containing cells (Figure 1C,D). Tethering the SA domain to the LCR (L-SA in Figure 1) or β-major promoter (Deng et al., 2012, and data not shown) did not activate βh1-globin expression, confirming that the effects of βh1-SA are specific. Basal level βh1-globin expression as well as βh1-SA-induced βh1-globin transcription were higher in GATA1 replete cells when compared to parental G1E cells, which is likely due to increased LCR and promoter activities in the presence of GATA1 (Figure S1A). The substantial increase in βh1-globin expression was associated with a trend towards lower β-major expression (see also below), but without significant changes of other erythroid genes, including α-globin and Kit, indicating that the effects of βh1-SA are not simply a consequence of differentiation induction (Figure 1D, Figure S1B).
Figure 1. Reactivation of the silenced βh1-globin gene in adult erythroid cells by the tethered Ldb1 SA domain.
(A) Experimental strategy. In normal adult erythroid cells, Ldb1 is part of a multi-protein complex at the LCR and the β-major promoter and contributes to LCR- β-major contacts. Zinc finger mediated tethering of the SA domain of Ldb1 to the βh1-globin promoter might juxtapose the βh1-globin promoter to the LCR at the expense of the βmaj-LCR interaction. (B) Anti-Flag ChIP in G1E cells expressing βh1-SA. (C) mRNA levels of βh1-globin in G1E cells (left) or G1E cells expressing activated GATA1-ER (G1E+GATA1, right) transduced with empty vector, or vector expressing SA fused to LCR-specific zinc finger (L-SA) or βh1-specific zinc finger (βh1-SA). (D) (left) βh1 mRNA levels plotted as percentage of total β-globin transcripts; (right) β-major expression in G1E+GATA1 cells transduced with indicated constructs. Expression is normalized to β-actin. Error bars denote SD. N=3. See also Figure S1.
βh1-SA-mediated activation of embryonic globin gene expression in primary adult erythroblasts
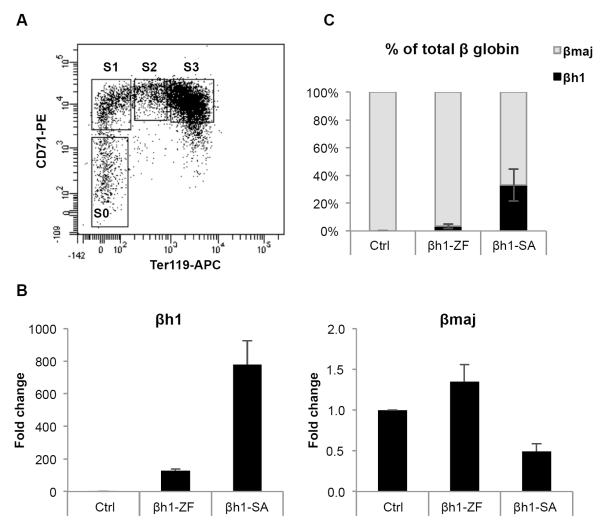
To address whether βh1-SA activates expression of βh1-globin in murine primary erythroid cells, we isolated immature definitive (adult type) erythroid progenitor cells from E13.5 wild type fetal livers by sorting for CD71 medium/low and Ter119 low populations (S0 cells) as described previously (Figure 2A) (Pop et al., 2010). Cells were infected with a retroviral vector expressing βh1-SA (infection rate >75%, data not shown) and expanded for 48 hours, followed by measurements of transcript levels by RT-qPCR. βh1-SA activated βh1-globin transcription by almost 800-fold compared to empty vector control (Figure 2B), exceeding 30% of total β-globin synthesis (Figure 2C). We noted that the zinc finger alone also raised βh1-globin expression but only to ~1/6 of the levels achieved by βh1-SA (Figure 2B,C). While the cause for this activity is unclear, we surmise that zinc finger binding might increase chromatin occupancy of nearby activators (Adams and Workman, 1995) that in turn contribute to LCR recruitment.
Figure 2. Reactivation of the silenced βh1-globin gene in primary erythroid progenitor cells by the tethered Ldb1 SA domain.
(A) Surface phenotypic staging of E13.5 fetal liver progenitor cells with Ter119 and CD71. Erythroid maturation occurs as cells progress from S0 to S3 populations (B) Fold-activation of βh1-globin and β-major mRNA levels relative to empty vector infected cells (MigR1) (C) βh1-globin mRNA levels plotted as percentage of total β-globin transcripts from S0 cells transduced with indicated constructs. Results were normalized to GAPDH. Error bars denote SEM. N=3. See also Figure S2.
We also examined the ability of βh1-SA to increase βh1-globin expression in erythroblasts that are further differentiated (S1 populations) and obtained similar results (Figure S2A,B). Importantly, β-major expression was slightly reduced in both βh1-SA expressing populations (Figure 2B,S2A), consistent with the idea that βh1-SA recruits the LCR towards the βh1-globin gene at the expense of the β-major gene. We noted that βh1-SA impaired expression of a few erythroid specific genes such as Band3, AHSP and Alas2 (Figure S2C). Since these genes are in part regulated by promoter proximal Ldb1-containing transcription factor complexes (Tripic et al., 2009), βh1-SA likely influences their expression in a dominant negative fashion, especially when expressed very highly (data not shown). However, in human erythroblasts, SA-fusion proteins did not inhibit the orthologous genes (see below), which might be due to species-specific effects and/or lower expression levels. Nevertheless, these events do not substantially impact overall erythroid maturation and α-globin gene expression, indicating that βh1-globin activation is gene specific and not the result of global differentiation induction.
Interestingly, the embryonic globin gene εy, which is located closer to the LCR than the βh1 gene, was also activated to some degree by βh1-SA expression in both S0 and S1 erythroid precursor cells (Figure S2D). Since βh1-SA does not detectably bind to the εy-globin gene (Figure 1B), this suggests that bringing the LCR closer into the vicinity of the εy-globin gene increases its likelihood of expression.
In sum, recruitment of the Ldb1 SA domain to the βh1-globin promoter is capable of substantially antagonizing the stringent developmental silencing mechanisms operational in definitive murine erythroid cells. If a similar degree of reactivation of fetal globin genes could be achieved in adult human erythroid cells, this would be highly therapeutically relevant (Platt, 2008).
