SUMMARY
Acquisition of the intestinal microbiota begins at birth and a stable microbial community develops from a succession of key organisms. Disruption of the microbiota during maturation by low-dose antibiotic exposure can alter host metabolism and adiposity. We now show that low-dose penicillin (LDP), delivered from birth, induces metabolic alterations and affects ileal expression of genes involved in immunity. LDP that is limited to early life transiently perturbs the microbiota, which is sufficient to induce sustained effects on body composition, indicating that microbiota interactions in infancy may be critical determinants of long-term host metabolic effects. In addition, LDP enhances the effect of high-fat diet induced obesity. The growth promotion phenotype is transferrable to germ-free hosts by LDP-selected microbiota, showing that the altered microbiota, not antibiotics per se, play a causal role. These studies characterize important variables in early-life microbe-host metabolic interaction, and identify several taxa consistently linked with metabolic alterations.
INTRODUCTION
Obesity, a complex disease, can increase the risk of diabetes, heart disease, and cancer (Vucenik and Stains, 2012). Along with dietary excess and genetic polymorphisms, the trillions of microbial cells in the intestinal microbiota can contribute to obesity by increasing energy extraction (Turnbaugh et al., 2006), or by altering metabolic signaling (Samuel et al., 2008) and inflammation (Cani et al., 2008; Henao-Mejia et al., 2012; Vijay-Kumar et al., 2010). Obese and lean humans differ in microbiota compositions and these phenotypes can be transferred to germ-free mice (Ley et al., 2005; Ridaura et al., 2013), highlighting the need for greater understanding of microbiota-host metabolic interactions.
Since early-life is a critical period for metabolic development (Cunningham et al., 2014; Dietz, 1994; Knittle and Hirsch, 1968), microbiota disruption during this window could lead to changes in body composition. In humans, early-life microbiota disruption, either due to delivery by Caesarian-section (Dominguez-Bello et al., 2010) or antibiotics, is associated with increased risk of overweight status later in childhood (Ajslev et al., 2011; Blustein et al., 2013; Huh et al., 2012; Murphy et al., 2013; Trasande et al., 2013); although in mice, antibiotic exposure can increase or decrease weight, depending on the dose, diet used, or strain (Dubos et al., 1963).
For decades, farmers have been exposing livestock to low doses of antibiotics to promote growth; the earlier in life that exposure begins, the more profound the effects (Cromwell, 2002; FDA, 2014). We have applied the agricultural model of growth promotion to interrogate fundamental host-microbe metabolic relationships. We previously showed that early-life sub-therapeutic antibiotic treatment increased fat mass, altered metabolic hormones, hepatic metabolism, and microbiota composition (Cho et al., 2012). Here, we extend these studies using low-dose penicillin (LDP) as a model agent disrupting the microbiota. We examined whether timing of LDP exposure is critical, whether synergies exist between penicillin- and dietary effects, and whether the altered microbiota is sufficient to yield metabolic phenotypes. Our experiments answered each of these questions in the affirmative. Remarkably, we found that microbial communities recovered after the cessation of antibiotics, yet the metabolic phenotypes persisted, highlighting the importance of the early-life microbiota in growth and development.
RESULTS
Effect of LDP commencement age on growth phenotypes
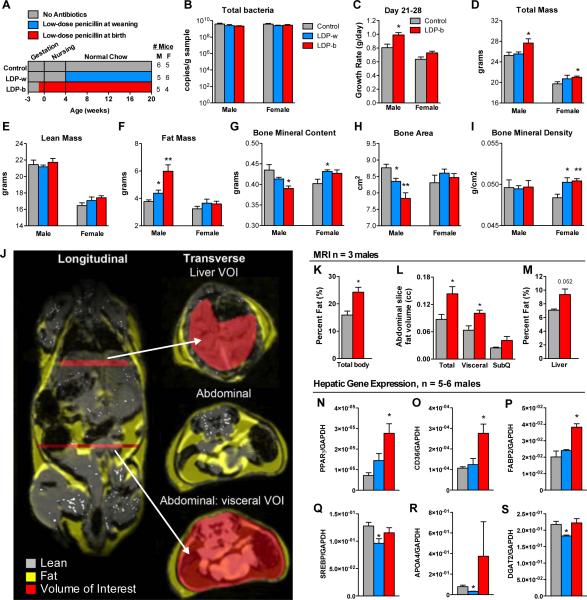
Introducing low-dose penicillin (LDP), chlortetracycline, or vancomycin to young mice at weaning increased subsequent fat mass (Cho et al., 2012). We now asked whether earlier exposure might have more substantial effects. C57BL/6J mice were exposed to LDP post-weaning (LDP-w), or their mother received LDP shortly before their pups’ birth (LDP-b) and through weaning, so that the pups would be initially colonized with an altered maternal microbiota and receive LDP while nursing. Control mice received no antibiotic (Figure 1A). LDP did not reduce microbial population size (Figure 1B), consistent with previous findings (Cho et al., 2012). Prior to weaning, male LDP-b mice had accelerated growth (Figure 1C), and adult male LDP-b mice had increased total and fat mass and decreased bone mineral content compared to controls (Figure 1D-I). LDP-w male mice showed similar trends, however, later LDP exposure had lesser effects on body composition. Female LDP-b mice, but not LDP-w, had significantly elevated total mass. Both female LDP groups had significantly elevated bone mineral density. Similar to DEXA, MRI showed increased body fat percentage in LDP-b male mice (Figure 1J-M), and also identified LDP-induced ectopic fat deposition including elevated total abdominal, visceral, and liver adiposity, which is linked with metabolic syndrome (Blüher, 2010; Rasouli et al., 2007). Associated with the trend of increased liver adiposity detected by MRI, LDP-b significantly increased hepatic expression of genes involved in adipogenesis, including PPARγ, CD36, and fatty acid binding protein 2 (FABP2) (Figure 1N-P), consistent with our prior observations (Cho et al., 2012). Contrastingly, the LDP-w mice had reduced expression of genes involved in cholesterol metabolism and fatty acid synthesis (Figure 1Q-S) compared to controls. This dichotomy may reflect the differences in phenotype development, or that down-regulation of genes involved in lipid metabolism in the LDP-w mice explains why these mice resist total and fat mass accumulation. In total, earlier penicillin exposure led to enhanced metabolic phenotypes, suggesting greater host vulnerability to microbiota disruption in infancy, with more pronounced effects in male mice.
Figure 1. Effect of timing of initial LDP exposure on body composition.
(A) Study design: C57B/L6J mice received low dose (1 μg/g body weight) penicillin (LDP) continuously for life, either beginning at weaning at day 28 of life (LDP-w, n = 5 male, 6 female), or from birth (LDP-b, n = 5 male, 4 female). Control mice had no penicillin exposure (n = 6 male, 5 female). (B) Levels of cecal bacteria at week 20 determined by quantitative PCR using universal bacterial 16S rRNA primers. (C) Early-life (peri-weaning: day 21 to day 28) growth rate. (D-I) Body composition and bone histomorphometry determined at week 20, by DEXA scanning. (J) MRI representation of lean (gray) and fat (yellow) tissue and regions that were analyzed for hepatic and visceral fat (red). (K) Adiposity (total body fat%) determined by MRI in three male mice each in LDP-b and control groups at week 18. (L) Total abdominal, visceral, and subcutaneous fat volume, and (M) liver adiposity (fat%). (N-S) Hepatic gene expression in week 20 male mice (n = 5-6/group). * p < 0.05, ** p < 0.01, *** p < 0.001, t-test compared to control. Graphs are displayed as mean ± SEM.
Dynamics of phenotype development and microbiota alterations
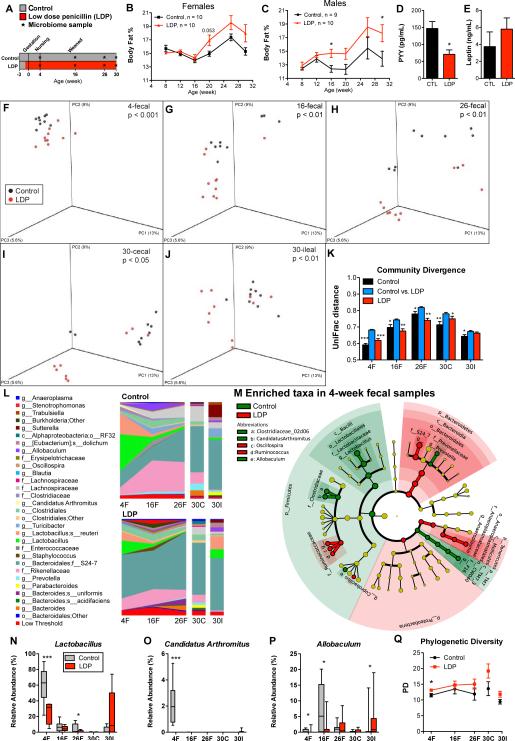
We next conducted longitudinal studies to permit a dynamic view of LDP-mediated phenotype development. Female and male LDP mice showed faster fat accumulation (rate differential = 0.13%/week female (p=0.002) and 0.15%/week male, (p=0.02)) and with significant increases in body fat percent detected earlier in males (Figure 2A-C). LDP reduced non-fasting serum peptide YY, and there was a trend of increased leptin (Figure 2D-E), alterations typically seen in obesity (Grudell and Camilleri, 2007; Morrison et al., 2009).
