Abstract
In this study, we investigated the capacity of chronic lymphocytic leukemia (CLL) B cells to undergo terminal differentiation into Ig-secreting plasma cells in T cell-independent and T cell-dependent responses. We used a two-step model involving stimulation with phorbol myristate acetate (PMA) and CD40L, together with cytokines (PMA/c and CD40L/c), for 7 days. We describe immunophenotypic modifications, changes in the levels of mRNA and protein for transcription factors and morphological and functional events occurring during the differentiation of CLL B cells into antibody-secreting cells (ASCs). The induction of differentiation differed significantly between the CD40L/c and PMA/c culture systems. The PMA/c culture system allowed CLL B cells to differentiate into IgM-secreting cells with an immunophenotype and molecular profile resembling those of preplasmablasts. By contrast, CD40L/c-stimulated cells had a phenotype and morphology similar to those of activated B cells and resembling those of the CLL B cells residing in the lymph node and bone marrow. These data suggest that the CLL B cells are not frozen permanently at a stage of differentiation and are able to differentiate into ASCs as appropriate stimulation are provided. The data presented here raise questions about the molecular processes and stimulation required for CLL B-cell differentiation and about the inability of CD40 ligand to induce differentiation of the CLL B cells.
B cells are important vectors of humoral immunity. Mature B cells differentiate into antibody-secreting cells (ASCs)—plasma cells, the terminal effector cells of the B-cell lineage. Mature B cells may follow two pathways after antigen engagement. The B cells may proliferate and then differentiate into plasmablasts, which produce antibodies (Abs) and then differentiate into short-lived (3–5 days) plasma cells in extrafollicular foci.1 These extrafollicular ASCs are generated at the start of the immune response and are a major source of germline IgM Abs.2 Alternatively, the B cells may differentiate into long-lived plasma cells, which are found preferentially in the bone marrow and are derived mainly from germinal centers.1, 2 B cells also leave the germinal center as memory B cells, which can rapidly differentiate into ASCs after re-exposure to antigen.1, 2
Plasma cells are generated by a highly regulated differentiation process, involving profound phenotypic, molecular and morphologic changes rendering the cell capable of producing large amounts of Abs.2, 3 The full plasma cell phenotype consists of a loss of B-cell markers (CD20, PAX5 and the products of the genes it regulates: BCL6, BACH2 and IRF8) and a gain of plasma cell markers (CD38, CD138, IRF4, BLIMP1 and XBP1s). PAX5 and BLIMP1 are the key transcription factors controlling the differentiation of B cells and plasma cells, respectively.2 PAX5 is essential for the development and maturation of B cells. It activates genes associated with B-cell function, while also repressing genes associated with plasma cell development and function, including the gene encoding the transcription factor XBP1, which is responsible for the formation of the machinery required for the production of large amounts of Abs.3, 4, 5 By contrast, BLIMP1 is essential for ASC differentiation and survival.3 BLIMP1 controls the genes involved in the secretion of immunoglobulins, such as IgH, IgL, the J chain and XBP1.3 BLIMP1 represses genes expressed in mature B cells, such as the PAX5, SPIB, CTIIA, AID and BCL6 genes.3, 6 The reciprocal inhibitory effects of PAX5 and BLIMP1 suggest that these two factors are at the heart of the molecular events occurring at the point of divergence of the B-lymphocyte and plasma cell lineages. Indeed, it has been shown that both naive and memory mature B cells can be induced to undergo terminal differentiation into Ig-secreting plasmablast/plasma cells by stimulation with CD40L and cytokines (in a CD40L system or with bystander help) (reviewed in Neron et al.7).8, 9, 10 Moreover, cytokines (such as interleukin (IL)-2,8, 11, 12 IL-4,8, 13 IL-6,12 IL-10,8, 12 and IL-128, 10) have an important role in the differentiation of B cells into Ig-secreting plasma cells.
Chronic lymphocytic leukemia B cells have been used as a model for studies of human B-cell differentiation.14 B-cell chronic lymphocytic leukemia (CLL) is the most frequent adult leukemia in the Western world. It is a heterogeneous disease, characterized by clonal proliferation and the accumulation of mature CD5+ B lymphocytes.15 The tissue microenvironment has a key role in the pathogenesis and progression of CLL.16 It is assumed that in vivo CLL B cells continuously transit from their niche in the lymph nodes and bone marrow to peripheral blood. Evidence from several studies suggests that CD40L, antigenic stimulation and microenvironment-derived cytokines are important factors in CLL.16, 17 The CLL microenvironment includes not only malignant B cells with an activated phenotype, but also networks of follicular dendritic cells and activated T helper cells (CD40L+ and IL4+), stromal cells and soluble factors.15, 18 Given the particular profile of the activated CLL B cells18 all of these elements may constitute a favorable environment for the terminal differentiation of these leukemic cells. By analogy to the situation occurring in classical lymphoid follicles, in which B cells are activated by antigen and T helper cells (CD40–CD40L interaction), we can imagine a scenario in which CLL B cells interact with CD40L+ activated T helper cells or bind to (auto)antigens,16, 19 then receive microenvironment-derived cytokines that induce their differentiation into ASCs.17 However, the impact of such a scenario on the differentiation of CLL B cells remains unclear. Nevertheless, the differentiation of CLL cells into Ig-secreting plasma cells has been shown to occur spontaneously in vivo.20, 21 In vitro activation of CD40 by CD40L protects CLL B cells from apoptosis by upregulation of the anti-apoptotic Bcl-2 family proteins Bcl-xL and Mcl-1, induction of Survivin and activation of the NFκB pathway.22, 23 In addition to maintaining CLL cell survival, CD40 signaling has been shown in many studies to induce proliferation of CLL B cells24, 25, 26 and to increase expression of CD80 and CD86.27, 28 Furthermore, it was very recently shown that the gene expression signature induced in CLL B cells by autologous activated T cells is very similar to that induced by CD40L stimulation.29
Protein kinase C (PKC) pathway is critically involved in B-cell function and development. During B-cell receptor (BCR) signaling, PKC contribute to the activation of NFκB and the RAS–MAPK–ERK signaling pathway.30 Signaling by BCR is crucial for the development, activation and differentiation of B cells.30 PKC is expressed in CLL and has critical roles in CLL pathogenesis.31 However, in CLL, BCR surface expression is usually low and BCR signaling is impaired in almost half of cases.32 Furthermore, IgM ligation of the BCR does not lead to phosphotyrosine induction or calcium flux.32 This finding is a characteristic of patients exhibiting indolent disease.32 Indeed, CD38-negative CLL B cells display reduced ability to transduce BCR-mediated signaling and to respond to BCR crosslinking.33 Phorbol myristate acetate (PMA) activates the PKC pathway by mimicking diacylglycerol, a natural ligand and activator of PKCs.34 The PMA is a mitogen and polyclonal activator of normal B cells and CLL B cells.35, 36 PMA induces the differentiation of CLL B cells in a T cell-independent manner.14, 36 It has a specific effect on CLL B cells, with no effect on other B-cell malignancies, and this effect requires the presence of IL-4.14, 37 However, to the best of our knowledge, a detailed characterization of resulting ASCs was not reported. Nonetheless, we showed in a previous study that several microenvironment components and effective factors involved in B-cell activation and differentiation, such as PMA and IL-4 in particular, support the survival of CLL B cells in vitro.17
In this study, we purified CD20-positive (CD20pos) CLL B cells from uniformly CD38-negative (CD38neg) patients, because (i) CD38pos CLL B cells are thought to express a more activated phenotype than CD38neg CLL B cells38 and (ii) the CD20posCD38neg phenotype corresponds to that of naive B cells (Bm1) and memory B cells (Bm5) described by Bohnhorst et al.,39 both these cell types being candidates for normal counterparts of CLL B cells.40 Chronic lymphocytic leukemia B cells were cultured in two different culture conditions: (i) the CD40L culture system, as described by Tarte et al.8 and referred to here as CD40L/c; (ii) a culture system in which CD40L was replaced with PMA (PMA/c). We sought (i) to characterize the ASCs generated from CLL B cells, and (ii) to investigate the fate of CLL B cells in terms of their differentiation in the CD40L culture system.
RESULTS
PMA/c-stimulated cells present a preplasmablast immunophenotype
PMA, alone or together with cytokines, has been shown to induce the differentiation of CLL B cells into plasmacytoid cells.14, 36, 37 However, to the best of our knowledge, the CD40L culture system has never before been used to study the differentiation of CLL B cells. A combination of CD40L and cytokines, as described by Tarte et al.,8 has been shown to induce the differentiation of mature B cells into CD20negCD38high plasmablasts/plasma cells. We validated this system for our cultures, using B cells from healthy donors (normal B cells) as a positive control. We stimulated CLL B cells and normal B cells with PMA or CD40L, in combination with the cytokines IL-2, IL-4, IL-10 and IL-12. On day (D)4, cells were harvested and incubated with IL-2, IL-6, IL-10 and IL-12 for 3 days.