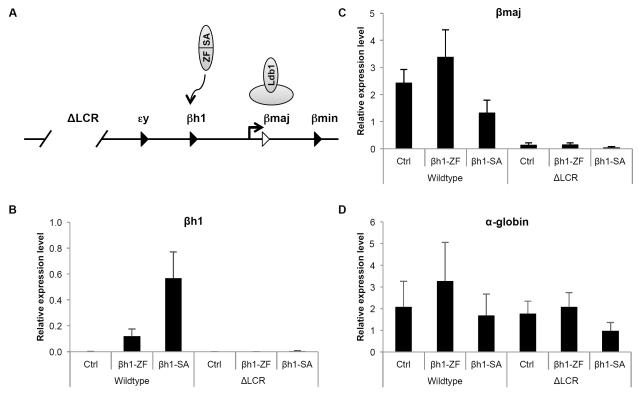
βh1-SA activation of βh1-globin expression is LCR-dependent
Based on our prior studies showing that tethering of Ldb1 to the β-globin promoter triggers looping (Deng et al., 2012), the mechanism by which βh1-SA activates gene expression most likely also involves promoting proximity between the βh1-globin gene and the LCR. To test this directly, we carried out chromosome conformation capture (3C) experiments in G1E cells expressing βh1-SA. However, the relatively short genomic distance between the LCR and the βh1-globin gene as well as the intervening εy-globin gene produced high basal level 3C signals. Moreover, since the activation of βh1-globin expression ranged around 15% of total β-globin synthesis, the signal to noise ratio of the 3C experiments was not high enough to allow reliable detection of changes in contact frequencies (data not shown). As an alternative approach we carried out genetic experiments using erythroid cells from mice lacking the LCR (ΔLCR) that contain a transgenic human β-globin locus to support erythropoiesis (Bender et al., 2000) (Figure 3A). If long-range LCR looping underlies βh1-SA function, the LCR deletion should impair βh1-SA activity. If on the other hand βh1-SA bypasses the requirement for the LCR by functioning locally as a strong transcriptional activator, its activity should be independent of the LCR (Figure 1A, 3A).
Figure 3. LCR dependence of βh1-globin induction by βh1-SA protein.
(A) Experimental concept. Testing the activity of βh1-SA in the absence of the LCR addresses whether it functions locally at the βh1-globin promoter or requires long range LCR looping for function. (B-D) mRNA levels of indicated genes were measured in S0 cells from wild type or ΔLCR/ΔLCR fetal liver cells expressing indicated constructs. Results were normalized to GAPDH. Error bars denote SEM. N=3. See also Figure S3.
βh1-SA was introduced into primary erythroid precursor cells (S0 populations, gated as in Figure 2A) isolated from E13.5 fetal livers derived from mice homozygous for the LCR deletion. While the loss of the LCR had no impact on basal level βh1-globin transcription it completely abrogated the induction of βh1-globin expression seen in the wild type mice (Figure 3B). As expected, β-major expression in the ΔLCR cells was low when compared with wild type cells (Figure 3C). Transcription of the α-globin gene that resides on a different chromosome was unaffected by the LCR deletion (Figure 3D). Adventitious activation of εy-globin expression was also abrogated in the absence of the LCR (Figure S3A), consistent with a model in which the LCR can form productive sporadic contacts with nearby genes (Noordermeer et al., 2011). Similar results were obtained using purified S1 cell populations (Figure S3B). In conclusion, βh1-SA mediated activation of βh1-globin transcription requires the LCR and by extension is dependent on chromatin looping. 3C experiments carried out in human erythroid cells further support the notion that zinc finger SA fusion proteins alter chromosome looping (see below).
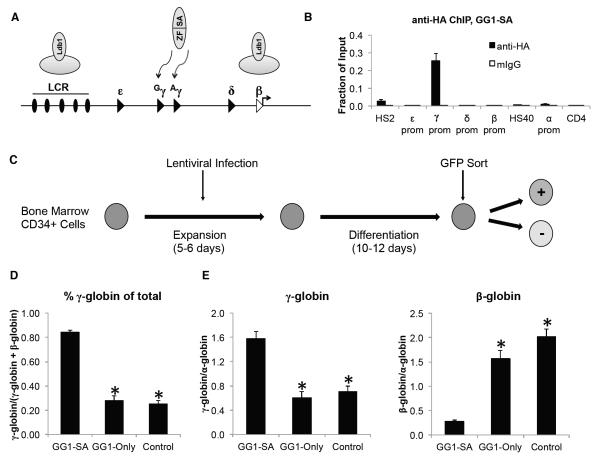
Zinc finger-SA mediated activation of γ-globin transcription in primary adult human erythroid cells
We next explored whether the strategy of forced chromatin looping could be employed to activate γ-globin expression in adult human erythroid cells (Figure 4A). To tether the Ldb1 SA domain to the two highly similar Gγ-globin and Aγ-globin genes, we fused it to a previously characterized artificial zinc finger protein (called GG1) that targets both promoters (Graslund et al., 2005). The fusion construct (GG1-SA) was HA-tagged and inserted upstream of an IRES-GFP cassette into a lentiviral vector containing the promoter of either the ankyrin or spectrin genes to drive erythroid-specific expression (Wilber et al., 2010). As a control, we used a construct containing just the γ-globin binding zinc finger protein (GG1-Only). To assess DNA binding specificity of GG1-SA in vivo, constructs were introduced into the human hematopoietic cell line K562 and analyzed by anti-HA ChIP. GG1-SA targeting was highly specific for the γ-globin promoters with little or no signal at the other globin promoters (Figure 4B).
Figure 4. Reactivation of the γ-globin gene in primary human erythroid cells by GG1-SA.
(A) Experimental concept. Note the presence of two fetal γ-globin genes (Gγ and Aγ) in humans. (B) Anti-HA ChIP analysis in K562 cells expressing GG1-SA. (C) Experimental outline. (D) Percentage of γ-globin and β-globin mRNA levels as determined by RT-qPCR. GG1-SA expression was driven by the ankyrin promoter and GG1-Only by the spectrin or ankyrin promoter. Control cells are sorted GFP negative cells. (E) Absolute γ-globin expression (left) and β-globin (right) relative to α-globin expression in adult erythroblasts. Error bars represent SEM. N = 15 for GG1-SA, 11 for GG1-Only, and 26 for control. Asterisks indicate statistically significant difference from GG1-SA by t-test. See also Figure S4.
To determine whether GG1-SA augments γ-globin expression in primary adult human erythroid cells we collected bone marrow-derived human CD34+ hematopoietic progenitor cells and differentiated them towards the erythroid lineage using a previously described two-phase liquid culture system involving an expansion and differentiation phase (Sankaran et al., 2008) (Figure 4C). Cells were infected with lentivirus containing GG1 constructs during the expansion phase and sorted for GFP expression on day 10-12 of erythroid differentiation. GFP negative cells served as controls. Sorting yielded pure populations of cells (Figure S4A) expressing comparable amounts of GG1-SA or GG1-Only mRNA (Figure S4B). Remarkably, GG1-SA expressing cells produced γ-globin approximating 85% of total globin synthesis (defined as γ-globin plus β-globin) compared with ~25% in GFP negative cells (Figure 4D).
Basal levels of γ-globin expression can rise upon exposure of human erythroblasts to in vitro culture conditions (Fibach et al., 1993). This is consistent with the human fetal genes being silenced less stringently compared to the murine embryonic globin genes (Sankaran et al., 2009). Nevertheless, GG1-SA-induced γ-globin levels that rival or exceed those achieved by pharmacological γ-globin inducers (Atweh and Fathallah, 2010; Bradner et al., 2010; Cao, 2004; Smith et al., 2000) or depletion of Bcl11a (Sankaran et al., 2008; Xu et al., 2013).