Figure 2. Dynamics of microbiota and adiposity changes over 30 weeks of life.
(A) Study design: Female and male C57B/L6 mice received low dose penicillin (LDP, n = 10 males and females), or no antibiotics (control, n = 9 males and 10 females) for 30 weeks. (B-C) Body fat percent over time, determined by DEXA scanning, in female and male mice. Serum levels of (D) peptide YY (PYY) and (E) leptin at week 30 in male mice. * p < 0.05 significantly different by t-test for B-E, and Q. (F-Q) Fecal specimens obtained at weeks 4, 16, and 26, and the terminal (week 30) cecal and ileal specimens from male mice were examined by sequencing the V1-2 region of the bacterial 16S rRNA gene. (F-J) Principal coordinate analysis (PCoA) of unweighted UniFrac distances of the intestinal microbiota at each sample point; p-values listed for differential clustering assessed by ADONIS test. (K) Divergence within (control vs control, LDP vs LDP) and between communities (control vs LDP) measured by unweighted UniFrac distance. The UniFrac matrix was permutated 10,000 times; p-values represent fraction of times permuted differences were greater than real distances; * p < 0.05 , **p < 0.01, *** p < 0.001 significantly different from intragroup distance. (L) Mean relative abundance of predominant bacteria (>1% in any sample) in control and LDP mice. Taxa are reported at the lowest identifiable level, indicated by the letter preceding the underscore: o, order; f, family, g, genus; s, species. (M) Cladograms, generated from LEfSe analysis, represent taxa enriched in control (green) or LDP (red) 4-week fecal microbiota. The central point represents the root of the tree (Bacteria), and each ring represents the next lower taxonomic level (phylum through genus). The diameter of each circle represents the relative abundance of the taxon. When full identification was not possible, g_ or s_ alone was used for genus or species, respectively. (N-P) Relative abundance of Lactobacillus, Candidatus Arthromitus (SFB), and Allobaculum; asterisks indicate level of significance by Mann-Whitney U test, see also Figure S1 and Table S1. (Q) Phylogenetic diversity in control and LDP male mice (Mean ± SEM), calculated at a uniform depth of 4,000 sequences/sample.
LDP significantly altered microbial community composition, measured by unweighted UniFrac distances (β-diversity), at all time points examined (Figures 2F-J). As the mice aged, positions of the control and LDP fecal samples changed unidirectionally on the principal coordinate analysis (PCoA) plots, with greater differences in LDP mice, suggesting acceleration of the normal age-related microbiota development. Intergroup β-diversity was significantly higher than either intragroup distances, showing the distinctive LDP and control microbiota community structures (Figure 2K).
The populations of mammalian microbiota show a pattern of succession in early life (Pantoja-Feliciano et al., 2013; Schaedler et al., 1965), and altered representations of particular taxa have been associated with the development of obesity (Kalliomäki et al., 2008). As expected, after nursing for 4 weeks, the control microbiota was dominated by Lactobacillus, diminishing as mice matured (Figure 2L-N). While LDP mice showed similar age-dependent population shifts, Lactobacillus levels were much lower at multiple time points than control. Focusing on early life (week 4), broad populations changes were seen from phylum to genus level (Figure 2M). In addition to Lactobacillus, LDP consistently reduced early-life Candidatus Arthromitus (segmented filamentous bacteria, SFB) and Allobaculum levels (Figure 2O-P) across multiple independent experiments (Table S1, Figure S1). As expected (Davis and Savage, 1976; Jiang et al., 2001), early life populations of Candidatus Arthromitus were highest in control pups and undetectable in older mice, but were significantly reduced by LDP (Figure 2O). These temporal changes indicate that LDP suppresses multiple taxa that typically peak early in life. Although high-dose (therapeutic) antibiotic exposures reduce microbiota diversity in animals and humans (Antonopoulos et al., 2009; Dethlefsen et al., 2008), LDP increased phylogenetic diversity at 4 weeks (Figures 2Q), consistent with decreases in particular prominent taxa, and detection of taxa with lower representation. By sampling across time, we identified microbes that were altered prior to the development of adiposity, suggesting roles in host metabolic development.
Interaction between LDP and dietary excess
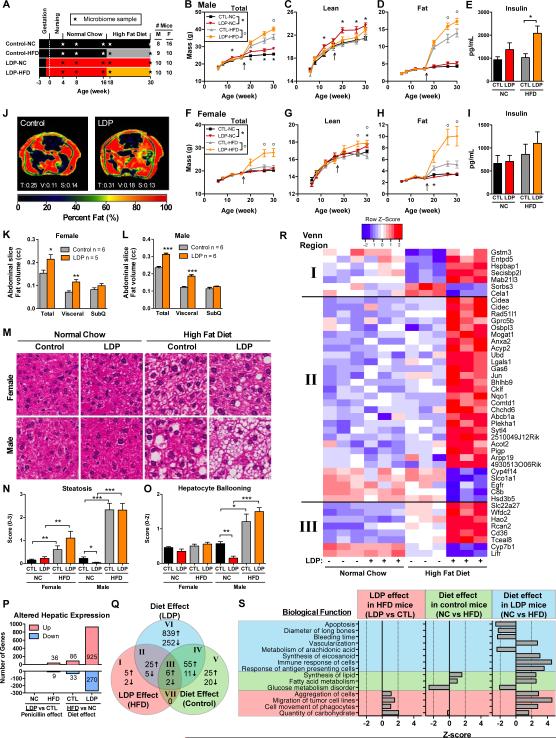
We next asked whether there were additive effects of early-life LDP and increased caloric intake. Mice received either LDP or not (control), were weaned to normal chow (NC), and at 17 weeks, half of the mice were switched to a high fat diet (HFD) (Figure 3A). In the males, total mass was increased by LDP, more so by HFD, and most by the combination (Figure 3B). In those fed NC, LDP increased lean mass, and in those given HFD, LDP markedly increased fat mass (Figure 3C-D). Fasting insulin was not altered by HFD alone, which may reflect that 45% kcal from fat does not produce as extensive metabolic effects as 60% kcal from fat (Pettersson et al., 2012). Insulin was slightly, but not significantly, increased by LDP, but significantly raised in mice receiving the combination (Figure 3E). In female mice, total mass, not increased by HFD alone, was increased by the combination of HFD and LDP, and lean mass was increased by LDP on either diet. Most notably, by 30 weeks, LDP approximately doubled the fat mass in females fed HFD (Figure 3F-H), but did not significantly alter fasting insulin (Figure 3I). In both male and female 26 week mice, LDP increased abdominal fat, localized to the visceral compartment (Figure 3J-L). In total, these studies provide evidence that LDP exposure enhances the HFD metabolic effects, and confirms prior observations (Goyal et al., 2012) that male mice are more predisposed to diet-induced obesity.
Figure 3. Effects of combining LDP and High Fat Diet.
(A) Study design: Male and female mice either received low dose penicillin (LDP) from birth or did not receive antibiotics (control, CTL). All mice were started on a normal (NC) chow diet, and then half were maintained on normal chow and half switched to a high fat (HFD) diet at week 17. (B-H) Body composition consisting of total, lean, and fat mass in male and female mice, determined by DEXA. (E, I) Fasting insulin in week 30 mice. (J-L) Distribution of abdominal body fat, determined by MRI at week 26 (n = 5-6 each). (J) Images show representative male mice; numbers represent the volume of total (T), visceral (V), and subcutaneous (S) fat mass in three consecutive slices. (K-L) Abdominal adiposity, quantified by total, visceral, and subcutaneous fat in three consecutive cross-sections in female and male mice. (M-O) Hepatic histopathology: (M) Representative hematoxylin and eosin-stained sections. (N-O) Steatosis and hepatocyte ballooning scores. (B-O) * p < 0.05, ** p < 0.01, *** p < 0.001, t-test for B-L; and Mann-Whitney U for N-O. Graphs are displayed as mean ± SEM. (P-S) Hepatic gene expression, measured by microarray in male mice at 30 weeks of age (n = 3 in each of the four exposure groups). (P) Number of genes that have significant (FDR-adjusted p < 0.05) hepatic expression differences comparing either LDP treatment or dietary changes, that are up- or down- regulated in the underlined group with respect to the non-underlined group. (Q) Comparative analysis of differential hepatic gene expression. Venn diagram indicates the number of overlapping genes in three separate pairwise comparisons, LDP effect in HFD mice (LDP vs CTL), diet effect in control mice (HFD vs NC) and diet effect in LDP mice (HFD vs NC). (R) Heatmaps depict gene expression values of Venn regions I-III (see also Figure S2 for regions IV-VI). (S) Predicted biological functions that are differentially represented, (p < 0.05, z-score > |2|) based on Ingenuity Pathway Analysis of microarray measurements of hepatic gene expression.