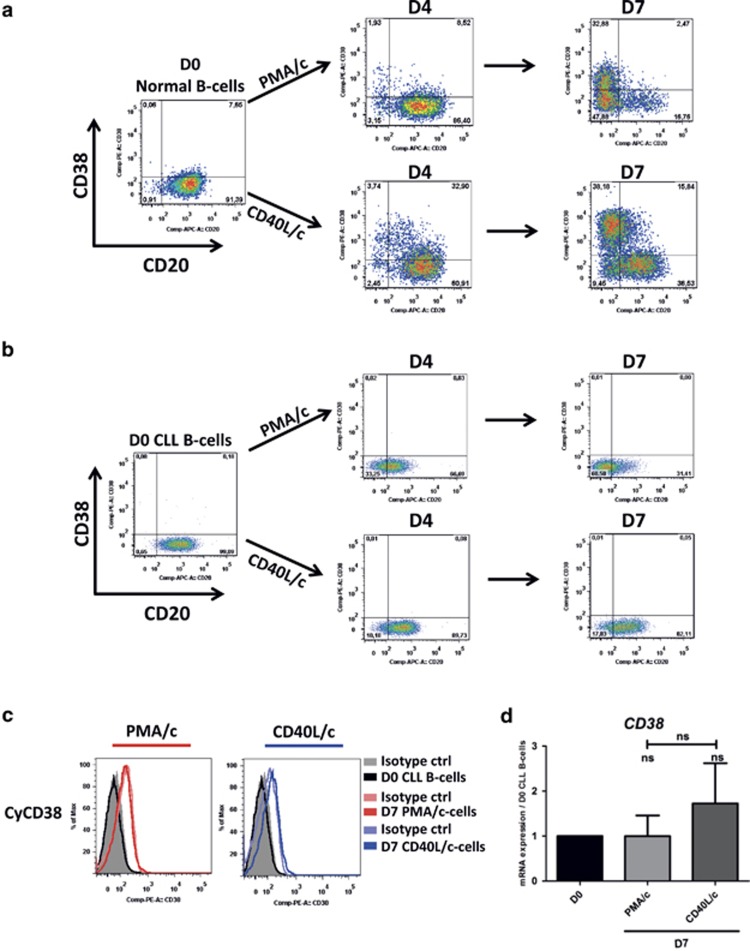
Double staining for CD20 and CD38 on the cell surface was performed and monitored by flow cytometry on D4 and D7. The expression of CD38, a marker of B-cell activation and plasma cell differentiation, on the surface of normal B cells increased with each step in the CD40L/c culture system (Figure 1a). On D7, a population of CD20negCD38high cells was detected, indicating the correct functioning of the CD40L/c culture system (Figure 1a). In PMA/c conditions, no CD38 expression was detected on D4 and the expression of this marker on D7 was weaker than that on cells in CD40L/c conditions (Figure 1a). However, for CLL B cells, no CD38 expression on the cell surface was detected in either PMA/c or CD40L/c conditions, on D4 or D7 (Figure 1b). We investigated the possibility that CD38 was expressed but not transferred to the cell surface, by evaluating the cytoplasmic expression of CD38 and CD38 mRNA levels. On D7, we found no CD38 in the cytoplasm and detected no mRNA for this marker in either PMA/c- or CD40L/c-stimulated cells (Figures 1c and d).
Figure 1.
CD20 and CD38 expression on normal and CLL B cells in the PMA/c and CD40L/c culture systems. Cells were stimulated with PMA or CD40L, in combination with the cytokines IL-2, IL-4, IL-10 and IL-12. On D4, cells were harvested and incubated with IL-2, IL-6, IL-10 and IL-12 for 3 days. (a, b) Cells were labeled with anti-CD20 and anti-CD38 mAbs to explore changes in the surface expression of these proteins between D0, D4 and D7 on normal (a) and CLL B cells (b). The cytometry plots are representative of two and twelve independent experiments for normal and CLL B cells, respectively. (c) Cells were labeled with anti-CD38 antibody after permeabilization. Cytometry plots for cytoplasmic CD38 are representative of twelve independent experiments. (d) The expression of the CD38 gene was evaluated by quantitative real-time RT-PCR on D0 CLL B cells and D7 stimulated cells. Results are expressed relative to gene expression in CLL B cells on D0, according to the 2−ΔΔCT method. Bars represent mean values±s.e.m. from five independent experiments. Statistical significance was calculated using the Wilcoxon test. ns, not significant. D, day. Cy, cytoplasmic.
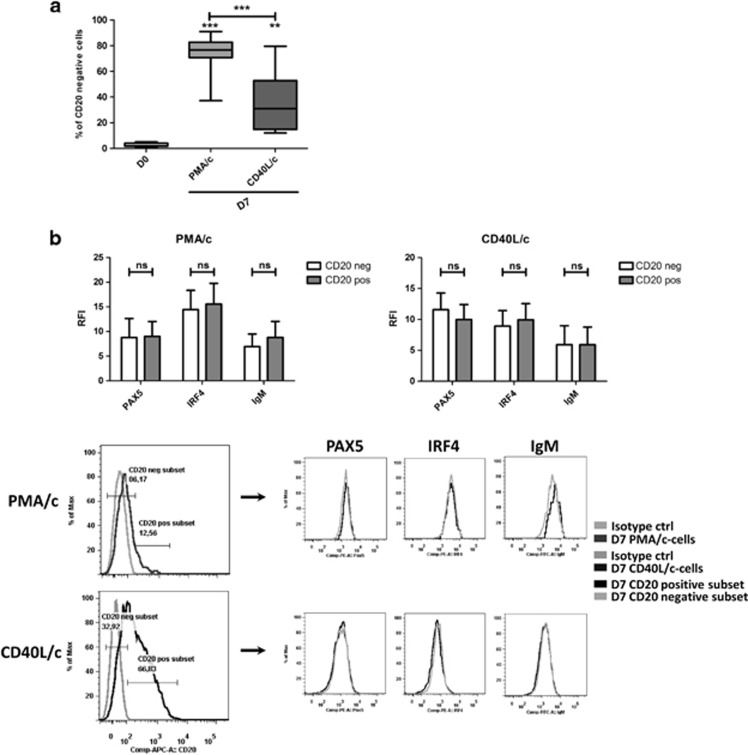
However, consistent with differentiation into plasma cells, which is characterized by a downregulation or lack of CD20 expression, both sets of conditions generated a CD20neg population. Significantly more of these cells were produced in PMA/c conditions (75%±13) than in CD40L/c conditions (36%±24) (Figure 2a). Indeed, ASC differentiation has been reported to be associated with an upregulation of IRF4, a downregulation of PAX5 and an increase in IgM synthesis.3, 41 For confirmation of the validity of the link established between the generation of a CD20-negative population and plasma cell differentiation in our system, we investigated the levels of expression of PAX5, IRF4 and IgM in the CD20-positive and -negative populations on D7, for both sets of conditions. On D7, cells were permeabilized and stained for CD20, PAX5, IRF4 and IgM. No significant difference in the expression of these molecules was observed between the CD20pos and CD20neg populations (Figure 2b), indicating that changes in CD20 expression are not a conclusive marker of differentiation, at least in our system. The expression of PAX5 by CD20neg cells was unexpected, but PAX5 expression has been detected in CD20neg B-cell lymphoma in the absence of plasmacytic differentiation.42
Figure 2.
CD20neg and CD20pos cells generated from CLL B cells in the PMA/c and CD40L/c culture system showed similar levels of expression of PAX5, IRF4 and IgM. (a) Percentages of CD20-negative cells on D7 in PMA/c and CD40L/c culture conditions. Data are represented as box and whisker (min to max) plots for 12 independent experiments. Statistical significance was calculated using the Wilcoxon test. (b) Cells were labeled with anti-PAX5, anti-IRF4 and anti-IgM Abs after permeabilization. Upper panel: Relative fluorescence intensities (RFIs were calculated as the ratio of the MFI of cells labeled with a specific Ab to that of cells labeled with a matched isotype control. Bars represent mean RFI values±s.e.m. for three independent experiments. Lower panel: Cytometry plots from a representative patient. Significance was calculated using the paired Student's t-test. **P<0.01, ***P<0.001. ns, not significant. D, day.
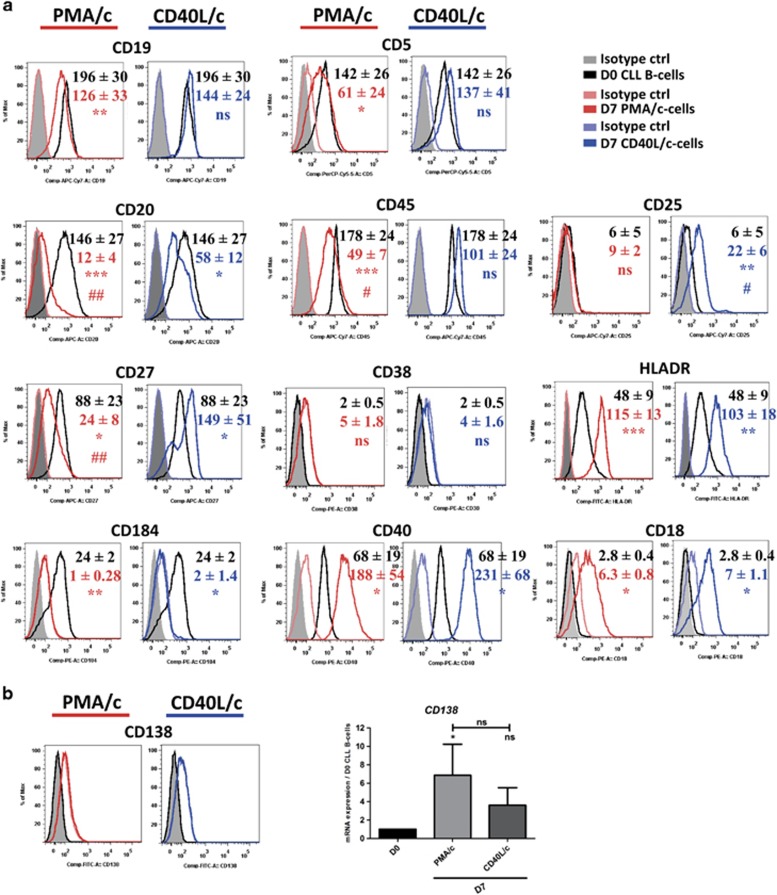
We addressed the question of changes in cell phenotype at D7, by staining cells for CD19, CD20, CD27, CD5, CD45, CD38, CD25, CD138, CD184, CD40, CD18 and HLA-DR (Figure 3a). Consistent with an ASC phenotype, PMA/c-stimulated cells had lower levels of CD19, CD20 and CD45 expression on their surface than on D0. CD40L/c-stimulated cells had significantly lower levels of CD20 expression than on D0, whereas the decrease in the levels of expression of CD19 and CD45 was not significant. A significant induction of CD25 and CD27 expression was observed on D7 CD40L/c-stimulated cells. By contrast, on D7 PMA/c-stimulated cells, CD27 was significantly downregulated and CD25 was not expressed. Cells in both sets of conditions displayed high levels of HL-ADR, CD40 and CD18 expression, but they also displayed a significant downregulation of CD184 (CXCR4). CD138 (also known as syndecan1), the hallmark of terminally differentiated human plasma cells, was not detected on the cell surface but an increase in mRNA levels for this marker was observed (Figure 3b). The immunophenotype, with an absence of documented B-cell and plasma cell marker (CD20, CD38, CD138) expression and weak expression of CD27, is consistent with the recently described phenotype of a transitional population of preplasmablasts generated by in vitro differentiation from human memory B cells.43
Figure 3.