We observed considerable variation in basal γ-globin expression among donors ranging from <10% to 50% of total globin synthesis (Figure S4C). As erythroid cells undergo maturation, the ratio of fetal to adult hemoglobin declines. To ensure that our observed increases in fetal globin production are not confounded by variable rates of erythroid maturation due to cell culture and donor variations, and/or secondary to GG1-SA expression, we normalized γ-globin transcripts against α-globin mRNA as an indicator of erythroid differentiation (Pope et al., 2000). GG1-SA induced a ~2.5-fold increase in γ-globin expression compared to GG1-Only and control (sorted GFP negative cells) populations (Figure 4E, S4D). Importantly, GG1-SA expression resulted in a marked reduction in β-globin transcription (13.5% vs. control and 18.1% vs. GG1-Only) (Figure 4E and S4D). Since the sum of γ-globin and β-globin transcripts remained similar vis-à-vis α-globin mRNA levels, this further supports the idea that GG1-SA functions via competition for the LCR. Similar reciprocal changes in globin transcription upon GG1-SA expression were observed when mRNA levels were normalized to GAPDH (Figure S4E). Finally, GG1-SA expression measurably increased the expression of the surface marker CD235a (Figure S4F), suggesting that GG1-SA might enhance erythroid maturation. This prompted us to assess a panel of erythroid genes and observed a modest (≤ 2-fold) increase of a subset of these (Figure S4G), suggesting that the increased γ- to β-globin ratio is not due to GG1-SA-mediated alterations in erythroid differentiation. Together these results demonstrate that GG1-SA strongly induces the expression of γ-globin with resulting loss in β-globin transcription.
GG1-SA mediated induction of fetal hemoglobin in peripheral blood derived erythroid precursor cells
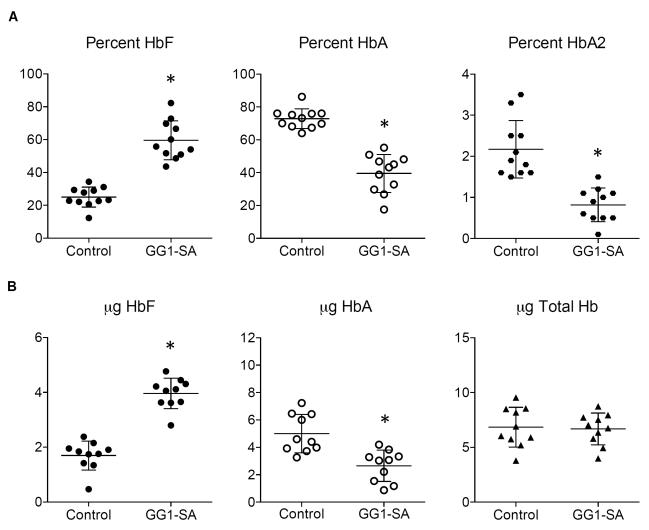
We next examined whether the GG1-SA-induced increase in γ-globin mRNA transcription is reflected in fetal hemoglobin production at the protein level. CD34+ progenitor cells isolated from peripheral blood were expanded, lentivirally infected, and differentiated towards the erythroid lineage as described (Breda et al., 2012). Hemoglobin profiles were determined by high-performance liquid chromatography (HPLC). GG1-SA expression led to a dramatic increase in the percentage of HbF compared to uninfected controls or GG1-Only (Figure 5A, S5A, and data not shown). Correspondingly, adult hemoglobin levels (HbA and HbA2) declined markedly. These results match those measuring γ-globin and β-globin mRNA levels from these populations (not shown). Similarly, we observed a dramatic rise in absolute HbF production (as measured in micrograms of protein) with a concomitant reduction in HbA levels (Figure 5B). This shift in hemoglobin production is likely an underestimate since GG1-expressing cells were not enriched by cell sorting in these experiments. Importantly, total hemoglobin production remained unchanged between controls and GG1-SA containing cells (Figure 5B), indicating that in these experiments GG1-SA expression did not significantly affect overall erythroid maturation.
Figure 5. GG1-SA activates HbF production in peripheral blood derived adult erythroid cells.
(A) Percent fetal hemoglobin (HbF, left) adult hemoglobin (HbA, middle), and hemoglobin delta (HbA2, right) in control peripheral blood derived erythroid cells versus GG1-SA containing cells as measured by HPLC. (B) Absolute level of HbF (left), HbA (middle), and total hemoglobin (right) as determined by HPLC. Data represent a total of 10 biological replicates from a total of three different healthy donors. Error bars represent SEM. Asterisks indicate statistically significant difference from control by t-test. See also Figure S5.
Finally, we followed the effects of GG1-SA at two stages of differentiation (day 4 and day 8) of peripheral blood-derived erythroid cells by analyzing globin mRNA levels and cell surface marker expression. At day 4 of erythroid differentiation GG1-SA strongly stimulated γ-globin transcription compared to controls with high levels being sustained at day 8 (Figure S5B). GG1-SA also attenuated β-globin transcription at both time points (Figure S5B). GG1-SA did not impair erythroid differentiation but slightly promoted it as measured by CD235a surface expression (Figure S5C), similar to the results obtained with bone marrow-derived cells. Taken together, these results demonstrate potent induction of HbF by GG1-SA at distinct stages of erythroid differentiation and independent of stem cell source.
Cellular distribution of γ-globin protein in adult erythroid cells containing GG1-SA
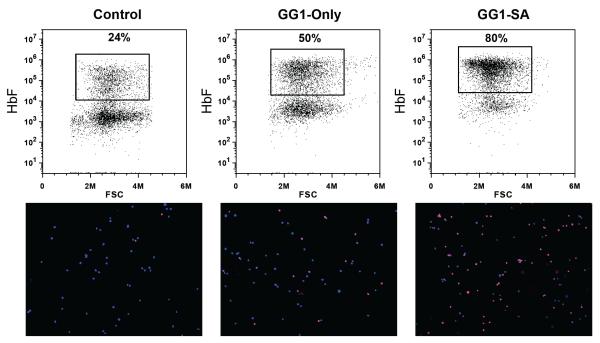
Fetal hemoglobin (HbF) production is heterocellular in normal individuals in whom a small proportion of cells with high level of HbF accounts for the overall low HbF levels (Boyer et al., 1975; Wood et al., 1975). We examined the cellular distribution of HbF in GG1-SA and GG1-Only expressing bone marrow-derived cultures by staining with an anti-HbF antibody followed by flow cytometry and immunofluorescence microscopy. GG1-SA resulted in nearly pancellular HbF expression whereas GG1-Only triggered expression merely in a subset of cells (Figure 6), consistent with the mRNA analyses. This observation further highlights the effectiveness of GG1-SA in stimulating fetal globin synthesis.
Figure 6. Cellular distribution of γ-globin protein in primary human erythroid cells.