Alterations in microbiota and diet can affect the liver (Henao-Mejia et al., 2012). HFD significantly increased hepatic steatosis in male and female mice at age 30 weeks, with greater increases in males, and increased hepatocyte ballooning in male mice. LDP reduced both metrics in male mice fed normal chow (Figure 3M-O), and had little additive effect in mice fed HFD. Neither LDP nor HFD strongly affected lobular inflammation or fibrosis (Figure S2A-B). However, although the magnitude of the effect was small, LDP significantly increased inflammation and fibrosis in female mice fed HFD. In total, histological analyses indicate that HFD is a stronger driver of non-alcoholic fatty liver disease (NAFLD) than LDP, and that the extent of HFD-mediated effects and LDP induced-inflammation differed by gender.
At the transcriptional level, HFD had a stronger effect on hepatic gene expression than LDP, and the dietary effect was greatest in mice receiving LDP (Figure 3P). Of the 1091 genes uniquely regulated by HFD in LDP mice, 980 (90%) had a non-significant trend in the same direction in control mice, indicating that microbiome disruption could amplify HFD-mediated changes. Through comparative analyses, we identified genes either uniquely regulated by LDP- or by HFD, requiring both interventions, or responding synergistically (Figures 3Q-R, S2O). In mice fed HFD, LDP altered genes related to carbohydrate metabolism and cell movement, which resembled the effects of HFD in LDP-treated mice, but not HFD alone (Figure 3S, Figure S2C-N). In contrast, HFD, alone or in combination with LDP, changed expression of genes related to glucose metabolism (decreased), and to fatty acid metabolism and lipid synthesis (increased). HFD-regulated genes in LDP mice were involved in functions linked to the microbiota (Figure 3S), such as increased immune and antigen presenting cell responses (Erturk-Hasdemir and Kasper, 2013), increased eicosanoid synthesis and decreased arachidonate metabolism (Antunes and Finlay, 2011), and increased vascularization (Stappenbeck et al., 2002).
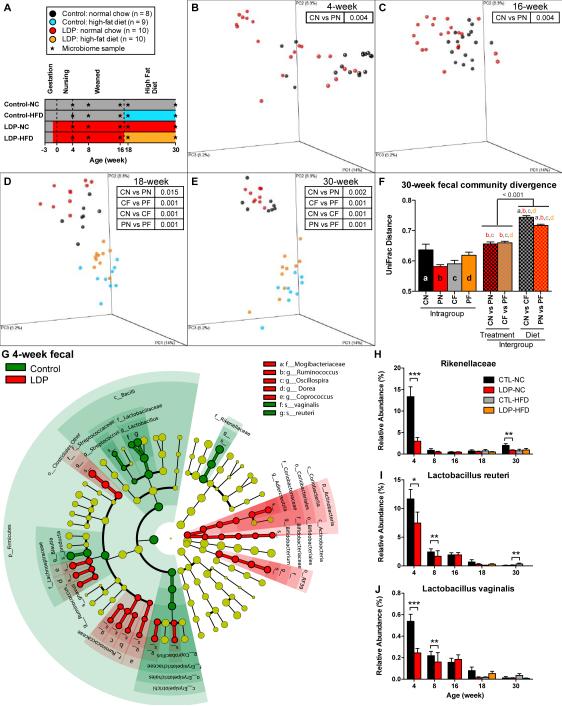
To examine microbiota changes, we studied serial fecal pellets and terminal cecal and ileal samples (Figure 4A). Based on unweighted UniFrac distances, both LDP and HFD significantly (p < 0.05, ADONIS test) altered community structure (Figure 4B-F), and HFD had larger effects on microbial community structure than LDP. Regardless of antibiotic exposure, HFD led to numerous enriched taxa in 30-week fecal samples, including the phylum Firmicutes and lower lineages, and decreased Bacteroidetes, as reported (Ley et al., 2005) (Figure S3A-B). Regardless of diet, LDP modulated multiple taxa (Figure S3C), including Lactobacillus. Focusing on early life (Figure 4G), LDP affected multiple taxa, some of which were consistently modulated across experiments (Table S1). Rikenellaceae levels, highest in control mice at 4 weeks, diminished with age, but were markedly reduced by LDP (Figure 4H). Levels of genus Lactobacillus, comprised predominantly of L. reuteri and L. vaginalis, also were reduced (Figure 4I-J), although less than prior (Figure 2N). These studies reveal that LDP and HFD have independent selective effects, with LDP consistently linked with specific microbial changes.
Figure 4. The effect of HFD and LDP on intestinal microbiota composition.
(A) Study design: control (C) mice did not receive antibiotics; LDP (P) mice received low-dose penicillin from birth. All mice were started on normal chow (N), and then half were switched to HFD (F) at week 17. Microbiota samples were collected longitudinally for 16S rRNA sequencing. (B-E) PCoA of fecal microbiota samples at weeks 4, 16, 18, and 30; p-values for differential clustering assessed by ADONIS test are indicated in the insets. (F) Mean pairwise intra- and intergroup unweighted UniFrac distances of week 30 fecal microbial, letters a-d indicate significant differences with respect to intragroup distances, p < 0.05 based on 10,000 permutations. (H) Cladogram representing taxa enriched in 4-week fecal samples in control or LDP mice, detected be the LEfSe tool. (H-J) Relative abundance of Rikenellaceae, Lactobacillus reuteri and Lactobacillus vaginalis in fecal samples from weeks 4-30, * p < 0.05, ** p < 0.01, *** p < 0.001, Mann-Whitney U test. Graphs are displayed as mean ± SEM. See also Figure S3.
Early-life metabolic programming: limited antibiotic exposures lead to durable phenotypes
Hypothesizing that early life was the critical period for programming host-microbe metabolic interactions, we sought to determine whether microbiota disruption limited to early life could suffice to induce metabolic effects. In addition to long-term (28-week) LDP or none (control), groups of mice received 4 or 8 weeks of LDP (Figure 5A), and to accelerate metabolic phenotypes, all were switched to HFD at 6 weeks of age. In female mice, all LDP groups developed elevated total, lean, and fat mass compared to controls, irrespective of LDP duration (Figure 5B). Compared to controls, following switch to HFD, female LDP mice had significantly elevated caloric intake (Figure 5C) and significantly faster total and fat mass accumulation rates from 6-20 weeks of age (Table S2). Later in life (weeks 20-28), all three LDP groups showed significantly slower rates in lean mass growth compared to controls, indicating catch-up by the control mice. Male LDP mice showed early elevations in total, lean, and fat mass, but did not have increased food intake or feed efficiency from 6-8 weeks of age (Figure S4A-D), and the early-life changes in body composition were lost with aging, consistent with increased early-life and gender-dependent sensitivity to HFD (Goyal et al., 2012; Pettersson et al., 2012), which may override microbe-mediated effects.
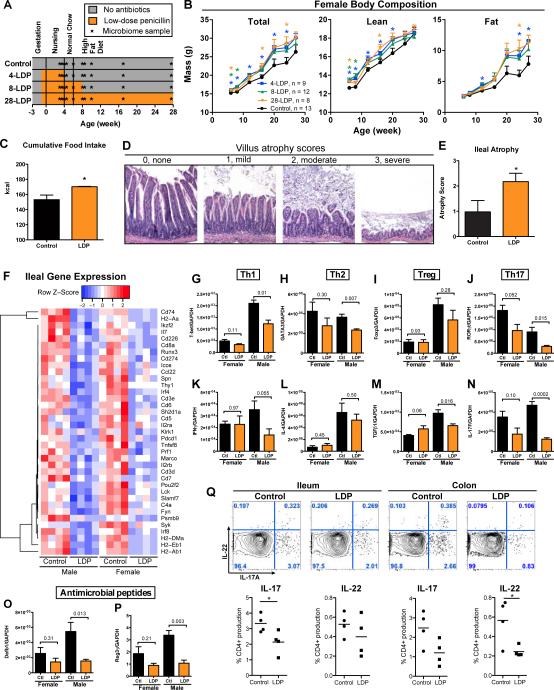
Figure 5. Early-life LDP induces lasting changes in adult body composition.