The immunophenotype of D7 cells. (A) On D0 and D7, cell immunophenotype was studied by direct labeling of CD19, CD20, CD27, CD184, CD5, CD45, CD38, CD40, CD25, HLA-DR, CD18 and CD138. (a) RFIs were calculated as the ratio of the MFI of cells labeled with a specific Ab to that of cells labeled with a matched isotype control. Mean RFI values±s.e.m. from eight independent experiments are represented with a color code: Black: D0, red: D7 PMA/c-stimulated cells, blue: D7 CD40L/-stimulated cells. Cytometry data are presented as plots for a representative patient. (b) Cytometry plot of CD138 expression for a representative patient. The expression of the CD138 gene was evaluated by quantitative real-time RT-PCR in D0 CLL B cells and D7 stimulated cells. Results are expressed relative to gene expression in CLL B cells on D0, according to the 2−ΔΔCT method. Bars represent mean values±s.e.m. from five independent experiments. Statistical significance was calculated using a the Wilcoxon test. *The D7 value is different from that in D0 CLL B cells. #The D7 value is different between PMA/c- and CD40L/-stimulated cells. *, #P<0.05, **, ##P<0.01, ***P<0.001. ns, not significant. D, day.
Unlike CD40L/c stimulation, PMA/c stimulation induces the differentiation of CLL B cells into IgM-secreting cells
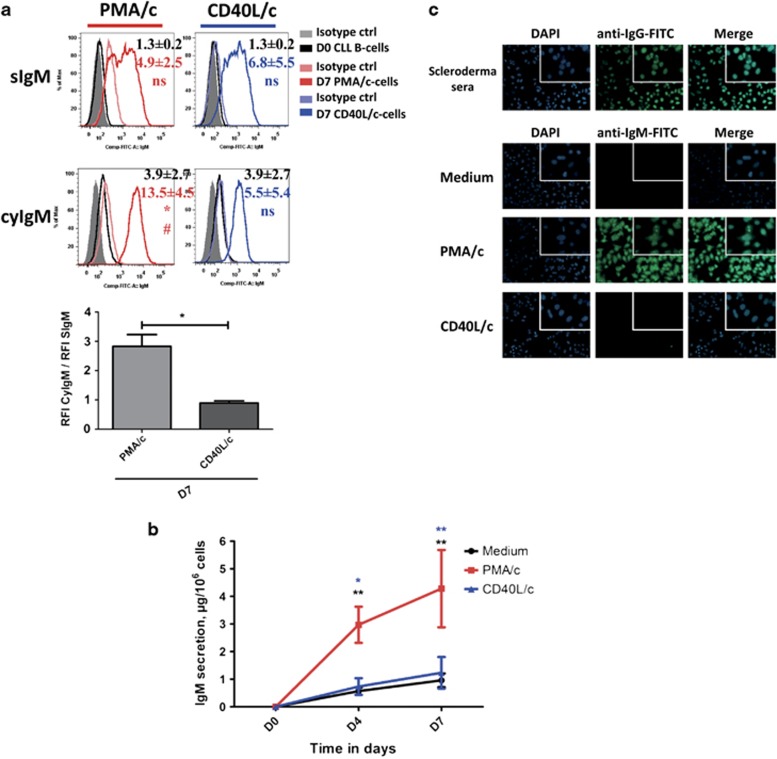
We investigated whether PMA/c-stimulated cells differentiated into functional ASCs, by assessing the secretion and surface/cytoplasmic expression of immunoglobulin. Figure 4a and Supplementary Figure 1A show the surface expression and (after permeabilization) cytoplasmic expression of IgM and IgG. On D0, IgM was absent from the surface, or present in only small amounts (as shown in other studies44, 45). However, IgM was detected in the cytoplasm, whereas no surface or cytoplasmic expression of IgG was detected. On D7, IgM expression on the cell surface and in the cytoplasm were detected for both PMA/c- and CD40L/c-stimulated cells (Figure 4a). Nevertheless, in PMA/c-stimulated cells, the relative fluorescence intensity (RFI) of IgM tripled after cell permeabilization, indicating that these cells were strongly involved in cytoplasmic Ig production (Figure 4a). No surface or cytoplasmic expression of IgG was detected (Supplementary Figure 1A). We used enzyme-linked immunosorbent assay to investigate IgM, IgG, and IgA secretion into the culture supernatants on D4 and D7 (Figure 4b and Supplementary Figure 1B). Unlike CD40L/c-stimulated cells or cells cultured in the absence of stimulation (medium only), PMA/c-stimulated cells produce substantial amounts of IgM (Figure 4b). Secreted IgM was detected as early as D4 (Figure 4b). The amounts of IgM produced by CD40L/c-stimulated cells were similar to those produced by control cells (medium-only control conditions). Very little secreted IgG and IgA was detected in cell culture supernatants for either set of conditions (Supplementary Figure 1B).
Figure 4.
IgM expression and secretion by PMA/c- and CD40L/c-stimulated cells. (a) D0 CLL B cells and D7 PMA/c- and CD40L/c-stimulated cells were labeled before and after permeabilization with fluorescein isothiocyanate-conjugated anti–human IgM mAbs or isotype control mAbs. The cytometry plots are representative of three independent experiments. The bar histogram shows the ratio of RFI for cytoplasmic IgM to that for surface IgM. RFI was calculated as the ratio of the MFI of cells labeled with a specific Ab to that of cells labeled with a matched isotype control. Data are representative of three independent experiments. Significance was calculated using the paired Student's t-test. (b) Culture supernatants were harvested on D4 and D7. IgM secretion was assessed with an enzyme-linked immunosorbent assay. The results are expressed as the mean±s.e.m. (in μg per 106 cells) for five independent experiments. Significance was calculated using the Wilcoxon test: *P<0.05, **P<0.01. S, surface, Cy, cytoplasmic, D, day. (c) Indirect immunofluorescence analysis showed that the IgM secreted into the culture supernatant of PMA/c-stimulated cells recognized autoantigens in HEp-2 cells. Magnification: × 200. Magnification for insets: × 630.
Chronic lymphocytic leukemia B cells are known to display a poly/autoreactive BCR.15 Indeed, several groups have reported that a large proportion of CLL cases have stereotyped BCRs with very similar complementarity-determining region 3 domains.15 The BCRs of unmutated CLL cells have been reported to display autoreactivity and polyreactivity, in contrast to those of mutated CLL cells. Several studies have demonstrated that CLL cells bind diverse autoantigens, including human IgG, cytoskeletal proteins, including vimentin, filamin B and cofilin-1, myosin heavy chain 2A, double-stranded DNA and self-antigens associated with apoptosis, such as oxidized LDL.46, 47 Thus, we determined whether the secreted IgM Abs were self-reactive, by performing indirect immunofluorescence assayss on fixed HEp-2 cells (Figure 4c). We found that, in three of the five CLLs, the IgM secreted by PMA/c-stimulated cells displayed a nuclear or nuclear/cytoplasmic staining pattern, whereas no staining was observed in untreated cells or CD40L/c-stimulated cells (Figure 4c).