Erythroid cells derived from peripheral blood CD34+ populations expressing indicated constructs stained with an anti-HbF antibody (red) and DAPI (blue), and analyzed by flow cytometry (upper). Cells analyzed by anti-HbF immunofluorescence microscopy (lower).
GG1-SA induces γ-globin promoter-LCR looping
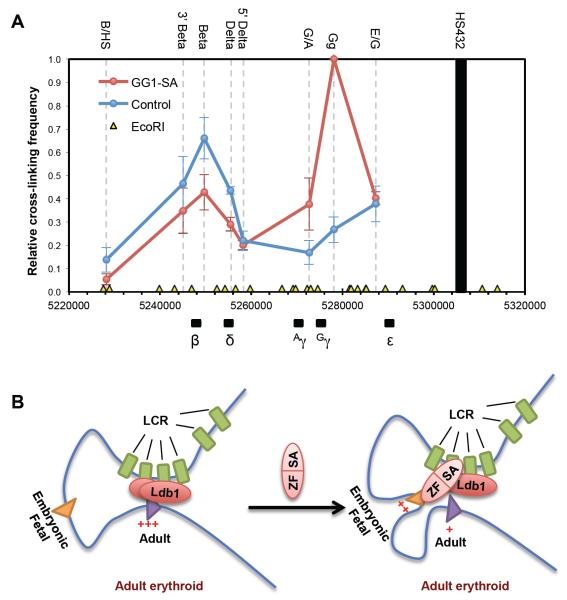
Several lines of evidence support the model that zinc finger-SA fusion proteins work via a looping mechanism, including their reciprocal effects on embryonic/fetal and adult globin expression, the dependence on the LCR for activity, and prior 3C studies demonstrating the ability of Ldb1 and its SA domain to trigger chromatin looping (Deng et al., 2012; Song et al., 2007). To directly assess the effects of GG1-SA expression on chromatin contacts within the β-globin locus, we carried out 3C experiments in adult erythroblasts. GG1-SA expression established a strong interaction between the LCR and the γ-globin genes that is absent in control cells (Figure 7). Please note that the primer pair (Gg) used for the two γ-globin promoters does not distinguish between these sequences, and the values represent the average normalized signals from both (Kiefer et al., 2011). The contact frequencies of the intervening restriction fragment (primer pair G/A) between the two γ-globin genes are also increased, albeit not as much as the γ-genes themselves. Additionally, interactions between the LCR and the adult globin genes (δ- and β-globin) were diminished, consistent with GG1-SA competitively reconfiguring looped contacts towards γ-globin promoter at the expense of the β-globin promoter. In light of the variation among donor samples leading to sizeable error bars, we plotted the individual 3C experiments showing essentially the same results in all samples (Figure S6A). Finally, results were robust towards different ways of normalizing the data (Figure S6B, and data not shown). In sum, tethering the SA portion of Ldb1 to the fetal globin promoters promotes their juxtaposition with the LCR leading to transcription activation with accompanying reduction in the expression of adult globin genes.
Figure 7. GG1-SA induces γ-globin promoter-LCR looping.
(A) 3C assay measuring locus-wide crosslinking frequencies in differentiated human erythroid cells expressing GG1-SA (red) or GFP negative control cells (blue). The EcoRI fragments containing HSs of the LCR (black bar) was used as the anchor region. Its cross-linking frequency with other indicated EcoRI fragments (names on the top of the graph) were assessed. The human β-globin genes are depicted on bottom of the graph, with chromosomal position coordinates. The EcoRI digestion sites are depicted as yellow triangles. Results were obtained using primary bone marrow-derived erythroid cells from three independent donors. 3C values were normalized to Tubulin, and the highest 3C value from the Gγ-containing fragment (Gg primer) in the GG1-SA sample, of each independent experiment was set as one. Error bars indicate SEM. N=3. (B) Model of controlling chromatin looping to reprogram the β-globin locus. See also Figure S6.
Discussion
Here, we reprogrammed a developmentally regulated gene locus by manipulating its chromatin organization. In adult erythroid cells, re-directing the LCR to the silenced embryonic or fetal β-type globin promoters by targeted tethering of the Ldb1 SA domain strongly activated transcription at the expense of the adult β-globin genes, thus at least partially reversing the developmental gene switch (Figure 7B). Our findings support that chromatin looping plays an essential role in controlling globin switching, and suggest a possible strategy for the treatment of those hemoglobinopathies that would benefit from the activation of alternative β-like globin genes. More broadly, they exemplify controlled manipulation of chromatin folding in the nucleus as a novel and powerful strategy to modulate, positively or negatively, the transcription of designated genes.
Central to the mechanism of globin gene switching is the exclusive interaction between the LCR and globin gene promoters at distinct developmental stages (Carter et al., 2002; Palstra et al., 2008; Tolhuis et al., 2002). In adult type murine erythroid cells, Ldb1 is required for the juxtaposition between the LCR and the adult β-major globin gene (Deng et al., 2012; Song et al., 2007). The present results indicate that the Ldb1 SA domain is also capable of forging a chromatin interaction between the LCR and globin genes that represent earlier stages of development, leading to marked increases in transcription.
Several lines of evidence strongly support loop formation as the underlying mechanism of transcriptional activation. First, deletion of the LCR essentially abrogated the effects of the tethered Ldb1 SA domain, indicating that it does not substitute for the LCR by simply enhancing promoter activity, but instead requires long-range LCR action. Second, increases in embryonic or fetal globin gene expression were accompanied by a reciprocal decrease in adult β-globin expression, consistent with competition for long-range LCR activity. Third, 3C experiments revealed that GG1-SA expression strongly stimulated γ-globin promoter-LCR interactions with a concomitant decrease in β-globin-LCR contacts. Finally, previous 3C experiments demonstrated the sufficiency of tethered Ldb1 for full reconstitution of an LCR-β-globin promoter loop in immature adult erythroid cells in which the globin locus is otherwise unlooped (Deng et al., 2012). Therefore, in aggregate these data strongly support that Ldb1 SA-induced transcriptional activation is a functional consequence of its impact on the higher order chromatin organization within the β-globin locus.
Prior studies involving transgenic mice suggested that genomic proximity to the LCR correlates with the ability to compete for LCR activity, presumably because closer genes have a higher probability of forging a productive interaction (Bauchwitz and Costantini, 2000; Hanscombe et al., 1991; Peterson and Stamatoyannopoulos, 1993; Tanimoto et al., 1999). We observed that βh1-SA not only activated βh1-globin expression but also to some degree the expression of εy-globin, which resides closer to the LCR than βh1-globin. Similar to βh1-globin, εy-globin up-regulation was dependent on the LCR. Therefore, the newly imposed spatial constraint of the LCR also increases the likelihood for interaction with a nearby gene. This lends further support to the idea that spatial proximity is a key determinant of enhancer activity.