(A) Study design: Both male and female mice received 4 (4-LDP), 8 (8-LDP), or 28 (28-LDP) weeks of low-dose penicillin (LDP), or no antibiotics (control). (B) Total, lean, and fat mass in female mice, measured by DEXA scanning. * p < 0.05 significant difference at a single time point measured by t-test. 4-, 8-, and 28-LDP mice had significantly increased rates of total and fat mass accumulation from 6-20 weeks of age (Table S2). (C) Cumulative food intake for female mice over 12 days during weeks 6 to 8 (n = 4 mice/group in metabolic cages). (D) Histopathology: representative H&E-stained ileal sections with villus atrophy scores 0-3. (E) Mean ± SEM ileal atrophy score in male control and LDP mice (n = 5 each), * p < 0.05 Mann-Whitney U for panels C&E. (F) Expression of ileal genes that are significantly altered by LDP in both male and female mice (n = 4 each) at 8 weeks of age, measured by the Nanostring Mouse Immunology Kit, genes shown p < 0.05, t-test (see also Table S3). Ileal expression of transcription factors (G-J) and cytokines (K-N) representative of four T-helper cell lineages, and expression of antimicrobial peptides (O-P) measured by qPCR; p-values shown for t-tests for 8-week male and female mice, n = 4 each. (Q) Flow cytometry of ileal and colonic tissue from male control and LDP mice at 8-weeks of age (n = 4) for IL-17 and IL-22, * p < 0.05 t-test. See also Figure S4. Graphs are displayed as mean ± SEM.
Disruption of defenses at the intestinal interface has been implicated in obesity (Cani et al., 2008; Henao-Mejia et al., 2012; Vijay-Kumar et al., 2010). By age 4 weeks, LDP induced substantial histopathologic effects in ileal tissues, notably shortened villi (Figure 5D-E), consistent with changed ileal architecture in LDP-mediated livestock growth promotion (Gaskins et al., 2002; Visek, 1978). Transcriptional profiling analysis of intestinal tissue by microarray (Figure S4E) and subsequent validation by Nanostring analysis revealed that the ileal atrophy from LDP was associated with a general decreased expression of genes involved in intestinal immune responses, with numerous consistencies across gender (Figure 5F and Table S3). LDP decreased expression of genes related to several biologic functions, such as differentiation, activation, recruitment, and adhesion of immune cells, and functions specifically related to antigen-presenting cells, T-cells, B-cells, and phagocytic cells (Figure S4F).
The gut microbiota influences the composition of CD4+ T cells within the intestinal mucosa (Hooper et al., 2012). LDP reduced expression of transcription factors and cytokines important for Th1 and Th17 cell differentiation and function (Figure 5 G-N), with significant effects in males and consistent trends in females. Antimicrobial peptides β-defensin 1 (Defb1) and regenerating islet-derived protein 3 gamma (RegIIIγ) also were down-regulated by LDP (Figure 5O-P). To confirm the Th17 cell alterations, we performed flow cytometric analysis on lamina propria cells isolated from the ileum and colon in 8-week old male mice. These showed reduced CD4+ IL-17- or IL-22-positive cell populations after LDP (Figure 5Q). Taken together, these results indicate that intestinal immune responses are globally reduced after LDP.
Next, we examined the effects of limited early-life LDP on the microbiota (Figure S5). By UniFrac analysis, the microbial communities in the females were distinct between control and LDP from the earliest time point studied (3 weeks) to the end of the experiment (28 weeks) (Figures 6A-C, ADONIS test, Table S4). However, after antibiotic cessation, community structure progressively reverted to normal in the fecal, cecal, and ileal samples. To assess community structure patterns according to age, we examined intragroup phylogenetic distances. In infancy, the microbiota of control mice are highly diverse, becoming more homogeneous as mice age, a pattern also observed in all LDP groups (Figure 6D, Table S5). After antibiotic cessation, microbiota from the 4-LDP and 8-LDP mice become no more distant from the control mice than the intragroup control variation, demonstrating recovery; however the 28- (continuous) LDP microbiota divergence never normalized (Figure 6E, Table S5), and in the complementary analyses, as expected, the opposite pattern was observed (Figure 6F). All told, the community analyses demonstrate early-life individual host microbiota variability that decreases during maturation, and that microbial populations are only transiently disrupted by limited duration LDP, yet the effects on phenotype are life-long (Figure 5B).
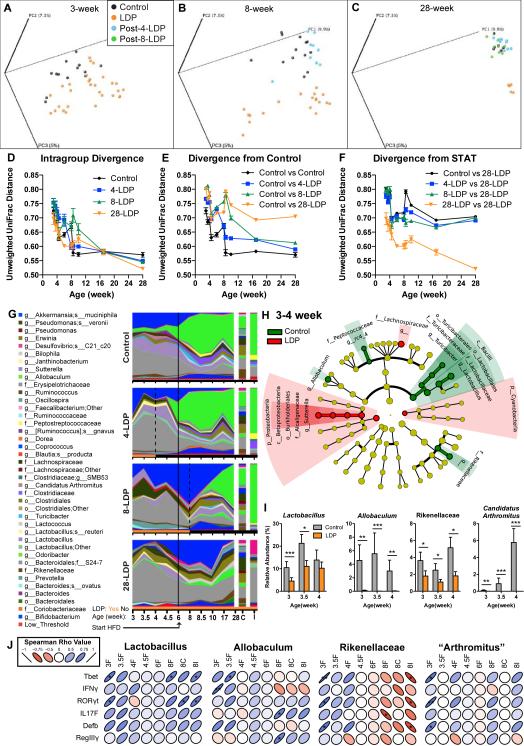
Figure 6. Effect of limited early-life LDP on the intestinal microbiota.
(A-C) PCoA of the unweighted UniFrac distances computed from 16S rRNA sequences from (A) week 3, (B) week 8, (C) and week 28 microbiota in female mice receiving either 4, 8, or 28 weeks of LDP or not receiving antibiotics (control). (D-F) Intragroup and intergroup divergence measured by unweighted UniFrac distances over time. (G) Relative abundance of the major taxa (>1% in any sample) in the intestinal microbiota. Dotted lines show cessation of antibiotics, and the solid line shows start of HFD. (H) Phylogenetic representation of the taxa in the week 3, 3.5, and 4 fecal samples significantly associated with control (green) or LDP (red), as determined by LEfSe. (I) Relative abundance of Lactobacillus, Allobaculum, Rikenellaceae, and Candidatus Arthromitus (SFB) in 3-4 week fecal samples. Graphs are displayed as mean ± SEM, * p < 0.05, ** p < 0.01, *** p < 0.001 Mann-Whitney U. (J) Spearman correlation of relative abundance of early-life (weeks 3-8) Lactobacillus, Allobaculum, Rikenellaceae, and Candidatus Arthromitus (SFB) with expression of ileal genes involved in intestinal defense. * p < 0.05, ** p < 0.01. See also Figure S6, and Table S4-5.
LDP induced numerous compositional changes in the microbiota (Figure 6G), and HFD had further effects. After HFD was started, control mouse Allobaculum markedly increased with concomitant decreases in Akkermansia mucinophila and an unidentified Bacteroidales S24-7 family member. After antibiotic cessation in the 4-LDP and 8-LDP mice, the patterns associated with HFD exposure of the control mice began to emerge, but were never present in the 28-LDP mice. The persistent phenotypes after LDP cessation, despite microbiota normalization, provide evidence that the early-life microbiota influences adult body composition. LDP suppressed early-life Lactobacillus, Allobaculum, Rikenellaceae, and Candidatus Arthromitus (SFB) (Figure 6H-I) as observed in the prior experiments (Table S1), further implicating them as candidate protective taxa; these organisms were positively correlated with markers of ileal immunity (Figure 5J).
LDP-altered microbiota are sufficient to induce metabolic changes
While the above observations suggest that the change in the microbiota is driving the metabolic and immunologic effects, they also could reflect the influence of the administered LDP on development. To eliminate the direct effects of penicillin, we transferred cecal microbiota from 18-week old female control or LDP mice (n = 3 each) (Figure 7A) to 3-week old female germ-free Swiss-Webster mice. While the strains of murine microbiota donor and germ-free recipient were not the same, this approach has been used to test the causality of altered microbiota between differing mouse strains (Vijay-Kumar et al., 2010) and host species (humans and mice) (Ridaura et al., 2013). The LDP-microbiota recipients increased total mass and fat mass at a faster rate (rate differential = 0.078 g/day total (p=0.01), 0.058 g/day fat (p=0.0012)) compared to controls; no changes were detected in lean mass (Figure 7B). These findings indicate that the host metabolic changes are driven by the LDP-altered microbiota and are not dependent on direct antibiotic effects on the host.
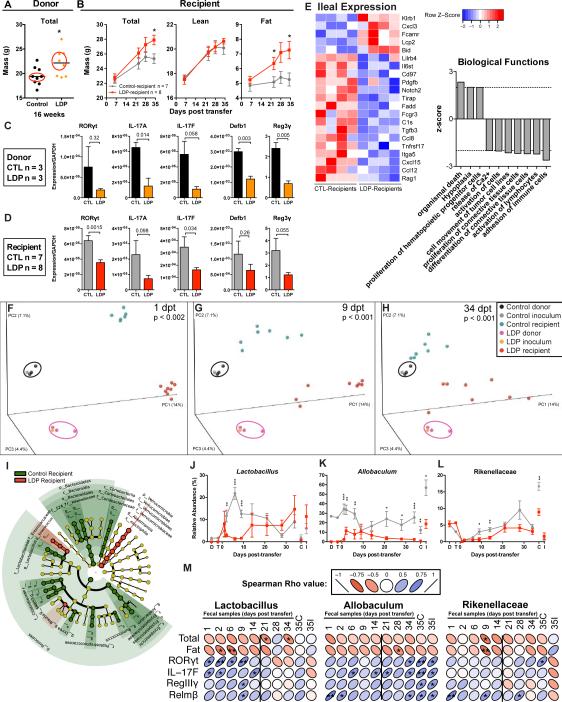
Figure 7. Metabolic, immunologic, and ecological consequences of transferring LDP-altered microbiota.