On D7, only PMA/c-stimulated CLL B cells have an ASC morphology
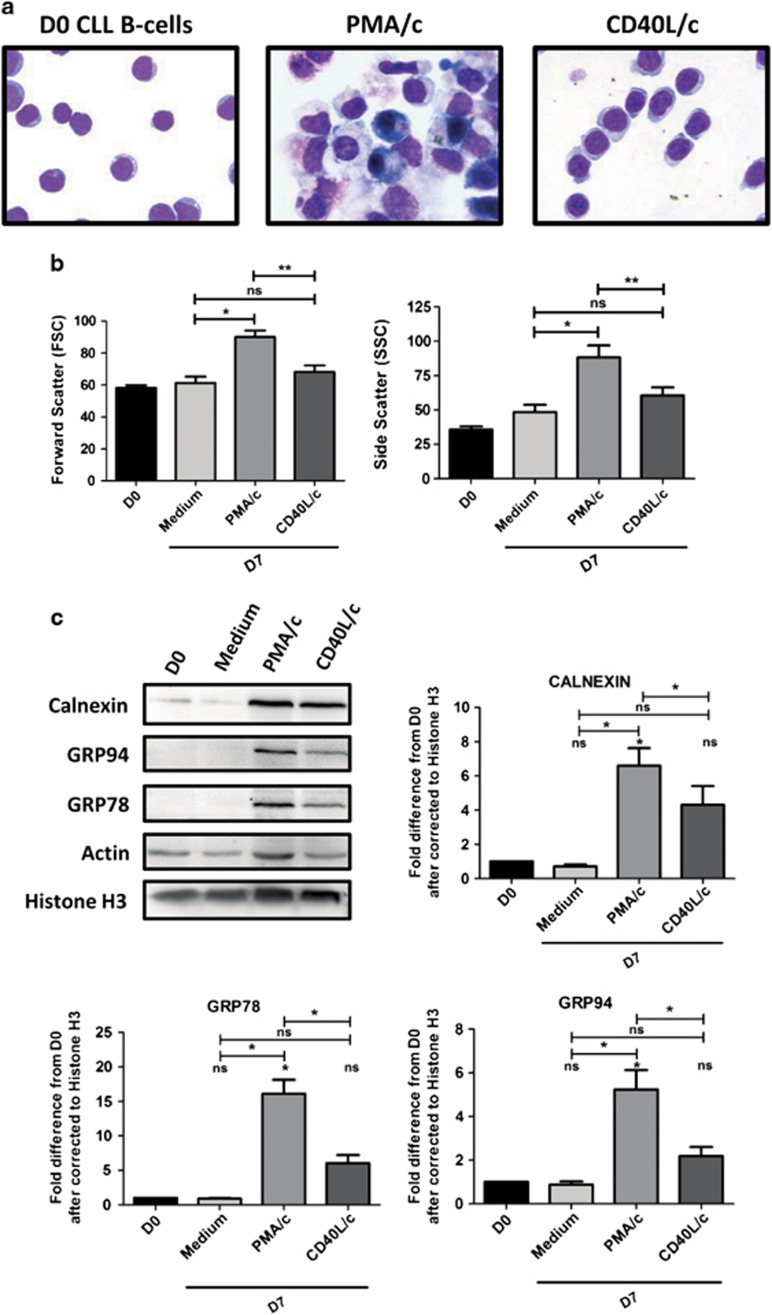
Having shown that PMA/c-stimulated cells presented a preplasmablast immunophenotype and secreted large amounts of Abs, we addressed the question of the changes in morphology and structure allowing them to secrete such large quantities of immunoglobulin. As shown by May–Grünwald–Giemsa staining (Figure 5a), on D0, CLL B cells had a small cell body, with a normal-shaped nucleus surrounded by a thin ring of cytoplasm. On D7, PMA/c-stimulated cells had a morphology typical of ASCs, with an eccentric nucleus and abundant cytoplasm, in which expansion of the endoplasmic reticulum (ER) and Golgi compartments was evident (Figure 5a). By contrast, CD40L/c-stimulated cells presented an activated B-cell or paraimmunoblast morphology (large lymphocytes), histological hallmarks specific to the CLL B cells resident in lymph nodes and bone marrow. However, the cytoplasm was clearly less well developed than that in PMA/c-stimulated cells (Figure 5a). Consistent changes in cell morphology and structure were also monitored by flow cytometry, on the basis of the relative sizes of the cells (forward scatter) and their granulometry (side scatter) (Figure 5b). In PMA/c-stimulated cells, the significant changes in light-scattering properties observed, together with the large increases in cell size, are consistent with the increase in cytoplasm content required for efficient antibody secretion. These data suggest that PMA/c-stimulated cells have the cytoplasmic structure required for Ig secretion.
Figure 5.
Morphological analysis and UPR induction. (a) CLL B cells on D0 and stimulated cells on D7 were stained with May-Grünwald-Giemsa reagent. Original magnification: × 1000. (b) Cell size and granularity were measured by flow cytometry. Relative cell size was determined by assessing the light diffracted at small angles (detected as forward scatter ). Granularity is proportional to the light diffracted at large angles (detected as side scatter). Bars represent mean values±s.e.m. from twelve independent experiments. Significance was calculated using the Wilcoxon test. (c) Immunoblot analysis and densitometry quantification of calnexin, GRP94, GRP78, actin, histone H3 protein in D0 CLL B cells, D7 non-stimulated cells (Medium), D7 PMA/c-stimulated cells and D7 CD40L/c-stimulated cells. Data are shown as mean values±s.e.m. for three independent experiments. Statistical significance was calculated using the Student's t-test. *P<0.05, **P<0.01, ns, not significant, D, day.
We studied the underlying molecular mechanism, by assessing the expression, on D7, by PMA/c-stimulated cells, of molecules implicated in the unfolded protein response (UPR).The accumulation of proteins in the lumen of the ER, whether physiological (secretory cells) or pathophysiological, triggers the UPR. The UPR is an elegant signaling system that regulates the folding, processing and transport of newly synthesized proteins (both surface and secreted proteins) across the ER membrane.48 Cells respond to the presence of unfolded proteins by the upregulation of ER-chaperones, such as GRP78/Bip and GRP94, which assist protein folding.49 The UPR is induced during the terminal differentiation of B cells.50, 51 We investigated whether the expression of components of the UPR pathway was induced by stimulation, in PMA/c- and CD40L/c-stimulated cells, the induction of the UPR targets GRP78 and GRP94, and induction of calnexin, a component of the protein folding machinery of the ER.49, 52 Compared to CLL B cells at D0, CD40L/c-stimulated cells had a slight increase in levels of calnexin (4±1.8 times higher), GRP78 (6±2 times higher) and GRP94 (2±0.7 times higher). However, PMA/c-stimulated cells had significantly higher levels of calnexin (7±2 times higher), GRP78 (16±4 times higher) and GRP94 (5±1.5-fold higher) than control cells and cells in CD40L/c conditions (Figure 5c). These data are consistent with an induction of the UPR, an important component of ASC differentiation.
IgM secretion by PMA/c-stimulated cells on D7 is dependent on transcriptional regulators previously identified in normal ASCs
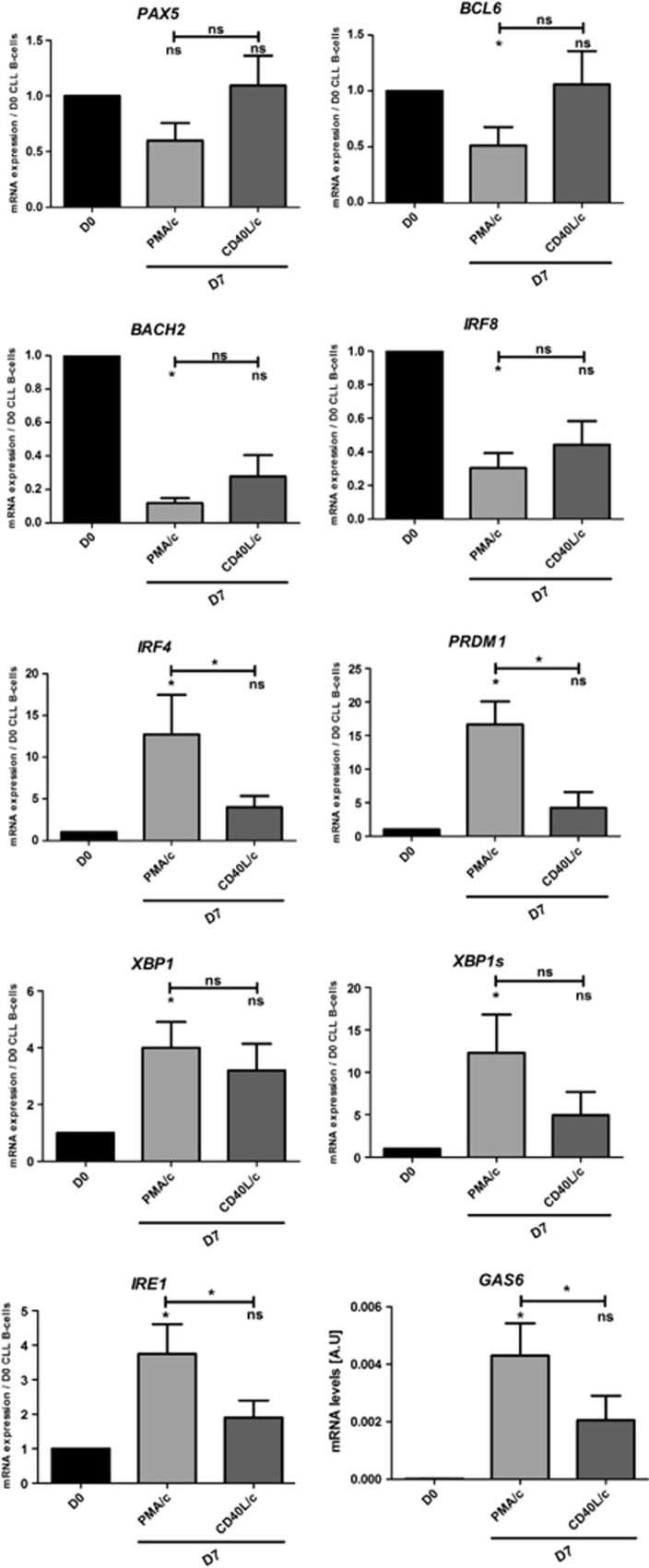
We investigated the molecular mechanism by which stimulation affected the transcriptional program involved in the terminal differentiation of CLL B cells into ASCs by quantitative reverse transcription-PCR (RT-PCR), immunoblotting and flow cytometry. We studied mRNA levels for several transcription factors found in B cells (PAX5, BCL6, BACH2 and IRF8) and involved in plasma cell differentiation (IRF4, PRDM1, XBP1 (unspliced and spliced form), IRE1 and GAS6). It has been shown that the repression of PAX5 and of the genes it regulates (IRF8 and BACH2) is an important step toward ASC differentiation.4, 53, 54 BCL6 has been shown to inhibit ASC differentiation by directly repressing BLIMP1.55 Conversely, upregulation of the transcription factor IRF4 and of BLIMP1 is required for complete ASC differentiation.6, 41 During ASC differentiation, the upregulation of IRF4 is associated with downregulation of the germinal center marker BCL6 and a loss of PRDM1 repression, resulting in the downregulation of PAX5 gene expression.41, 56 Moreover, high levels of IRF4 expression induce the expression of PRDM1 and XBP1.41, 56 Both these genes encode transcription factors involved in ASC differentiation and Ig secretion.5 Consistent with the molecular phenotype of ASCs, our quantitative RT-PCR data show that PMA/c-stimulated cells display a downregulation of B-cell transcription factor genes, including PAX5, IRF8, BACH2 and BCL6, and a significant upregulation of the plasma cell marker genes IRF4 (13±10-fold), PRDM1 (17±8-fold) and XBP1 (4±2-fold) (Figure 6). XBP1 is a transcription factor involved in plasma cell differentiation that also plays a role in the UPR and is essential for the secretion of large amounts of Ig by plasma cells.5, 50 Only the spliced form of XBP1 (XBP1s) can activate the UPR efficiently.51 Our data show that XBP1s was significantly upregulated (12±11-fold) by stimulation. Moreover, mRNA levels for IRE1 (encoding ERN1, which induces XBP1 mRNA splicing5), were also significantly increased, by a factor of 4±2-fold (Figure 6). We also studied mRNA levels for growth arrest-specific gene 6 (GAS6). Gene expression profiling data showed that GAS6 was more strongly expressed in plasmablasts and plasma cells than in B cells.57 GAS6 mRNA is present at negligible levels in precursor B cells and B-cell lines but is detected in terminally differentiated plasma cell lines.58 In our study, on D0, no mRNA for GAS6 was detected in any of the CLL B cells. On D7, PMA/c-stimulated cells contained substantial amounts of GAS6 mRNA (Figure 6), indicating that these cells had switched to the plasma cell compartment.