Since numerous protein complexes orchestrate the developmental switch in β-globin gene expression (Sankaran et al., 2010), it came as a surprise that a single molecule could exert such strong activating effects on the embryonic or fetal globin genes in adult erythroid cells. This indicates that direct recruitment of the active LCR to the γ-globin genes can effectively override its developmental repression. As a corollary, it is plausible that regulation of chromatin looping might also underlie the function of factors normally involved in globin gene switching. Indeed, Bcl11a, a potent silencing factor of the γ-globin genes in human cells, does not detectably bind to the γ-globin gene promoters but appears to regulate the looped configuration of the locus (Xu et al., 2010). Future studies analyzing the effects of forced LCR-γ-globin looping on the various postulated γ-globin repression mechanisms are expected to provide additional insights into the control of normal developmental globin switching.
Tethering the SA domain did not activate murine embryonic genes to the same level as the human fetal genes. This is consistent with a more stringent developmental repression within the murine environment (Sankaran et al., 2009). Exposure of developing human erythroid cells to in vitro culture conditions variably elevates basal fetal globin gene expression, suggesting that the human γ-globin genes are in a poised and therefore more responsive state when compared to the murine embryonic genes. (Fibach et al., 1993). Hence GG1-SA might act in an environment that is more favorable for further activation. It is also possible that the chromatin state at the fetal globin genes facilitates more efficient access of GG1-SA to its target site. However, even under the most restrictive conditions as are found in murine cells, the silent βh1-gene promoter remains accessible to βh1-SA and allows for activation of βh1-globin expression up to ~30 % of total β-globin synthesis. In fact, such levels, if achieved at the fetal globin genes in vivo in human sickle cell anemia patients would be sufficient to ameliorate the clinical course of their disease (see also below) (Noguchi et al., 1988; Platt et al., 1991). If desired, means to further increase the efficiency of embryonic or fetal globin transcription induction could be conceived. Improvements in ZF design, or the use of multiple ZF proteins could potentially enhance the specificity and efficiency to raise the competitiveness of the targeted gene for enhancer activity. Alternative approaches for recruiting the SA domain such as through TALEs or catalytically inactive CRISPR-Cas9 systems could also be tested (Perez-Pinera et al., 2013; Sun et al., 2012).
Currently, numerous strategies involving molecular genetic tools are being pursued with the goal to ameliorate sickle cell anemia. These include the transgenic expression of γ-globin or anti-sickling forms of β-globin and genome editing to repair the sickle cell mutation or to diminish the expression of Bcl11a (Chandrakasan and Malik, 2014; Bauer et al., 2013; Hardison and Blobel, 2013). These approaches, including the one presented here have specific strengths but also weaknesses that must be considered prior to their potential use in human patients. Several insights from our studies are relevant when considering a forced looping approach for therapeutic intervention. First, increases in fetal γ-globin expression were accompanied by reduced adult β-globin expression. This agrees with the notion that the LCR is incapable of interacting with multiple globin promoters at once but instead contacts either the fetal or adult genes (Enver et al., 1990; Wijgerde et al., 1995). The reduction in adult β-globin expression might be an added therapeutic benefit since diminished synthesis of defective β-globin chains, as occurs in sickle cell anemia, would further improve the ratio of γ-globin chains available to pair with the α-globin chains. Second, in normal adults, total fetal hemoglobin synthesis ranges below 3% but is concentrated in a small fraction of cells with high fetal hemoglobin expression (F-cells). However, GG1-SA expression raised fetal globin expression in the vast majority of transduced cells. Therefore, it is expected that in a therapeutic setting fewer dysfunctional erythroid cells would remain to contribute to disease. Finally, marked effects in fetal globin reactivation were achieved with relatively low levels of GG1-SA transgene expression (<1% of globin genes), which may be a more easily achievable and sustainable compared to what would be required if expressing a γ-globin transgene.
In summary, we have shown that forced long-range chromatin interactions can be used to reactivate a developmentally silenced gene. When employed at a complex gene locus at which promoters compete for enhancer activity this strategy can lead to reciprocal reduction in the expression of alternate genes. Therefore, manipulation of chromatin looping can in principle be applied for gene activation or repression at any gene that is controlled by a looped distal enhancer. Refinements in targeted tethering technologies and the exploration of additional nuclear factors involved in chromatin looping are expected to increase the appeal of this technology for a wider range of applications.
EXPERIMENTAL PROCEDURES
Artificial Zinc Finger Design
Custom ZF proteins each containing six tandem Cys2-His2 domains were designed to target 18-19 base pairs at the βh1-globin promoter. ZFs were assembled from two-finger units and screened for DNA binding by ELISA assay in vitro as described (Bartsevich et al., 2003). Chosen target sequences were within the DNase I hypersensitive region to facilitate access but avoid interference with known transcription factor binding sites. Five ZFs were FLAG tagged and screened by anti-FLAG ChIP for in vivo binding to the βh1 promoter in G1E cells. Two of these produced sufficiently high ChIP signals with the desired specificity. The better one two with the target sequence AAGGGGAGCAAGGTCCAG was used for experiments in this report. The lesser one also activated βh1 expression albeit to a lower degree (not shown). The ZFs targeting HS2 of the mouse LCR and the human γ-globin promoters have been described (Deng et al., 2012) (Graslund et al., 2005).
Constructs
The βh1-globin zinc finger cDNA was cloned into MigR1 retroviral vector with three Flag tags and a nuclear localization signal (NLS) at its N-terminus. The SA domain containing amino acids 1-200 of Ldb1 was inserted C-terminal to the ZF. The lentiviral vectors pCL20 expressing the GG1 from the spectrin or ankyrin promoter were gifts from Andrew Wilber (Wilber et al., 2010). The SA domain was attached C-terminal to GG1 tagged with HA. GG1-Only contains GG1 with an attached HA-tag.
Cell Culture
G1E cells were cultured as described (Weiss et al., 1997). G1E cells containing GATA1-ER were treated with 100 nM estradiol for indicated periods of time (denoted as G1E+GATA1). K562 cells were grown in RPMI medium supplemented with 10% FBS serum and 1% penicillin-streptomycin.
Mouse Primary Erythroblasts
Fetal liver erythroid cells were isolated from strain-matched E13.5 wild type or ΔLCR/ΔLCR embryos (129 strain) (Bender et al., 2000), according to protocol 2012-7-660. Single cells suspensions were reacted with PE-conjugated anti-CD71 and APC-conjugated anti-Ter119 antibodies and sorted by FACS. S0 and S1 populations (Figure 2A) were infected with retrovirus, and cultured for 43-48 h in medium containing Iscove’s MDM with 15% fetal bovine serum, 1% penicillin-streptomycin, 1% glutamine, 10 ng/mL mIL3, 20 ng/mL mIL6, 50 ng/mL mSCF, 10 ng/mL mFLT3L, 0.5 U/mL erythropoietin (Amgen) and 1 μM dexamethasone (Sigma).