Cecal microbiota from 3 control and 3 LDP 18-week old female C57B/L6J mice were collected, pooled, and transferred to 3-week old germ-free female Swiss-Webster mice by oral gavage. (A) Microbiota donors were selected based on the median total mass determined by DEXA scanning at week 16. (B) Total, lean, and fat mass in the now-conventionalized germ-free control- and LDP-microbiota recipient mice (n = 7 and 8, respectively), measured by DEXA scanning over the 35-day experiment. (C-D) Ileal gene expression in microbiota donor (n = 3 CTL, LDP) (C) and recipient (n = 7 & 8, CTL, LDP, respectively) (D) mice measured by qPCR, p-values listed from t-tests. (E) Expression of ileal genes significantly different between control- and LDP recipients (n = 4 each), measured by the Nanostring Mouse Immunology Kit, (p < 0.05, t-test). Biological functions predicted to be significantly increased (p< 0.05, z > |2|) in LDP mice based on Ingenuity Pathway Analysis of Nanostring expression values. (F-H) Community structure assessed by PCoA of unweighted UniFrac distances of microbiota samples: donor cecum; the transferred inoculum; and the recipient mouse fecal samples at days 1, 9, and 34 post-transfer (dpt). Significant differences in clustering between control and LDP were calculated by ADONIS test. (I) LEfSe cladogram depicting taxa in fecal samples from 1-14 dpt that are significantly increased in control (green) or LDP (red) specimens. Relative abundance of (J) Lactobacillus, (K) Allobaculum, and (L) Rikenellaceae, significant differences assessed by Mann-Whitney U. Graphs are displayed as mean ± SEM. (M) Spearman correlations between host phenotypic measurements and candidate protective taxa found in multiple experiments. The vertical lines separate the early (pre-phenotype) time-points. * p < 0.05. See also Figure S6-7 and Table S6.
As we now expected, the LDP microbiota donor mice had decreased ileal expression of genes involved in Th17 populations and antimicrobial peptides (Figure 7C). At 8 weeks of age, the LDP-microbiota recipients showed similar trends to the donor mice (Figure 7D). The LDP-altered microbiota also changed immunological gene expression compared to the recipients of the control microbiota (Figure 7E), and these changes were related to predicted biological functions similarly altered in the 8-week old male and female mice directly exposed to LDP (Figure S4F), including decreased differentiation, activation, adhesion, recruitment, and quantity of immune cells. These studies provide evidence that altered microbiota can mediate changes in intestinal immune gene expression and are consistent with the global reduction in intestinal immune responses observed in the LDP-exposed microbiota donors (Fig 5F-Q).
In contrast to the conserved ileal gene expression functional changes with LDP, hepatic gene expression was more specific to the experimental conditions (Figure S4G-H, Table S3). Nevertheless, we found consistencies, such as genes related to organismal death in males and females directly exposed to LDP, but few such differences also were observed in the transfer experiment (Figure S4I).
Microbe-host associations in microbe-induced obesity
In the absence of further antibiotic selective pressure, the recipient-microbiota communities remained distinct, based on their respective donors (Figure 7F-I, Table S6). Next, we asked whether specific taxa were differentially represented in the transfer recipients in the early time points (1-14 days post-transfer), prior to detected phenotype development. LEfSe analysis (Figure 7I) identified many of the same taxa as in prior experiments (Table S1), including Lactobacillus, Allobaculum, and Rikenellaceae (Figure 7J-L). These organisms were negatively correlated with total and/or fat mass (Figure 7M), and positively correlated with expression of the Th17 transcription factor RORγt and cytokine IL-17F and antimicrobial peptides RegIIIγ and Relmβ, consistent with prior experiments.
Predicted metagenomic analysis, performed using PICRUSt (Langille et al., 2013) showed substantial commonalities at multiple time points and across experiments (Figure S6). In particular, recipients of LDP or of LDP-microbiota showed increased representation of predicted KEGG pathways involved in protein and lipid biosynthesis, whereas the control and control-microbiota recipient mice were predicted to have increased carbohydrate and terpenoid biosynthesis pathway representation. These results indicate potential functional shifts reflecting the compositional differences.
Microbe-induced obesity is limited in sequential transfers
Finally, to address whether the LDP growth promotion effect would continue after a period of recovery from antibiotic selective pressure, a second microbiota transfer to germ-free mice was performed. Cecal microbiota were collected from three control-microbiota recipients (CR1) or LDP-microbiota recipients (PR1) at 8 weeks of age, pooled, and transferred to new groups of germ-free mice (CR2, PR2) (Figure S7A). At 23 days post-transfer, both the CR2 and PR2 mice were lean compared to PR1 mice, with no significant adiposity differences over two months (Figure S7B-E). While the CR2 and PR2 mice continued to have distinct fecal microbial community structures, intergroup distances were decreased compared to the first transfer, and the candidate protective taxa were not suppressed in the PR2 recipients (Figure S7F-L, Table S1). Thus, in a sequential transfer without direct antibiotic exposure, there was normalization of the metabolic and the ecologic phenotypes; the restoration of Lactobacillus, Allobaculum, and Rikenellaceae populations further strengthens the evidence for a putative protective role.
DISCUSSION
We have proposed microbe-induced obesity (MIO) as a condition in which excessive adiposity results from primary perturbation of the microbiota, in contrast to diet-induced obesity (DIO) or gene-induced obesity (GIO) (Cox and Blaser, 2013). We had developed a mouse model of MIO, showing that sub-therapeutic antibiotic treatment (including LDP) delivered at weaning led to increased adiposity (Cho et al., 2012). We now indicate five characteristics of MIO. First, early life is the critical window of host-microbe metabolic interaction, in which pre-weaning microbiota perturbation promotes growth, and is sufficient for lasting metabolic changes. Second, even with exposures limited to infancy, the elevated adiposity emerges later, in early- to mid-adulthood. Third, penicillin exposures amplify diet-induced obesity and effects on hepatic gene expression, metabolic hormone levels, and visceral fat accumulation. Fourth, the altered microbiota alone, not continued LDP exposures, is causal. That germ-free chickens do not gain weight in response to low-dose penicillin also provides evidence that the penicillin-altered microbiota drives metabolic changes (Coates et al., 1963). Fifth, the transmissibility of the phenotype is lost with successive transfers, indicating that phenotypic recovery is possible when antibiotic exposure stops. In total, our work extends the MIO concept and defines key features of the model.
We performed our studies using penicillin, a β-lactam antibiotic, the class most frequently prescribed to children in the US and world-wide (Chai et al., 2012; Clavenna and Bonati, 2011). As in other animal models of disease, we found variability in the timing and extent of phenotype emergence with notable gender differences (Markle et al., 2013; Pettersson et al., 2012; Yurkovetskiy et al., 2013). Nevertheless, LDP consistently enhanced growth, affecting lean, fat, or bone, or a combination thereof.
We also have identified key microbiota characteristics in this MIO model. First, consistent with our earlier findings (Cho et al., 2012), LDP exposure was not associated with reduced microbial abundance or taxonomic diversity, indicating that compositional differences drive MIO. Second, after antibiotic cessation, the microbiota returns to a normal population structure; however, metabolic consequences last, providing evidence that early-life microbiota participate in long-term metabolic programming. Third, particular organisms were consistently underrepresented in LDP mice prior to phenotype development (Table S1), suggesting that their loss during a critical window is detrimental. These findings are consistent with prior studies showing that early life changes in metabolism or exposure to antibiotics predict later adiposity in rodents (Knittle and Hirsch, 1968) and humans (Ajslev et al., 2011; Murphy et al., 2013; Trasande et al., 2013).
Among the four candidate protective organisms, modulation of host metabolism by Lactobacillus (Beijerinck, 1901), is known (Million et al., 2012), whereas the associations with Allobaculum, Rikenellaceae, and Candidatus Arthromitus (SFB) are novel. Finding associations with the recently described Allobaculum (Greetham et al., 2004), and recently classified family Rikenellaceae (Collins et al., 1985), and with the long-known (Leidy, 1849) but not yet cultured SFB, indicate the ability of sequence-based approaches to identify new host-microbe interactions. All microbiota members do not equally impact the host; prior studies indicate that these four organisms have either metabolic and/or immunologic interactions (Daniel et al., 2013; Ivanov et al., 2009; Karlsson et al., 2011; Martinez et al., 2009; Ravussin et al., 2012), which may contribute to the observed protection from weight gain in the control animals.