Figure 6.
Day 7 analysis of the mRNAs for transcription factors involved in B cell-to-plasma cell differentiation. (a) The expression of the PAX5, BCL6, BACH2, IRF8, IRF4, PRDM1, IRE1 and XBP1s genes was evaluated by quantitative real-time RT-PCR on D0 and D7. Results are expressed relative to gene expression in CLL B cells on D0, according to the 2−ΔΔCT method. Bars represent mean values±s.e.m. from five independent experiments. Statistical significance was calculated using the Wilcoxon test: *P<0.05. ns, not significant.
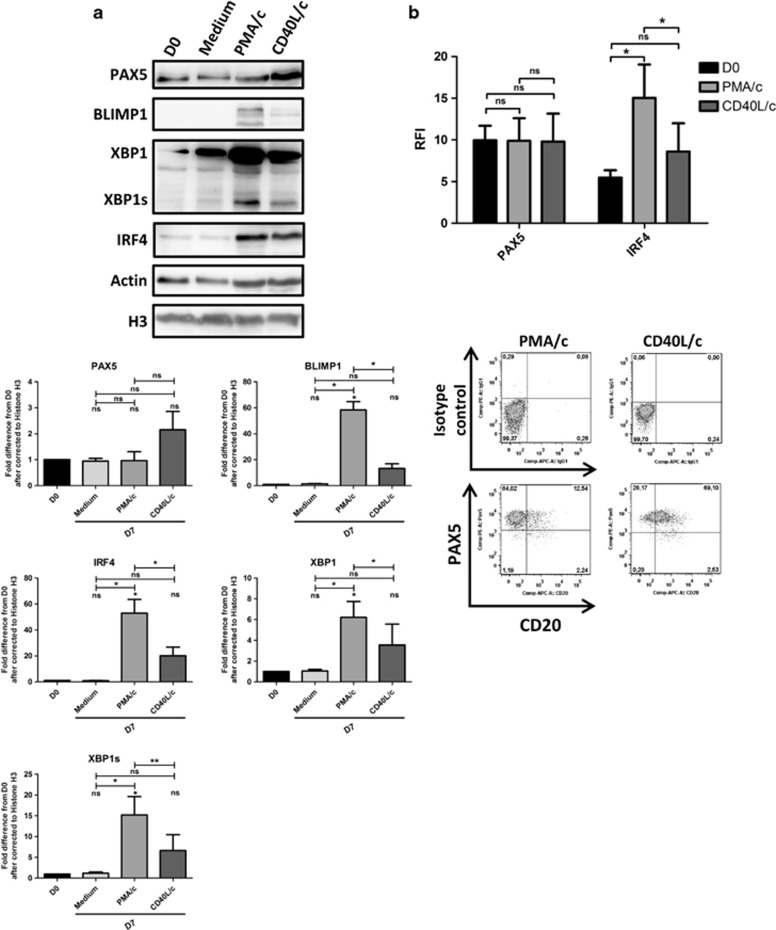
For confirmation of the changes in the transcription of key transcription factors involved in terminal B-cell differentiation, we used immunoblotting and flow cytometry to study the levels of the corresponding proteins (Figure 7). PAX5, IRF4, XBP1, XBP1s and BLIMP1 protein levels are shown in Figure 7a. Densitometry analysis of PMA/c-stimulated cells showed a significant increase in the amounts of IRF4 (53±18-fold), XBP1 (6±2-fold), XBP1s (15±7-fold) and BLIMP1 (58±11-fold) in response to stimulation (Figure 7a). The changes in protein levels observed for PMA/c-stimulated cells were > those observed for CD40L/c-stimulated cells (Figure 7a). However, no change in PAX5 levels was observed in PMA/c-stimulated cells (Figure 7a). This result was confirmed by flow cytometry (Figure 7b). The labeling of D7 PMA/c-stimulated cells with an antibody directed against PAX5 after permeabilization showed that almost all cells were PAX5-positive (Figure 7b). These data are consistent with recent findings on the characterization of the transitional preplasmablast stage, which weakly expresses PAX5, IRF8, BACH2 and BCL6 and expresses the plasma cell transcription factors gene PRDM1 and, predominantly, the unspliced form of XBP1.43
Figure 7.
Day 7 proteomic analysis of transcription factors involved in B cell-to-plasma cell differentiation. (a) Immunoblot analysis and densitometry quantification of PAX5, IRF8, IRF4, XBP1, XBP1s and BLIMP1 in D0 CLL B cells, D7 non-stimulated cells (Medium), D7 PMA/c-stimulated cells and D7 CD40L/c-stimulated cells. The data shown are representative of three independent experiments. Statistical significance was calculated with using Student's t-test. (b) Cells were labeled with anti-PAX5 and anti-IRF4 Abs after permeabilization. Upper panel: relative fluorescence intensities (RFIs) were calculated as the ratio of the MFI of cells labeled with a specific Ab to that of cells labeled with a matched isotype control. Bars represent RFI mean values±s.e.m. from three independent experiments. Lower panel: cytometry plots for a representative patient. Significance was calculated using a paired Student's t-test. *P<0.05, **P<0.01. ns, not significant. D, day.
Discussion
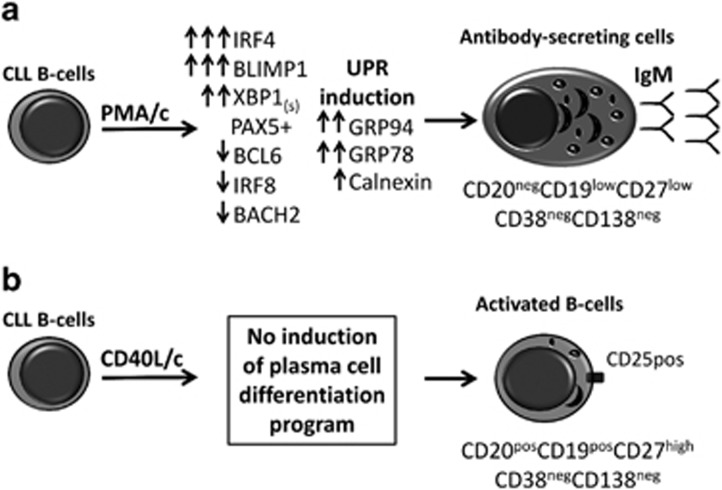
In this study, the D7 PMA/c-stimulated cells acquired the morphology of ASCs, as attested by their well-developed cytoplasm and an eccentrically placed nucleus. These cells displayed (i) high levels of cytoplasmic IgM expression, (ii) induction of the UPR, as indicated by the upregulation of UPR targets and of the protein folding machinery of the ER: GRP78/Bip and GRP94 and calnexin and (iii) the secretion of IgM (Figure 8a). Overall, these data suggest that the CLL B cells are not frozen permanently at a stage of differentiation and are able to differentiate into ASCs as appropriate stimulation are provided. By contrast, in the CD40L/c system (Figure 8b), D7-generated cells did not secrete Abs and had an activated B-cell morphology, resembling that of CLL B cells in the lymph nodes and bone marrow.59, 60
Figure 8.