Two Phase Differentiation of Bone Marrow Derived Human CD34+ Cells
Human CD34+ cells were obtained from de-identified healthy bone marrow donors after informed consent by the University of Pennsylvania Stem Cell Core. Cells were expanded in StemSpan SFEM medium with 1X StemSpan CC100 cytokine cocktail (STEMCELL Technologies) and 1% penicillin-streptomycin, followed by differentiation in medium consisting of SFEM medium, 1U/mL erythropoietin, 5ng/mL IL3, 20ng/mL SCF, 1μM estradiol, and 2μM dexamethasone. On day 10-12 of differentiation GFP+ cells were sorted on a MoFlo XDP (Beckman Coulter) or an Arial (BD Biosciences) cell sorter. Cell purity was confirmed by post sort flow cytometry.
Two Phase Differentiation of Peripheral Blood Derived Human CD34+ Cells
Peripheral blood was obtained from healthy donors after obtaining written consent, according to protocol 05070077971 from the Weill Cornell Medical College Institutional Review Board. The mononuclear fraction from a Ficoll-Hypaque density gradient was enriched for CD34+ cells by immunomagnetic separation using the CD34 microbeads kit (Miltenyi Biotec Inc., Auburn, CA). Expansion and differentiation were achieved using a protocol as described (Breda et al., 2012). Briefly, cells were expanded in StemSpan SFEM medium, 1X StemSpan CC100 cytokine cocktail, 2U/mL Erythropoietin, 1 μM dexamethasone (Sigma) and 1% penicillin-streptomycin. Differentiation was induced in StemSpan SFEM medium, 10ng/mL SCF, 10 μM β-mercaptoethanol, 10 U/mL erythropoietin and 1% penicillin-streptomycin.
Viral Infections
Retroviral infections of G1E cells have been described (Tripic et al., 2009). For primary fetal liver cells, spin-infection conditions were modified to 2,000 rpm at room temperature for 1 hour with appropriate cytokines added, and cells were switched to fresh medium immediately after infection. Lentivirus was concentrated using Peg-it Virus Precipitation Solution (System Biosciences) or ultracentrifugation. Primary human cells were infected on two consecutive days starting at the second or third day of the expansion phase with freshly prepared virus. Cells were spin-infected at 2,250 rpm at room temperature for 1.5 hours with appropriate cytokines. K562 cells were spin-infected once using the same conditions.
Chromatin immunoprecipitation
Anti-HA (clone 12CA5) and anti-Flag (clone M2, F3165, Sigma) ChIP was performed as described (Tripic et al., 2009) using primers listed in the supplement.
RT-qPCR
RNA from 105-106 cells was extracted with Trizol (Invitrogen). Reverse transcription reactions were performed with random hexamers using Superscript II (Invitrogen) or iScript (Bio-Rad). cDNA samples were quantified by SybrGreen qPCR.
Immunostaining
Human primary cells were stained with an APC-conjugated fetal hemoglobin antibody (Invitrogen, MHFH05) using the BD Cytofix/Cytoperm kit (BD Biosciences) per manufacturers protocol. Surface staining was performed with PE-labeled antiCD71 and APC-labeled antiCD235a (BD Biosciences).
HPLC Analyses
Hemolysis of cells was achieved in ice water for 30 minutes and two cycles of freeze thawing. Hemoglobin content and quality were analyzed by HPLC as described (Ou and Rognerud, 1993), using a PolyCAT A column (cat # 3.54CT0315; 3μ, 1500Å PolyLC Inc., MD) on a System Gold 126 Solvent Module instrument (Beckman Coulter, Fullerton, CA). Hemoglobins were measured at a wavelength of 415 nm and compared to hemoglobin standards with known concentration (Analytical Control System).
Chromosome Conformation Capture (3C)
3C assays were performed as described (Hagège et al., 2007) with the following modifications. 2×106 cells were crosslinked with 1% formaldehyde at room temperature for 10 min, followed by glycine quenching, cell lysis, EcoRI digestion, and T4 ligation. 3C ligation products were quantified in triplicates by quantitative SYBR Green real-time PCR as described (Kiefer et al., 2011). The 3C signals were normalized to template standards generated from a locus-specific BAC and to 3C signals from the tubulin locus. To eliminate variability between donor samples, interaction frequencies between the anchor fragment and the fragment encompassing the Gγ-globin gene (Figure 7A) or the β-globin (Figure S6B) from the GG1-SA sample of each donor were set to one. Primer sequences are listed in (Hou et al., 2010; Kiefer et al., 2011) and in the supplement.
Supplementary Material
Research Highlights.
Tethering Ldb1 to embryonic or fetal globin genes activates their expression in adult erythroblasts.
Activation of embryonic/fetal globin genes is associated with reduced adult globin expression.
Tethered Ldb1 promotes looped contacts with the fetal enhancers at the expense of the adult ones.
Forced chromatin looping presents a novel therapeutic strategy for sickle cell anemia.
Acknowledgements
We thank Andrew Wilber for the GG1 constructs. We are grateful to Hongxin Wang for technical assistance, to Jongjoo Lee for help with mouse experiments, and members of the laboratory for valuable discussions. This work was supported by NIH grants 5R37DK058044 and 1RO1HL119479 to G.A.B. J.W.R. was supported by an American Heart Association postdoctoral fellowship 13POST16950014. A.D. and I.K. were supported by the Intramural program of the NIDDK, NIH (DK015508) P.D.G., and A.R. were employees of Sangamo Biosciences Inc. when involved in this work. Additional support was provided by the Associazione Veneta per la Lotta alla Talassemia (AVLT, IT) to L.B., NIH grant NHLBI-5R01HL102449 to S.R., and the European Community grant FP7-HEALTH-2012-INNOVATION to L.B. and S.R.
Footnotes
SUPPLEMENTAL INFORMATION Supplemental figures and experimental materials are available online.