MIO could be mediated by alterations in the liver or the intestine. While our results confirm the previously observed late life hepatic effects (Cho et al., 2012), such as altered expression of genes involved in fatty acid metabolism (e.g. CD36, PPARγ, and FABP2), there were few changes conserved in young males and females and in microbiota recipients. This suggests that the large number of hepatic gene expression changes in the 30-week males may be a consequence, rather than a cause, of obesity. Nevertheless, our studies show that LDP increased liver adiposity, visceral fat accumulation, and fasting insulin, indicating that microbiota disruption can worsen obesity-related complications.
Ileal changes are associated with altered hepatic gene expression and increased adiposity; reduced epithelial tight junction proteins (Cani et al., 2008), loss of inflammasome function (Henao-Mejia et al., 2012), lowered microbial surveillance [in TLR5-/- mice; (Vijay-Kumar et al., 2010)], or thinning of the intestinal mucus layer (Everard et al., 2013) can enhance translocation of bacterial products and drive systemic inflammation and obesity. The associations we observed of reduced ileal integrity and expression of intestinal defense genes are consistent with the prior studies, and provide the complementary model of MIO, in addition to models that manipulate host genetic or dietary factors.
LDP suppressed genes involved in antibacterial responses, with processes involved in antigen presentation, Th17 cell differentiation, and bacterial killing. One explanation invokes inhibited colonization by highly interactive bacteria such as SFB, known to be penicillin-sensitive (Davis and Savage, 1976), that promote Th17 gut responses (Ivanov et al., 2009). Regardless of SFB involvement in initial immunologic programming, other organisms also stimulate Th17 responses, since SFB were absent from both control-and LDP-microbiota recipients, which show altered Th17 markers. Two (Lactobacillus and Allobaculum) of the three other candidate protective taxa positively correlate with RORγT and IL-17 expression, suggesting roles in intestinal Th17 differentiation. However, while there are fewer Th17 cells and diminished expression of genes involved in intestinal immune responses after LDP, our observations do not provide causal evidence that LDP is directly altering intestinal immune responses. Nevertheless, these findings are consistent with prior studies that have identified impaired intestinal barrier function and immunity as important factors in the development of metabolic syndromes (Cani et al., 2008; Henao-Mejia et al., 2012; Vijay-Kumar et al., 2010).
The metabolic consequences observed indicate that microbiota disruption during a critical developmental window can explain later life phenomena, including adult obesity. LDP may decrease particular early life-protective bacterial populations, consistent with co-evolution with microbes having specific interactions promoting metabolic fitness (Blaser, 2006). As “keystone” species, their reduction could lead to broad intestinal ecologic changes, permitting increases in taxa having alternate roles in metabolic or immunologic development (Chung et al., 2012). That the microbiota recovers, yet the phenotype remains, suggests that microbiota changes can affect metabolic programming; studies examining epigenetic regulation could help elucidate mechanisms of host-microbe interaction.
Epidemiologic studies provide evidence that antibiotic exposures in human infancy can lead to increased weight later in life (Ajslev et al., 2011; Murphy et al., 2013; Trasande et al., 2013). Combining multiple mouse models with high-throughput microbial 16S rRNA profiling, we identified key microbiota members impacted by antibiotic treatment and linked with host physiologic changes. If humans have analogous organisms, their loss in early life due to collateral antibiotic effects may have life-long consequences. Restoration of lost taxa following early-life antibiotic exposures is a potential strategy to reverse MIO and related sequelae. Nevertheless, humans have greater dietary and lifestyle variation than do experimental mice, which introduces factors that affect recovery from metabolic disruption, or susceptibility to weight gain. Further studies are needed to examine specific metabolic interactions of key members of the early life microbiota and to determine whether these changes are important in humans.
EXPERIMENTAL PROCEDURES
Animals
All animal experiments were performed according to IACUC-approved protocols. Low dose penicillin (LDP) was delivered via drinking water as described (Cho et al., 2012). Control mice did not receive antibiotics. Diets used in the studies were either normal chow (NC, Purina Mills International Diet # 5001, 13.5% kcal from fat), or high fat diet (HFD, Rodent Diet D12451, Research Diets, 45% kcal from fat). For the microbiota transfer experiments, cecal contents were collected from 3 microbiota donors from each group and placed in a pre-reduced anaerobically sterilized liquid dental transport medium (Anaerobe Systems, Morgan Hill CA), pooled in sterile pre-reduced saline under anaerobic conditions, and then transferred to 3- to 4-week old germ-free Swiss Webster mice via oral gavage (control-recipients n = 7, LDP-recipients n = 8). The microbiota-recipient mice were housed in autoclaved cages, under specific pathogen-free conditions, and fed irradiated HFD, as the donor mice had been. See Extended Experimental Procedures for details on individual experiments.
Body composition analysis
Body composition was determined using dual energy X-ray absorptiometry (DEXA) with a Lunar PIXImus II mouse densitometer (GE Medical Systems, Waukesha WI) and by magnetic resonance imaging on a 7-tesla MRI system (Bruker Biospin MRI, Ettlingen, Germany). For the MRI, fat and lean tissue were determined using the iterative decomposition of water and fat with an echo asymmetry and least-squares (IDEAL) method (Reeder et al., 2005; Reeder et al., 2004).
Hepatic and ileal gene expression profiling
Total RNA was extracted by the RNeasy Mini Kit (Qiagen, Valencia CA). For microarrary analysis, expression profiling was performed using the Affymetrix Genechip Chip Mouse 430_2 system (Affymetrix, Santa Clara CA). The raw microarray data was normalized using the Robust Multi-Array (RMA) method (Irizarry, 2003). The Limma package in R was used to fit a linear model to the expression data using empirical Bayes moderated t-statistics (Smyth, 2004). RNA-seq libraries were generated using the TruSeq v2 RNA sample prep (Illumina, San Diego, CA); libraries were sequenced with 2x50bp paired-end reads in an Illumina HiSeq 2500. Fastq files were processed as follows: alignment was done with STAR (Dobin et al., 2012), with an index build from the mm10 reference sequence. The genes annotated in UCSC mm10 were quantified with HTSeq-count v0.5.3p9 (Anders et al., 2014). Differential expression analysis was performed with the Bioconductor package DESeq (Anders and Huber, 2010). Ileal gene expression was measured by the nCounter GX Mouse Immunology Kit (Nanostring Technologies, Seattle, WA, USA).
Flow cytometry
Lamina propria mononuclear cells were isolated from the ileum and colon. Cells were stimulated with 50 ng/ml phorbol 12-myristate 13-acetate (PMA) and 500 ng/ml Ionomycin for 4 hours at 37°C in the presence of brefeldin A (GolgiPlug, Becton-Dickinson, Franklin Lakes NJ). Following this in vitro stimulation, cells were stained with LIVE/DEAD Fixable Aqua (LifeTechnologies), anti-CD3, and anti-CD4 and fixed in 4% paraformaldehyde in PBS, then permeabilized in Perm/Wash buffer (Becton-Dickinson) and stained with anti-IL-17A and anti-IL-22 (eBioscience, San Diego CA). Cells were acquired on an LSRII (Becton-Dickinson) and analyzed with FlowJo (Tree Star, Ashland OR) software.
Microbial community analysis
DNA was extracted using the MoBio PowerSoil DNA Extraction Kit and the microbial 16S rRNA gene was amplified with barcoded fusion primers, targeting either the V1-2 (Fierer et al., 2008) or the V4 (Caporaso et al., 2012) region (Figure S5, Table S7). The QIIME pipeline (Caporaso et al., 2010) was used for quality filtering of DNA sequences, demultiplexing, taxonomic assignment, and calculating α- and β diversity. Metagenomic content of the microbiota samples was predicted from the 16S rRNA profiles and KEGG pathway functions were categorized at level 3 using the phylogenetic investigation of communities by reconstruction of unobserved states (PICRUSt) tool (Langille et al., 2013). Linear discriminant analysis effect size (LEfSe) (Segata et al., 2011) was used to detect significant changes in relative abundance of microbial taxa and predicted KEGG pathways between control and LDP mice.
Supplementary Material
RESEARCH HIGHLIGHTS.