PMA, but not CD40L, triggers CLL B cells to differentiate into ASCs. (A) CLL B cells cultured in PMA/c system acquired the morphology of ASCs, as evidenced by an eccentrically placed nucleus and a well-developed cytoplasm due to an extensive ER and Golgi apparatus. ASCs/plasma cells are specialized cells capable of secreting large amounts of Ig, and therefore require a functional UPR. The UPR is induced when the endoplasmic reticulum is overloaded with unfolded proteins. Activation of the UPR leads to a decrease in the amount of new unfolded proteins and an enhanced production of chaperones (GRP78/Bip, GRP94 and Calnexin), which facilitate protein folding. These morphological and molecular modifications primed the CLL B cells to differentiate into IgM-secreting cells. These cells presented an immunophenotype (CD20negCD19lowCD27lowCD38negCD138neg) and transcription factor profile (co-expression of B cell (PAX5) and plasma cell transcription (BLIMP1 and XBP1)) of preplasmablast. (b) CD40L/c system fail to induce plasma cell differentiation program and CLL B cells were unable to differentiate into ASCs. However, generated cells had an activated B-cell morphology and phenotype (CD25+CD27+) and an increased expression of IRF4 resembling that of CLL B cells in the lymph nodes and bone marrow. The capacity of CLL B cells to differentiate into ASCs in PMA/c system and not in CD40L/c may result from the activation requirements and the intrinsic mechanism of CLL B-cells differentiation.
In our study D7 PMA/c-stimulated cells displayed a significant increase in the expression of BLIMP1, IRF4, XBP1 and XBP1s (as shown by quantitative RT-PCR and immunoblotting) and a significant decrease in the amounts of mRNA for BCL6, BACH2 and IRF8. Thus, consistent with morphology and Ig secretion results, these data indicate that D7 PMA/c-stimulated cells had differentiated into ASCs with a transcriptional program recapitulating that described for plasma cell differentiation. Despite quantitative RT-PCR results showing a downregulation of PAX5 transcription in D7 PMA/c-stimulated cells, PAX5 protein continued to be detected. However, concomitant expression of BLIMP1 and PAX5 has been shown in tonsil plasma cells.61, 62 Moreover, PAX5 expression has been detected in some IgM-secreting multiple myeloma and plasma cells in Waldenstrom's macroglobulinemia.63, 64 In addition, an upregulation of GAS6 transcription was observed on D7 in PMA/c-stimulated cells. GAS6 upregulation begins in CD20neg/low preplasmablasts, with expression reaching particularly high levels in normal plasmablasts, before being downregulated when these cells differentiate into plasma cells.54
Immunophenotyping data for D7 PMA/c- and CD40L/c-stimulated cells revealed a significant downregulation of the surface expression of CD20, but with no surface expression or mRNA for CD38. The expression of CD38 is tightly regulated in normal B-cell differentiation. CD38 is induced in mature naive B cells after activation, with higher levels of expression attained when these cells enter the germinal center. CD38 expression then decreases during centrocyte/centroblast maturation. It is completely absent in memory B cells, but CD38 re-expression is observed in terminally differentiated plasma cells.65 The absence of this marker despite the presence of factors (such as IL-2 and IL-10) involved in regulating CD38 expression66 may result from an intrinsic feature of malignant cells. This conclusion is supported by the results of a study in which the stimulation, with interferons (which are known to stimulate CD38), of CD38neg CLL B cells from CD38-negative patients or after sorting from CD38-positive patients, did not result in the induction of CD38 expression.67 Furthermore, the ectopic expression of CD38 in CD38neg CLL B cells has no effect on cell differentiation or morphology.68 Nevertheless, the molecular and epigenetic mechanisms that control CD38 expression in CLL still unknown. CD20 is found in all B lymphocytes, but is not present in plasma cells.39, 54 The downregulation of CD20 expression on PMA/c-stimulated cells was systematic and highly significant with respect to CD40L/c-stimulated cells. The downregulation of CD20 observed on D7 CD40L/c-stimulated cells can provide a biological explanation for the low levels of CD20 expression on CLL B cells. Indeed, it has been shown that CD20 expression is stronger on circulating CLL B cells than on CLL B cells in the bone marrow and lymph nodes.69 The activation of CLL B cells by T cells and cytokines in the lymph nodes can lead to the downregulation of CD20. Thus, the partial or complete loss of this surface B-cell marker may be due to the influence of the microenvironment. However, exceptional populations with unusual immunophenotypes have also been reported. Several studies have identified CD38neg ASCs as a possible transitional stage or precursor of plasma cells.9, 43, 62, 70 Moreover, Avery et al.,9 has shown that CD38neg ASCs and CD27low populations generated in vitro concomitantly express BLIMP1, XBP1 and PAX5. However, these authors suggested that the CD27high population provides the precursors of ASC, independently of CD38 expression. A population of CD27neg/low malignant plasma cells has been reported in multiple myeloma and was associated with high-risk disease.71 Arce et al.70 identified CD38low Ig-secreting cells as early plasma cells found exclusively in the tonsils and expressing low levels of CD27. Indeed, it is now assumed that the differential expression of CD20, CD38 and CD138 on the surface of human ASCs may correspond to an alternative or additional ASC subsets derived from independent differentiation pathways.72 Moreover, human ASCs/plasma cells have a heterogeneous phenotype that is related to their anatomic localization and degree of maturation.62, 72 Thereby, the molecular profile and immunophenotype of the ASCs generated in our culture may be the signature of an alternative ASC subsets or a transitional stage toward plasma cell differentiation. A preplasmablast population was recently reported to lack documented B-cell and plasma cell markers (CD20, CD38, CD138), in addition to displaying weak expression of CD27 and the B-cell transcription factor PAX5.43 These cells were generated by in vitro differentiation from human memory B cells and were shown to have an in vivo counterpart present in human tonsils and lymph nodes.43 It has recently been suggested that a CD27low population generated by the activation in vitro of human memory B cells constitutes a transient preplasmablast population.73 These cells express PAX5, GRP78/Bip and the plasma cell transcription factors IRF4, BLIMP1, XBP1.73 In addition, four secreting human myeloma cell lines (K620, U266, XG6, NAN6) have been shown to be CD20negCD38negCD27neg.74 Overall, these findings indicate that the PMA/c culture system allows CLL B cells to differentiate into antigen-secreting early plasma cells with an immunophenotype (CD20neg/lowCD38negCD27lowCD138neg) and molecular profile similar to those of preplasmablasts.
Indeed, D7 CD40L/c-stimulated cells appeared to be activated B cells, as shown by their upregulation of CD27 and CD25, activated B-cell morphology and non-antibody-secreting phenotype. CD27 was shown to be progressively upregulated on activated B cells and to be strongly expressed on plasma cells that had differentiated from naive, germinal center and memory B cells.9 CD25, the alpha chain of the IL-2 receptor, has been shown to be upregulated on activated B cells and absent from plasmablasts and plasma cells.12, 75 Moreover, in a study comparing the phenotypes and functions of CD25neg and CD25pos B cells, the CD25-positive cells were larger and more granulated and were found to be highly specialized, professional antigen-presenting B cells that were unable to secrete Ig,76 consistent with our observations of CD40L-stimulated cells on D7. CD40L/c-stimulated cells also display significant expression of CD40, CD18 and HLA-DR and a significant downregulation of CD184 (CXCR4). CXCR4, the receptor of SDF1 (CXCL12), plays an important role in survival, growth and chemoresistance in B-CLL. Its expression was shown to be lower on CLL B cells from lymph node and bone marrow than on circulating CLL B cells.19 Furthermore, we observed an increase of IRF4 expression in D7 CD40L/c-stimulated cells. Indeed, IRF4 expression was shown to be found mainly in proliferation centers microenvironment in CLL.77 Overall, this pattern of expression, together with the low level of CD20 expression and cell morphology, results in a strong resemblance between CD40L/c-stimulated cells and the resident CLL B cells located in the lymph node and bone marrow.15, 16, 19, 59, 60, 69, 78 Thus, we could suggest the in vitro CD40L/c system as a model for the CLL microenvironment. However, validation of this interpretation will require further research. Nonetheless, the question raised here is why CD40 ligand, a strong activator of NFκB signaling and well known to induce B cell differentiation, is unable to induce differentiation of the CLL cells. In order to understand this unanswered question we investigate the expression levels of STAT3 and STAT5 in both PMA/c- and CD40L/c-stimulated cells using immunoblot analysis. We found that PMA/c-stimulated levels express higher levels of these molecules than CD40L/c-stimulated cells (data not shown).
In summary, unlike B-cells,7, 8, 79 CLL B cells are not able to differentiate into ASCs in the CD40L system. We propose that the CLL B cells activation requirements to differentiate into ASCs seem more stringent. BCR signaling or TLR stimulation could be a candidate. However, the activation of CLL B cells by PMA, an activator of PKC, led to their differentiation into ASCs. It is important to note that PKC, some isoforms of which are overexpressed in CLL, is a common mediator of multiple signaling pathways relating to CLL survival, including antigenic stimulation and signaling through BCR, an important component of the CLL microenvironment.31 Thus, the response of CLL cells to PMA stimulation may also have physiological significance in CLL. The role of PKC pathway in CLL B cells differentiation needs further investigations. However, the molecular mechanism by which PMA induces differentiation of CLL B cells, shown to be specific to malignant CLL B cells,14 is to be explored. We also suggest that ASC differentiation can occur in the concomitant presence of PAX5 and execution of the transcriptional program of plasma cells (IRF4, BLIMP1 and XBP1). This molecular phenotype, associated with a CD20negCD38negCD27low immunophenotype, may constitute the signature of a stage of transition toward plasma cell differentiation: the preplasmablast. Finally, extrapolating from the observation made for most in vitro terminal differentiation models for memory B cells, in which these cells give rise to predominantly IgG and IgA class-switched Ab-secreting cells, the predominant secretion of IgM by ASCs differentiated from CLL B cells in our culture system is not consistent with germinal center Ag-experienced B cells being the normal counterpart of these cells.However, IgM+ memory B cells as the normal counterpart of CLL B cells is still plausible. Moreover, our data suggest that the normal counterpart of CLL B cells is likely to be a B-cell compartment producing poly/autoreactive Abs (such as marginal zone B cells or B1 cells40). Furthermore, the inability of CLL B cells to differentiate into ASCs in the CD40L culture system provides additional support for this hypothesis. However, we speculate that the CD40L/c system may reproduce some of the events occurring in the CLL microenvironment.