Author contributions W.D., J.W.R. S.R., A.D. and G.A.B. conceived the study, designed experiments and wrote the paper; W.D., J.W.R., I.K., L.B., I.M. and K.J. performed experiments; G.A.B., A.R. and P.D.G. designed and supervised ZF assembly and in vitro characterization.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adams CC, Workman JL. Binding of disparate transcriptional activators to nucleosomal DNA is inherently cooperative. Molecular and Cellular Biology. 1995;15:1405–1421. doi: 10.1128/mcb.15.3.1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atweh G, Fathallah H. Pharmacologic induction of fetal hemoglobin production. Hematol. Oncol. Clin. North Am. 2010;24:1131–1144. doi: 10.1016/j.hoc.2010.08.001. [DOI] [PubMed] [Google Scholar]
- Bartsevich VV, Miller JC, Case CC, Pabo CO. Engineered Zinc Finger Proteins for Controlling Stem Cell Fate. Stem Cells. 2003;21:632–637. doi: 10.1634/stemcells.21-6-632. [DOI] [PubMed] [Google Scholar]
- Bauchwitz R, Costantini F. Developmentally distinct effects on human ε-, γ-and Δ-globin levels caused by the absence or altered position of the human β-globin gene in YAC transgenic mice. Human Molecular Genetics. 2000;9:561–574. doi: 10.1093/hmg/9.4.561. [DOI] [PubMed] [Google Scholar]
- Bauer DE, Kamran SC, Lessard S, Xu J, Fujiwara Y, Lin C, Shao Z, Canver MC, Smith EC, Pinello L, et al. An Erythroid Enhancer of BCL11A Subject to Genetic Variation Determines Fetal Hemoglobin Level. Science. 2013;342:253–257. doi: 10.1126/science.1242088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bender MA, Bulger M, Close J, Groudine M. β- globin Gene Switching and DNase I Sensitivity of the Endogenous β- globin Locus in Mice Do Not Require the Locus Control Region. Molecular Cell. 2000;5:387–393. doi: 10.1016/s1097-2765(00)80433-5. [DOI] [PubMed] [Google Scholar]
- Boyer SH, Belding TK, Margolet L, Noyes AN. Fetal Hemoglobin Restriction to a Few Erythrocytes (F Cells) in Normal Human Adults. New Series. 1975;188:361–363. doi: 10.1126/science.804182. [DOI] [PubMed] [Google Scholar]
- Bradner JE, Mak R, Tanguturi SK, Mazitschek R, Haggarty SJ, Ross K, Chang CY, Bosco J, West N, Morse E, et al. Chemical genetic strategy identifies histone deacetylase 1 (HDAC1) and HDAC2 as therapeutic targets in sickle cell disease. Proceedings of the National Academy of Sciences. 2010;107:12617–12622. doi: 10.1073/pnas.1006774107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breda L, Casu C, Gardenghi S, Bianchi N, Cartegni L, Narla M, Yazdanbakhsh K, Musso M, Manwani D, Little J, et al. Therapeutic Hemoglobin Levels after Gene Transfer in β-Thalassemia Mice and in Hematopoietic Cells of β-Thalassemia and Sickle Cells Disease Patients. PLoS ONE. 2012;7:e32345. doi: 10.1371/journal.pone.0032345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao H. Induction of human γ-globin gene expression by histone deacetylase inhibitors. Blood. 2004;103:701–709. doi: 10.1182/blood-2003-02-0478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter D, Chakalova L, Osborne CS, Dai Y-F, Fraser P. Long-range chromatin regulatory interactions in vivo. Nat Genet. 2002;32:623–626. doi: 10.1038/ng1051. [DOI] [PubMed] [Google Scholar]
- Chandrakasan S, Malik P. Gene Therapy for Hemoglobinopathies. Hematol. Oncol. Clin. North Am. 2014;28:199–216. doi: 10.1016/j.hoc.2013.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng W, Lee J, Wang H, Miller J, Reik A, Gregory PD, Dean A, Blobel GA. Controlling Long-Range Genomic Interactions at a Native Locus by Targeted Tethering of a Looping Factor. Cell. 2012;149:1233–1244. doi: 10.1016/j.cell.2012.03.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drissen R. The active spatial organization of the β-globin locus requires the transcription factor EKLF. Genes & Development. 2004;18:2485–2490. doi: 10.1101/gad.317004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enver T, Raich N, Ebens AJ, Papayannopoulou T, Costantini F, Stamatoyannopoulos G. Developmental regulation of human fetal-to-adult globin gene switching in transgenic mice. Nature. 1990;344:309–313. doi: 10.1038/344309a0. [DOI] [PubMed] [Google Scholar]
- Fibach E, Burke KP, Schechter AN, Noguchi CT, Rodgers GP. Hydroxyurea increases fetal hemoglobin in cultured erythroid cells derived from normal individuals and patients with sickle cell anemia or beta-thalassemia. Blood. 1993;81:1630–1635. [PubMed] [Google Scholar]
- Graslund T, Li X, Magnenat L, Popkov M, Barbas C. Exploring Strategies for the Design of Artificial Transcription Factors. Journal of Biological Chemistry. 2005;280:3707–3714. doi: 10.1074/jbc.M406809200. [DOI] [PubMed] [Google Scholar]
- Hagège H, Klous P, Braem C, Splinter E, Dekker J, Cathala G, de Laat W, Forné T. Quantitative analysis of chromosome conformation capture assays (3C-qPCR) Nat Protoc. 2007;2:1722–1733. doi: 10.1038/nprot.2007.243. [DOI] [PubMed] [Google Scholar]
- Hanscombe O, Whyatt D, Fraser P, Yannoutsos N, Greaves D, Dillon N, Grosveld F. Importance of globin gene order for correct developmental expression. Genes & Development. 1991;5:1387–1394. doi: 10.1101/gad.5.8.1387. [DOI] [PubMed] [Google Scholar]
- Hardison RC, Blobel GA. GWAS to Therapy by Genome Edits? Science. 2013;342:206–207. doi: 10.1126/science.1245813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou C, Dale R, Dean A. Cell type specificity of chromatin organization mediated by CTCF and cohesin. Proceedings of the National Academy of Sciences. 2010;107:3651–3656. doi: 10.1073/pnas.0912087107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiefer CM, Lee J, Hou C, Dale RK, Lee YT, Meier ER, Miller JL, Dean A. Distinct Ldb1/NLI complexes orchestrate γ-globin repression and reactivation through ETO2 in human adult erythroid cells. Blood. 2011;118:6200–6208. doi: 10.1182/blood-2011-06-363101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noguchi CT, Rodgers GP, Serjeant G, Schechter AN. Levels of Fetal Hemoglobin Necessary for Treatment of Sickle Cell Disease. N. Engl. J. Med. 1988;318:96–99. doi: 10.1056/NEJM198801143180207. [DOI] [PubMed] [Google Scholar]
- Noordermeer D, de Wit E, Klous P, van de Werken H, Simonis M, Lopez-Jones M, Eussen B, de Klein A, Singer RH, de Laat W. Variegated gene expression caused by cell-specific long-range DNA interactions. Nat Cell Biol. 2011;13:944–951. doi: 10.1038/ncb2278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou CN, Rognerud CL. Rapid analysis of hemoglobin variants by cation-exchange HPLC. Clin. Chem. 1993;39:820–824. [PubMed] [Google Scholar]
- Palstra R-J, de Laat W, Grosveld F. Beta-globin regulation and long-range interactions. Adv. Genet. 2008;61:107–142. doi: 10.1016/S0065-2660(07)00004-1. [DOI] [PubMed] [Google Scholar]