Early-life is a critical window of host-microbe metabolic interaction
Low-dose penicillin treatment amplifies diet-induced obesity
A transient early microbiota perturbation leads to long-term increased adiposity
The penicillin-altered microbiota has a causal role in inducing metabolic changes
ACKNOWLEDGMENTS
We acknowledge assistance with sequencing from Argonne National Laboratory and NYULMC Genome Technology Center, and help from the NYULMC Histology Core, and the New York Genome Center. These studies were supported by NIH grants R01 DK 090989 and 1UL1RR029893, the Diane Belfer Program in Human Microbial Ecology, Knapp Family Foundation, and Daniel Ziff Foundation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
AUTHOR CONTRIBUTIONS
M. J. Blaser, L. M. Cox, and I. Cho designed experiments and interpreted data. L. M. Cox performed experiments and participated in analysis of the data. S. Yamanishi, N. Robine contributed to gene expression studies. J. Sohn performed experiments and contributed to data analysis. A.V. Alekseysenko advised on microbiome analytical methods and performed data analyses. H. Li performed longitudinal modeling and other statistical tests. J. M. Leung and P. Loke performed flow cytometry and interpretation of immunologic data. S. Kim designed and performed MRI experiments and advised in data analysis. Z. Gao participated in gene expression studies and performed PICRUSt analyses. A. B. Rogers, D. Mahana, and J. G. Zarate Rodriguez performed histological analyses. L. M. Cox and M. J. Blaser were responsible for writing this manuscript, as reviewed by all authors
REFERENCES
- Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biology. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S, Pyl PT, Huber W. HTSeq - A Python framework to work with high-throughput sequencing data. Journal Article. 2014 doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ajslev TA, Andersen CS, Gamborg M, Sørensen TIA, Jess T. Childhood overweight after establishment of the gut microbiota: the role of delivery mode, pre-pregnancy weight and early administration of antibiotics. Int J Obes. 2011;35:522–529. doi: 10.1038/ijo.2011.27. [DOI] [PubMed] [Google Scholar]
- Antonopoulos DA, Huse SM, Morrison HG, Schmidt TM, Sogin ML, Young VB. Reproducible community dynamics of the gastrointestinal microbiota following antibiotic perturbation. Infect Immun. 2009;77:2367–2375. doi: 10.1128/IAI.01520-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antunes LCM, Finlay BB. A comparative analysis of the effect of antibiotic treatment and enteric infection on intestinal homeostasis. Gut Microbes. 2011;2:105–108. doi: 10.4161/gmic.2.2.15610. [DOI] [PubMed] [Google Scholar]
- Beijerinck MW. Sur les ferments lactiques de l'industrie. Archives Néerlandaises des Sciences Exactes et Naturelles. 1901;6:212–243. [Google Scholar]
- Blaser MJ. Who are we? Indigenous microbes and the ecology of human diseases. EMBO Rep. 2006;7:956–960. doi: 10.1038/sj.embor.7400812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blüher M. The distinction of metabolically ‘healthy’ from ‘unhealthy’ obese individuals. Curr Opin Lipidol. 2010;21:38–43. doi: 10.1097/MOL.0b013e3283346ccc. [DOI] [PubMed] [Google Scholar]
- Blustein J, Attina T, Liu M, Ryan AM, Cox LM, Blaser MJ, Trasande L. Association of caesarean delivery with child adiposity from age 6 weeks to 15 years. Int J Obes. 2013;37:900–906. doi: 10.1038/ijo.2013.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cani PD, Bibiloni R, Knauf C, Waget A, Neyrinck AM, Delzenne NM, Burcelin R. Changes in gut microbiota control metabolic endotoxemia-induced inflammation in high-fat diet-induced obesity and diabetes in mice. Diabetes. 2008;57:1470–1481. doi: 10.2337/db07-1403. [DOI] [PubMed] [Google Scholar]
- Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pena AG, Goodrich JK, Gordon JI, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Huntley J, Fierer N, Owens SM, Betley J, Fraser L, Bauer M, et al. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 2012;6:1621–1624. doi: 10.1038/ismej.2012.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chai G, Governale L, McMahon AW, Trinidad JP, Staffa J, Murphy D. Trends of outpatient prescription drug utilization in US children, 2002–2010. Pediatrics. 2012;130:23–31. doi: 10.1542/peds.2011-2879. [DOI] [PubMed] [Google Scholar]
- Cho I, Yamanishi S, Cox L, Methe BA, Zavadil J, Li K, Gao Z, Mahana D, Raju K, Teitler I, et al. Antibiotics in early life alter the murine colonic microbiome and adiposity. Nature. 2012;488:621–626. doi: 10.1038/nature11400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung H, Pamp SJ, Hill JA, Surana NK, Edelman SM, Troy EB, Reading NC, Villablanca EJ, Wang S, Mora JR, et al. Gut immune maturation depends on colonization with a host-specific microbiota. Cell. 2012;149:1578–1593. doi: 10.1016/j.cell.2012.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clavenna A, Bonati M. Differences in antibiotic prescribing in paediatric outpatients. Arch Dis Child. 2011;96:590–595. doi: 10.1136/adc.2010.183541. [DOI] [PubMed] [Google Scholar]
- Coates ME, Fuller R, Harrison GF, Lev M, Suffolk SF. A comparison of the growth of chicks in the Gustafsson germ-free apparatus and in a conventional environment, with and without dietary supplements of penicillin. Br J Nutr. 1963;17:141–150. doi: 10.1079/bjn19630015. [DOI] [PubMed] [Google Scholar]
- Collins MD, Shah HN, Mitsuoka T. Reclassification of Bacteroides microfusus (Kaneuchi and Mitsuoka) in a New Genus Rikenella, as Rikenella microfusus comb. nov. Syst Appl Microbiol. 1985;6:79–81. [Google Scholar]
- Cox LM, Blaser MJ. Pathways in microbe-induced obesity. Cell Metab. 2013;17:883–894. doi: 10.1016/j.cmet.2013.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cromwell G. Why and how antibiotics are used in swine production. Anim Biotechnol. 2002;13:7–27. doi: 10.1081/ABIO-120005767. [DOI] [PubMed] [Google Scholar]
- Cunningham SA, Kramer MR, Narayan KMV. Incidence of childhood obesity in the United States. N Engl J Med. 2014;370:403–411. doi: 10.1056/NEJMoa1309753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniel H, Gholami AM, Berry D, Desmarchelier C, Hahne H, Loh G, Mondot S, Lepage P, Rothballer M, Walker A, et al. High-fat diet alters gut microbiota physiology in mice. ISME J. 2013;8:295–308. doi: 10.1038/ismej.2013.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis CP, Savage DC. Effect of penicillin on the succession, attachment, and morphology of segmented, filamentous microbes in the murine small bowel. Infect Immun. 1976;13:180–188. doi: 10.1128/iai.13.1.180-188.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dethlefsen L, Huse S, Sogin ML, Relman DA. The pervasive effects of an antibiotic on the human gut microbiota, as revealed by deep 16S rRNA sequencing. PLoS Biol. 2008;6:e280. doi: 10.1371/journal.pbio.0060280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietz WH. Critical periods in childhood for the development of obesity. Am J Clin Nutr. 1994;59:955–959. doi: 10.1093/ajcn/59.5.955. [DOI] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2012 doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez-Bello MG, Costello EK, Contreras M, Magris M, Hidalgo G, Fierer N, Knight R. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc Natl Acad Sci USA. 2010;107:11971–11975. doi: 10.1073/pnas.1002601107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubos R, Schaedler RW, Costello RL. The effect of antibacterial drugs on the weight of mice. J Exp Med. 1963;117:245–257. doi: 10.1084/jem.117.2.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erturk-Hasdemir D, Kasper DL. Resident commensals shaping immunity. Curr Opin Immunol. 2013;25:450–455. doi: 10.1016/j.coi.2013.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Everard A, Belzer C, Geurts L, Ouwerkerk JP, Druart C, Bindels LB, Guiot Y, Derrien M, Muccioli GG, Delzenne NM, et al. Crosstalk between Akkermansia muciniphila and intestinal epithelium controls diet-induced obesity. Proc Natl Acad Sci USA. 2013;110:9066–9071. doi: 10.1073/pnas.1219451110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FDA, U.S. Food a Drug Administration Approved animal drug products online (Green Book) 2014.