Methods
Patients
CLL B cells were obtained from the peripheral blood of 13 patients (four women and nine men; median age (range): 67(48–82)) diagnosed in accordance with international guidelines (Table 1). All patients provided written informed consent for participation in the study. All procedures involving samples from patients were approved by the local institutional review board (Comité de Protection des Personnes (CPP) Nord-Ouest, Amiens, France). All cases presented a clonal expansion of small lymphocytes with high nucleus/cytoplasm ratios, co-expressing CD19, CD5 and CD23 and uniformly negative for CD38. None of the patients was on treatment at the time of the analysis.
Table 1. Patient characteristics.
| Patient | Sex | Age | Binet stage | Matutes score | CD38 | Cytogenetics |
|---|---|---|---|---|---|---|
| 1 | M | 69 | A | 5 | — | 17p del |
| 2 | M | 80 | A | 5 | — | ND |
| 3 | F | 59 | B | 5 | — | 13q14 del |
| 4 | F | 58 | A | 4 | — | ND |
| 5 | F | 80 | A | 5 | — | ND |
| 6 | M | 82 | A | 5 | — | NORMAL |
| 7 | F | 68 | A | 4 | — | ND |
| 8 | M | 65 | A | 5 | — | 13q14 del |
| 9 | M | 76 | A | 5 | — | Trisomy 12 Monosomy 9 |
| 10 | M | 55 | A | 5 | — | 13q14 del |
| 11 | M | 48 | B | 5 | — | 13q14, 11q del |
| 12 | M | 65 | A | 5 | — | Trisomy 12 |
| 13 | M | 75 | A | 4 | — | ND |
Abbreviations: del, deletion; F, female; M, male; ND, not determined.
Cell isolation and culture
Peripheral blood mononuclear cells were isolated by the Ficoll density gradient centrifugation of heparin-treated venous blood samples from B-CLL patients. CD19+ CD5+ CLL B cells were purified by negative selection, by magnetic bead-activated cell sorting with a B-cell (B-CLL) isolation kit (Miltenyi Biotec, Paris, France). All preparations had a purity of at least 98% and the cells co-expressed CD19 and CD5 on their surface (as assessed by flow cytometry). We checked that the purified CLL B cells were not contaminated with other immune cells, by direct labeling with anti-CD2, CD14 and CD56 Abs. Cells were cultured in RPMI 1640 medium (Gibco-Invitrogen, Saint Aubin, France) supplemented with 10% fetal calf serum, (PAA, Velizy-Villacoublay, France), 100 IU ml−1 penicillin, 100 μg ml−1 streptomycin and 2 mM L-glutamine, at 37 °C, under a humidified atmosphere containing 5% CO2.
On D0, purified CLL B cells were used to seed cultures at a density of 2 × 106 cells/ml and were stimulated for 4 days with PMA (1 μg ml−1, Santa Cruz Biotechnology, Heidelberg, Germany) or histidine-tagged soluble recombinant human CD40L (50 ng ml−1), anti-polyhistidine monoclonal antibody (mAb) (5 μg ml−1; R&D Systems, Abingdon, UK) and a combination of cytokines IL-2 (50 ng ml−1), IL-4 (50 ng ml−1), IL-10 (50 ng ml−1) and IL-12 (2 ng ml−1). The cells were cultured in 5 ml wells, in six-well, flat-bottomed culture plates.
On D4, the cells were harvested, washed and used to seed cultures at a density of 106 cells per ml in the presence of IL-2 (50 ng ml−1), IL-6 (50 ng ml−1), IL-10 (50 ng ml−1), and IL-12 (2 ng ml−1). The cultures were incubated for a further 3 days. On D7, cells were harvested, washed and analyzed. All human recombinant cytokines were purchased from PeproTech EC (Neuilly-Sur-Seine, France).
Normal B-cell cultures were carried out as described by Tarte et al.8
Immunophenotypic analysis
Cells were stained with the appropriate combinations of fluorochrome-conjugated Abs, in a three- to five-color direct immunofluorescence staining protocol. The sources and specificities of the mAbs used and of the murine isotype-matched controls are listed in Supplementary Table 1. The Cytofix/Cytoperm kit (BD Biosciences, Le Pont de Claix, France) was used for the intracellular staining of immunoglobulin M (IgM), IgG, CD38, PAX5 and IRF4, according to the manufacturer's recommendations. Flow cytometry analysis was performed with a FACSCantoII flow cytometer (BD Biosciences). FlowJo software (Tree Star, Ashland, OR, USA) was used for data analysis. Antigen density was expressed as RFI, that is the ratio of the mean fluorescence intensity (MFI) of cells labeled with a specific Ab to that of cells labeled with a matched isotype control.
Morphological analysis
For the morphological study on D7, cells were spun in a Cytospin machine (500 r.p.m., 5 min). Cytospin smears were stained with May–Grünwald–Giemsa reagent. Cells were viewed under a light microscope (Axio Imager.M2; Zeiss, Le Pecq, France) equipped with an AxioCam MRc5 microscope digital camera and images were acquired with ZEN pro Software (Zeiss). We used ImageJ (NIH, http://rsbweb.nih.gov/ij/) and GIMP software (http://www.gimp.org) to render the backgrounds of all images uniform and to improve image clarity.
Quantitative RT-PCR analysis
On D0, D4 and D7, we isolated total RNA from cultured cells with the RNeasy Mini kit (Qiagen, Courtaboeuf, France). We then used 1 μg of total RNA for cDNA synthesis with the High Capacity cDNA Archive kit (Applied Biosystems, Courtaboeuf, France). For transcript detection by quantitative real-time PCR, we used primers, probes and TaqMan Universal PCR Master Mix as recommended by the manufacturer (Applied Biosystems) and PCR was run on a StepOnePlusTM Real-time PCR System (Applied Biosystems). The TaqMan Gene Expression assays for PRDM1 (BLIMP1) (Assay ID Hs00153357_m1), PAX5 (Hs00172003_m1), BCL6 (Hs00277037_m1), XBP1 (Hs00231936_m1), XBP1s (Hs03929085_g1), IRF4 (Hs01056533_m1), IRF8 (Hs01128710_m1), BACH2 (Hs00222364_m1), ERN1 (Hs00176385_m1), GAS6 (Hs01090305_m1), CD38 (Hs01120071_m1) and CD138 (Hs00896423_m1) were purchased from Applied Biosystems. For quantification, β2-microglobulin (B2M, 4333766F) was used as endogenous control. Differences in mRNA levels were assessed by the 2−ΔΔCT quantitative RT-PCR method, where ΔΔCT=(ΔCT D7−ΔCT D0).
For GAS6, differences in mRNA levels were assessed by the 2−ΔCT method, with ΔCT=(CT of target gene−CT of B2M).
Immunoblot analysis
Sodium dodecyl sulfate polyacrylamide gel electrophoresis and immunoblotting were performed according to standard procedures. The Abs used are listed in Supplementary Table 1. A pellet of 2 × 106 cells was solubilized in 50 μl of Laemmli buffer. We then resolved 40 μl of the lysate by sodium dodecyl sulfate polyacrylamide gel electrophoresis in a 10% polyacrylamide gel. After electrophoresis, the proteins were transferred to polyvinylidene difluoride membranes (Thermo Scientific, Courtaboeuf, France), which were then blocked by incubation in 0.02% Tween 20 and 5% non-fat dry milk in Tris-buffered saline. The membranes were then probed with the appropriate primary Abs and antibody binding was detected by incubation with a horseradish peroxide-conjugated secondary Ab and an enhanced chemiluminescence substrate (Clarity Western ECL substrate, Bio-Rad, Marnes-la-Coquette, France). The results were visualized on a ChemiDocTM MP Imaging System (Bio-Rad). Densitometric quantification was performed with ImageJ analysis software (NIH) (http://rsbweb.nih.gov/ij/).
Analysis of IgM, IgG and IgA secretion
The levels of human IgM, IgG, and IgA in the culture supernatants were quantified with the corresponding ELISA kit (Bethyl Laboratories, Montgomery, TX, USA). Immunoglobulin production (in μg per 106 cells) was estimated by dividing the amount of Ig in the culture supernatant by the number of live cells.
Indirect immunofluorescence assays
For IFAs, HEp-2 cell-coated slides (INOVA Diagnostics, Inc., San Diego, CA, USA) were incubated at room temperature with culture supernatant for 30 min, washed in PBS, incubated with an fluorescein isothiocyanate-conjugated anti-human IgM antibody and viewed under a fluorescence microscope (Axio Imager.M2; Zeiss) equipped with an AxioCam MRc5 microscope digital camera. Images were acquired with ZEN pro Software (Zeiss). Positive control staining with serum samples from patients with scleroderma, an autoimmune disease, and negative control staining with culture medium from the cultures of cells of healthy subjects, were included in all experiments.