- Palstra R-J, Tolhuis B, Splinter E, Nijmeijer R, Grosveld F, de Laat W. The β-globin nuclear compartment in development and erythroid differentiation. Nat Genet. 2003;35:190–194. doi: 10.1038/ng1244. [DOI] [PubMed] [Google Scholar]
- Perez-Pinera P, Kocak DD, Vockley CM, Adler AF, Kabadi AM, Polstein LR, Thakore PI, Glass KA, Ousterout DG, Leong KW, et al. RNA-guided gene activation by CRISPR-Cas9-based transcription factors. Nature Methods. 2013;10:973–976. doi: 10.1038/nmeth.2600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson KR, Stamatoyannopoulos G. Role of gene order in developmental control of human gamma- and beta-globin gene expression. Molecular and Cellular Biology. 1993;13:4836–4843. doi: 10.1128/mcb.13.8.4836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platt OS. Hydroxyurea for the treatment of sickle cell anemia. N. Engl. J. Med. 2008;358:1362–1369. doi: 10.1056/NEJMct0708272. [DOI] [PubMed] [Google Scholar]
- Platt OS, Brambilla DJ, Rosse WF, Milner PF, Castro O, Steinberg MH, Klug PP. Mortality in sickle cell disease. Life expectancy and risk factors for early death. N. Engl. J. Med. 1994;330:1639–1644. doi: 10.1056/NEJM199406093302303. [DOI] [PubMed] [Google Scholar]
- Platt OS, Thorington BD, Brambilla DJ, Milner PF, Rosse WF, Vichinsky E, Kinney TR. Pain in sickle cell disease. N. Engl. J. Med. 1991;325:11–16. doi: 10.1056/NEJM199107043250103. [DOI] [PubMed] [Google Scholar]
- Pop R, Shearstone JR, Shen Q, Liu Y, Hallstrom K, Koulnis M, Gribau J, Socolovsky M. A Key Commitment Step in Erythropoiesis Is Synchronized with the Cell Cycle Clock through Mutual Inhibition between PU.1 and S-Phase Progression. PLoS Biol. 2010;8 doi: 10.1371/journal.pbio.1000484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pope SH, Fibach E, Sun J, Chin K, Rodgers GP. Two-phase liquid culture system models normal human adult erythropoiesis at the molecular level. Eur J Haematol. 2000;64:292–303. doi: 10.1034/j.1600-0609.2000.90032.x. [DOI] [PubMed] [Google Scholar]
- Sankaran VG, Menne TF, Xu J, Akie TE, Lettre G, Van Handel B, Mikkola HKA, Hirschhorn JN, Cantor AB, Orkin SH. Human Fetal Hemoglobin Expression Is Regulated by the Developmental Stage-Specific Repressor BCL11A. Science. 2008;322:1839–1842. doi: 10.1126/science.1165409. [DOI] [PubMed] [Google Scholar]
- Sankaran VG, Xu J, Orkin SH. Advances in the understanding of haemoglobin switching. Br J Haematol. 2010;149:181–194. doi: 10.1111/j.1365-2141.2010.08105.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sankaran VG, Xu J, Ragoczy T, Ippolito GC, Walkley CR, Maika SD, Fujiwara Y, Ito M, Groudine M, Bender MA, et al. Developmental and species-divergent globin switching are driven by BCL11A. Nature. 2009;460:1093–1097. doi: 10.1038/nature08243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith RD, Li J, Noguchi CT, Schechter AN. Quantitative PCR analysis of HbF inducers in primary human adult erythroid cells. Blood. 2000;95:863–869. [PubMed] [Google Scholar]
- Song S-H, Hou C, Dean A. A Positive Role for NLI/Ldb1 in Long-Range β-Globin Locus Control Region Function. Molecular Cell. 2007;28:810–822. doi: 10.1016/j.molcel.2007.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun N, Liang J, Abil Z, Zhao H. Optimized TAL effector nucleases (TALENs) for use in treatment of sickle cell disease. Molecular BioSystems. 2012;8:1255–1263. doi: 10.1039/c2mb05461b. [DOI] [PubMed] [Google Scholar]
- Tanimoto K, Liu Q, Bungert J, Engel JD. Effects of altered gene order or orientation of the locus control region on human β-globin gene expression in mice. Nature. 1999;398:344–348. doi: 10.1038/18698. [DOI] [PubMed] [Google Scholar]
- Tolhuis B, Palstra R-J, Splinter E, Grosveld F, de Laat W. Looping and interaction between hypersensitive sites in the active beta-globin locus. Molecular Cell. 2002;10:1453–1465. doi: 10.1016/s1097-2765(02)00781-5. [DOI] [PubMed] [Google Scholar]
- Tripic T, Deng W, Cheng Y, Zhang Y, Vakoc CR, Gregory GD, Hardison RC, Blobel GA. SCL and associated proteins distinguish active from repressive GATA transcription factor complexes. Blood. 2009;113:2191–2201. doi: 10.1182/blood-2008-07-169417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vakoc CR, Letting DL, Gheldof N, Sawado T, Bender M, Groudine M, Weiss MJ, Dekker J, Blobel GA. Proximity among distant regulatory elements at the β-globin locus requires GATA-1 and FOG-1. Molecular Cell. 2005;17:453–462. doi: 10.1016/j.molcel.2004.12.028. [DOI] [PubMed] [Google Scholar]
- Weatherall DJ. Phenotype[mdash]genotype relationships in monogenic disease: lessons from the thalassaemias : Article : Nature Reviews Genetics. Nat. Rev. Genet. 2001;2:245–255. doi: 10.1038/35066048. [DOI] [PubMed] [Google Scholar]
- Weiss MJ, Yu C, Orkin SH. Erythroid-cell-specific properties of transcription factor GATA-1 revealed by phenotypic rescue of a gene-targeted cell line. Molecular and Cellular Biology. 1997;17:1642–1651. doi: 10.1128/mcb.17.3.1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch JJ, Watts JA, Vakoc CR, Yao Y, Wang H, Hardison RC, Blobel GA, Chodosh LA, Weiss MJ. Global regulation of erythroid gene expression by transcription factor GATA-1. Blood. 2004;104:3136–3147. doi: 10.1182/blood-2004-04-1603. [DOI] [PubMed] [Google Scholar]
- Wijgerde M, Grosveld F, Fraser P. Transcription complex stability and chromatin dynamics in vivo. Nature. 1995;377:209–213. doi: 10.1038/377209a0. [DOI] [PubMed] [Google Scholar]
- Wilber A, Tschulena U, Hargrove PW, Kim YS, Persons DA, Barbas CF, Nienhuis AW. A zinc-finger transcriptional activator designed to interact with the γ-globin gene promoters enhances fetal hemoglobin production in primary human adult erythroblasts. Blood. 2010;115:3033–3041. doi: 10.1182/blood-2009-08-240556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood WG, Stamatoyannopoulos G, Lim G, Nute PE. F-cells in the adult: normal values and levels in individuals with hereditary and acquired elevations of Hb F. Blood. 1975;46:671–682. [PubMed] [Google Scholar]
- Xu J, Sankaran VG, Ni M, Menne TF, Puram RV, Kim W, Orkin SH. Transcriptional silencing of γ-globin by BCL11A involves long-range interactions and cooperation with SOX6. Genes & Development. 2010;24:783–798. doi: 10.1101/gad.1897310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Bauer DE, Kerenyi MA, Vo TD, Hou S, Hsu Y-J, Yao H, Trowbridge JJ, Mandel G, Orkin SH. Corepressor-dependent silencing of fetal hemoglobin expression by BCL11A. Proc. Natl. Acad. Sci. U.S.a. 2013;110:6518–6523. doi: 10.1073/pnas.1303976110. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.