- Fierer N, Hamady M, Lauber CL, Knight R. The influence of sex, handedness, and washing on the diversity of hand surface bacteria. Proc Natl Acad Sci USA. 2008;105:17994–17999. doi: 10.1073/pnas.0807920105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaskins H, Collier C, Anderson D. Antibiotics as growth promotants: mode of action. Anim Biotechnol. 2002;13:29–42. doi: 10.1081/ABIO-120005768. [DOI] [PubMed] [Google Scholar]
- Goyal A, Dureja AG, Sharmaρ DK, Dhiman K. A comprehensive insight into the development of animal models for obesity research. Global Journal of Medical Research. 2012;12:39–44. [Google Scholar]
- Greetham HL, Gibson GR, Giffard C, Hippe H, Merkhoffer B, Steiner U, Falsen E, Collins MD. Allobaculum stercoricanis gen. nov., sp. nov., isolated from canine feces. Anaerobe. 2004;10:301–307. doi: 10.1016/j.anaerobe.2004.06.004. [DOI] [PubMed] [Google Scholar]
- Grudell ABM, Camilleri M. The role of peptide YY in integrative gut physiology and potential role in obesity. Curr Opin Endocrinol Diabetes Obes. 2007;14:52–57. doi: 10.1097/MED.0b013e3280123119. [DOI] [PubMed] [Google Scholar]
- Henao-Mejia J, Elinav E, Jin C, Hao L, Mehal WZ, Strowig T, Thaiss CA, Kau AL, Eisenbarth SC, Jurczak MJ, et al. Inflammasome-mediated dysbiosis regulates progression of NAFLD and obesity. Nature. 2012;482:179–185. doi: 10.1038/nature10809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooper LV, Littman DR, Macpherson AJ. Interactions between the microbiota and the immune system. Science. 2012;336:1268–1273. doi: 10.1126/science.1223490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh SY, Rifas-Shiman SL, Zera CA, Edwards JWR, Oken E, Weiss ST, Gillman MW. Delivery by caesarean section and risk of obesity in preschool age children: a prospective cohort study. Arch Dis Child. 2012;97:610–616. doi: 10.1136/archdischild-2011-301141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry RA. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 2003;31:15e–15. doi: 10.1093/nar/gng015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov II, Atarashi K, Manel N, Brodie EL, Shima T, Karaoz U, Wei D, Goldfarb KC, Santee CA, Lynch SV, et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell. 2009;139:485–498. doi: 10.1016/j.cell.2009.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang HQ, Bos NA, Cebra JJ. Timing, localization, and persistence of colonization by segmented filamentous bacteria in the neonatal mouse gut depend on immune status of mothers and pups. Infect Immun. 2001;69:3611–3617. doi: 10.1128/IAI.69.6.3611-3617.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalliomäki M, Collado MC, Salminen S, Isolauri E. Early differences in fecal microbiota composition in children may predict overweight. Am J Clin Nutr. 2008;87:534–538. doi: 10.1093/ajcn/87.3.534. [DOI] [PubMed] [Google Scholar]
- Karlsson CLJ, Molin G, Fåk F, Johansson Hagslätt M-L, Jakesevic M, Håkansson Å, Jeppsson B, Weström B, Ahrné S. Effects on weight gain and gut microbiota in rats given bacterial supplements and a high-energy-dense diet from fetal life through to 6 months of age. The British journal of nutrition. 2011;106:887–895. doi: 10.1017/S0007114511001036. [DOI] [PubMed] [Google Scholar]
- Knittle JL, Hirsch J. Effect of early nutrition on the development of rat epididymal fat pads: cellularity and metabolism. J Clin Invest. 1968;47:2091–2098. doi: 10.1172/JCI105894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langille MGI, Zaneveld J, Caporaso JG, Mcdonald D, Knights D, Reyes JA, Clemente JC, Burkepile DE, Thurber RLV, Knight R, et al. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nature Biotechnol. 2013;31:814–821. doi: 10.1038/nbt.2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leidy J. On the existence of entophyta in healthy animals, as a natural condition. Proc Acad Natl Sci Phila. 1849;4:225–233. [Google Scholar]
- Ley RE, Bäckhed F, Turnbaugh P, Lozupone CA, Knight RD, Gordon JI. Obesity alters gut microbial ecology. Proc Natl Acad Sci USA. 2005;102:11070–11075. doi: 10.1073/pnas.0504978102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markle JGM, Frank DN, Mortin-Toth S, Robertson CE, Feazel LM, Rolle-Kampczyk U, von Bergen M, McCoy KD, Macpherson AJ, Danska JS. Sex differences in the gut microbiome drive hormone-dependent regulation of autoimmunity. Science. 2013;339:1084–1088. doi: 10.1126/science.1233521. [DOI] [PubMed] [Google Scholar]
- Martinez I, Wallace G, Zhang C, Legge R, Benson AK, Carr TP, Moriyama EN, Walter J. Diet-induced metabolic improvements in a hamster model of hypercholesterolemia are strongly linked to alterations of the gut microbiota. Appl Environ Microbiol. 2009;75:4175–4184. doi: 10.1128/AEM.00380-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Million M, Angelakis E, Paul M, Armougom F, Leibovici L, Raoult D. Comparative meta-analysis of the effect of Lactobacillus species on weight gain in humans and animals. Microb Pathog. 2012;53:100–108. doi: 10.1016/j.micpath.2012.05.007. [DOI] [PubMed] [Google Scholar]
- Morrison CD, Huypens P, Stewart LK, Gettys TW. Implications of crosstalk between leptin and insulin signaling during the development of diet-induced obesity. Molec Basis Dis. 2009;1792:409–416. doi: 10.1016/j.bbadis.2008.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy R, Stewart AW, Braithwaite I, Beasley R, Hancox RJ, Mitchell EA. Antibiotic treatment during infancy and increased body mass index in boys: an international cross-sectional study. Int J Obes. 2013 doi: 10.1038/ijo.2013.218. doi: 10.1038/ijo.2013.1218. [DOI] [PubMed] [Google Scholar]
- Pantoja-Feliciano IG, Clemente JC, Costello EK, Perez ME, Blaser MJ, Knight R, Dominguez-Bello MG. Biphasic assembly of the murine intestinal microbiota during early development. ISME J. 2013;7:1112–1115. doi: 10.1038/ismej.2013.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersson US, Waldén TB, Carlsson P-O, Jansson L, Phillipson M. Female mice are protected against high-fat diet induced metabolic syndrome and increase the regulatory T cell population in adipose tissue. PLoS ONE. 2012;7:e46057. doi: 10.1371/journal.pone.0046057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasouli N, Molavi B, Elbein SC, Kern PA. Ectopic fat accumulation and metabolic syndrome. Diabetes Obes Metab. 2007;9:1–10. doi: 10.1111/j.1463-1326.2006.00590.x. [DOI] [PubMed] [Google Scholar]
- Ravussin Y, Koren O, Spor A, LeDuc C, Gutman R, Stombaugh J, Knight R, Ley RE, Leibel RL. Responses of Gut Microbiota to Diet Composition and Weight Loss in Lean and Obese Mice. Obesity. 2012;20:738–747. doi: 10.1038/oby.2011.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeder SB, Pineda AR, Wen Z, Shimakawa A, Yu H, Brittain JH, Gold GE, Beaulieu CH, Pelc NJ. Iterative decomposition of water and fat with echo asymmetry and least-squares estimation (IDEAL): application with fast spin-echo imaging. Magn Reson Med. 2005;54:636–644. doi: 10.1002/mrm.20624. [DOI] [PubMed] [Google Scholar]
- Reeder SB, Wen Z, Yu H, Pineda AR, Gold GE, Markl M, Pelc NJ. Multicoil Dixon chemical species separation with an iterative least-squares estimation method. Magn Reson Med. 2004;51:35–45. doi: 10.1002/mrm.10675. [DOI] [PubMed] [Google Scholar]
- Ridaura VK, Faith JJ, Rey FE, Cheng J, Duncan AE, Kau AL, Griffin NW, Lombard V, Henrissat B, Bain JR, et al. Gut microbiota from twins discordant for obesity modulate metabolism in mice. Science. 2013;341:1079. doi: 10.1126/science.1241214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuel BS, Shaito A, Motoike T, Rey FE, Backhed F, Manchester JK, Hammer RE, Williams SC, Crowley J, Yanagisawa M, et al. Effects of the gut microbiota on host adiposity are modulated by the short-chain fatty-acid binding G protein-coupled receptor, Gpr41. Proc Natl Acad Sci USA. 2008;105:16767–16772. doi: 10.1073/pnas.0808567105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaedler R, Dubos R, Costello R. The development of the bacterial flora in the gastrointestinal tract of mice. J Exp Med. 1965;122:59–66. doi: 10.1084/jem.122.1.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, Garrett WS, Huttenhower C. Metagenomic biomarker discovery and explanation. Genome Biol. 2011;12:R60. doi: 10.1186/gb-2011-12-6-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth GK. Linear Models and Empirical Bayes Methods for Assessing Differential Expression in Microarray Experiments. Stat Appl Genet Mol Biol. 2004;3 doi: 10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]
- Stappenbeck TS, Hooper LV, Gordon JI. Developmental regulation of intestinal angiogenesis by indigenous microbes via Paneth cells. Proc Natl Acad Sci USA. 2002;99:15451–15455. doi: 10.1073/pnas.202604299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trasande L, Blustein J, Liu M, Corwin E, Cox LM, Blaser MJ. Infant antibiotic exposures and early-life body mass. Int J Obes. 2013;37:16–23. doi: 10.1038/ijo.2012.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444:1027–1131. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- Vijay-Kumar M, Aitken JD, Carvalho FA, Cullender TC, Mwangi S, Srinivasan S, Sitaraman SV, Knight R, Ley RE, Gewirtz AT. Metabolic syndrome and altered gut microbiota in mice lacking Toll-like receptor 5. Science. 2010;328:228–231. doi: 10.1126/science.1179721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visek W. The mode of growth promotion by antibiotics. J Anim Sci. 1978;46:1447. [Google Scholar]
- Vucenik I, Stains JP. Obesity and cancer risk: evidence, mechanisms, and recommendations. Ann NY Acad Sci. 2012;1271:37–43. doi: 10.1111/j.1749-6632.2012.06750.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yurkovetskiy L, Burrows M, Khan AA, Graham L, Volchkov P, Becker L, Antonopoulos D, Umesaki Y, Chervonsky AV. Gender bias in autoimmunity is influenced by microbiota. Immunity. 2013;39:400–412. doi: 10.1016/j.immuni.2013.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.