Statistical analysis
All statistical analyses were performed with Prism 5 software (GraphPad Software, La Jolla, CA, USA). The statistical significance of differences between groups was determined using the Wilcoxon test or the Student's t-test, as appropriate; P values <0.05 were considered to be statistically significant and P values <0.01 were considered to be highly statistically significant. Differences are denoted as follows: *P<0.05, **P<0.01 and ***P<0.001.
Acknowledgments
We would like to thank Dr Paulo Marcelo for his material support (ICAP plateform, Flow Cytometry, UFR Santé, Amiens, France), Dr Hakim Houchi (GRAP, INSERM ERI24) for his assistance with quantitative real-time PCR experiments, Eliane Bissac and Véronique Debuysscher for their assistance with western blotting experiments, and the Hematology Department for their assistance with MGG staining. We also thank the Centre Hospitalo-Universitaire d'Amiens (CHU d'Amiens), Conseil Régional de Picardie and the French National Institute of Health and Medical Research (INSERM) for their financial support.
The authors declare no conflict of interest.
Footnotes
The Supplementary Information that accompanies this paper is available on the Immunology and Cell Biology website (http://www.nature.com/icb)
Supplementary Material
References
- Manz RA, Hauser AE, Hiepe F, Radbruch A. Maintenance of serum antibody levels. Annu Rev Immunol. 2005;23:367–386. doi: 10.1146/annurev.immunol.23.021704.115723. [DOI] [PubMed] [Google Scholar]
- Goodnow CC, Vinuesa CG, Randall KL, Mackay F, Brink R. Control systems and decision making for antibody production. Nat Immunol. 2010;11:681–688. doi: 10.1038/ni.1900. [DOI] [PubMed] [Google Scholar]
- Schmidlin H, Diehl SA, Blom B. New insights into the regulation of human B-cell differentiation. Trends Immunol. 2009;30:277–285. doi: 10.1016/j.it.2009.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nera KP, Lassila O. Pax5—a critical inhibitor of plasma cell fate. Scand J Immunol. 2006;64:190–199. doi: 10.1111/j.1365-3083.2006.01809.x. [DOI] [PubMed] [Google Scholar]
- Shaffer AL, Shapiro-Shelef M, Iwakoshi NN, Lee AH, Qian SB, Zhao H, et al. XBP1, downstream of Blimp-1, expands the secretory apparatus and other organelles, and increases protein synthesis in plasma cell differentiation. Immunity. 2004;21:81–93. doi: 10.1016/j.immuni.2004.06.010. [DOI] [PubMed] [Google Scholar]
- Turner CA, Jr., Mack DH, Davis MM. Blimp-1, a novel zinc finger-containing protein that can drive the maturation of B lymphocytes into immunoglobulin-secreting cells. Cell. 1994;77:297–306. doi: 10.1016/0092-8674(94)90321-2. [DOI] [PubMed] [Google Scholar]
- Neron S, Nadeau PJ, Darveau A, Leblanc JF. Tuning of CD40-CD154 interactions in human B-lymphocyte activation: a broad array of in vitro models for a complex in vivo situation. Arch Immunol Ther Exp (Warsz) 2011;59:25–40. doi: 10.1007/s00005-010-0108-8. [DOI] [PubMed] [Google Scholar]
- Tarte K, De Vos J, Thykjaer T, Zhan F, Fiol G, Costes V, et al. Generation of polyclonal plasmablasts from peripheral blood B cells: a normal counterpart of malignant plasmablasts. Blood. 2002;100:1113–1122. [PubMed] [Google Scholar]
- Avery DT, Ellyard JI, Mackay F, Corcoran LM, Hodgkin PD, Tangye SG. Increased expression of CD27 on activated human memory B cells correlates with their commitment to the plasma cell lineage. J Immunol. 2005;174:4034–4042. doi: 10.4049/jimmunol.174.7.4034. [DOI] [PubMed] [Google Scholar]
- Dubois B, Massacrier C, Vanbervliet B, Fayette J, Briere F, Banchereau J, et al. Critical role of IL-12 in dendritic cell-induced differentiation of naive B lymphocytes. J Immunol. 1998;161:2223–2231. [PubMed] [Google Scholar]
- Le Gallou S, Caron G, Delaloy C, Rossille D, Tarte K, Fest T. IL-2 requirement for human plasma cell generation: coupling differentiation and proliferation by enhancing MAPK-ERK signaling. J Immunol. 2012;189:161–173. doi: 10.4049/jimmunol.1200301. [DOI] [PubMed] [Google Scholar]
- Jego G, Bataille R, Pellat-Deceunynck C. Interleukin-6 is a growth factor for nonmalignant human plasmablasts. Blood. 2001;97:1817–1822. doi: 10.1182/blood.v97.6.1817. [DOI] [PubMed] [Google Scholar]
- Hasbold J, Corcoran LM, Tarlinton DM, Tangye SG, Hodgkin PD. Evidence from the generation of immunoglobulin G-secreting cells that stochastic mechanisms regulate lymphocyte differentiation. Nat Immunol. 2004;5:55–63. doi: 10.1038/ni1016. [DOI] [PubMed] [Google Scholar]
- van Kooten C, Rensink I, Aarden L, van Oers R. Differentiation of purified malignant B cells induced by PMA or by activated normal T cells. Leukemia. 1993;7:1576–1584. [PubMed] [Google Scholar]
- Zenz T, Mertens D, Kuppers R, Dohner H, Stilgenbauer S. From pathogenesis to treatment of chronic lymphocytic leukaemia. Nat Rev Cancer. 2010;10:37–50. doi: 10.1038/nrc2764. [DOI] [PubMed] [Google Scholar]
- Ghia P, Chiorazzi N, Stamatopoulos K. Microenvironmental influences in chronic lymphocytic leukaemia: the role of antigen stimulation. J Intern Med. 2008;264:549–562. doi: 10.1111/j.1365-2796.2008.02030.x. [DOI] [PubMed] [Google Scholar]
- Ghamlouch H, Ouled-Haddou H, Damaj G, Royer B, Gubler B, Marolleau JP. A Combination of cytokines rescues highly purified leukemic CLL B-cells from spontaneous apoptosis in vitro. PLoS ONE. 2013;8:e60370. doi: 10.1371/journal.pone.0060370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damle RN, Calissano C, Chiorazzi N. Chronic lymphocytic leukaemia: a disease of activated monoclonal B cells. Best Pract Res Clin Haematol. 2010;23:33–45. doi: 10.1016/j.beha.2010.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herishanu Y, Perez-Galan P, Liu D, Biancotto A, Pittaluga S, Vire B, et al. The lymph node microenvironment promotes B-cell receptor signaling, NF-kappaB activation, and tumor proliferation in chronic lymphocytic leukemia. Blood. 2011;117:563–574. doi: 10.1182/blood-2010-05-284984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu SM, Winchester RJ, Feizi T, Walzer PD, Kunkel HG. Idiotypic specificity of surface immunoglobulin and the maturation of leukemic bone-marrow-derived lymphocytes. Proc Natl Acad Sci USA. 1974;71:4487–4490. doi: 10.1073/pnas.71.11.4487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubartelli A, Sitia R, Zicca A, Grossi CE, Ferrarini M. Differentiation of chronic lymphocytic leukemia cells: correlation between the synthesis and secretion of immunoglobulins and the ultrastructure of the malignant cells. Blood. 1983;62:495–504. [PubMed] [Google Scholar]
- Granziero L, Ghia P, Circosta P, Gottardi D, Strola G, Geuna M, et al. Survivin is expressed on CD40 stimulation and interfaces proliferation and apoptosis in B-cell chronic lymphocytic leukemia. Blood. 2001;97:2777–2783. doi: 10.1182/blood.v97.9.2777. [DOI] [PubMed] [Google Scholar]
- Tromp JM, Tonino SH, Elias JA, Jaspers A, Luijks DM, Kater AP, et al. Dichotomy in NF-kappaB signaling and chemoresistance in immunoglobulin variable heavy-chain-mutated versus unmutated CLL cells upon CD40/TLR9 triggering. Oncogene. 2010;29:5071–5082. doi: 10.1038/onc.2010.248. [DOI] [PubMed] [Google Scholar]
- Fluckiger AC, Rossi JF, Bussel A, Bryon P, Banchereau J, Defrance T. Responsiveness of chronic lymphocytic leukemia B cells activated via surface Igs or CD40 to B-cell tropic factors. Blood. 1992;80:3173–3181. [PubMed] [Google Scholar]
- Crawford DH, Catovsky D. In vitro activation of leukaemic B cells by interleukin-4 and antibodies to CD40. Immunology. 1993;80:40–44. [PMC free article] [PubMed] [Google Scholar]
- Plander M, Seegers S, Ugocsai P, Diermeier-Daucher S, Ivanyi J, Schmitz G, et al. Different proliferative and survival capacity of CLL-cells in a newly established in vitro model for pseudofollicles. Leukemia. 2009;23:2118–2128. doi: 10.1038/leu.2009.145. [DOI] [PubMed] [Google Scholar]
- Van den Hove LE, Van Gool SW, Vandenberghe P, Bakkus M, Thielemans K, Boogaerts MA, et al. CD40 triggering of chronic lymphocytic leukemia B cells results in efficient alloantigen presentation and cytotoxic T lymphocyte induction by up-regulation of CD80 and CD86 costimulatory molecules. Leukemia. 1997;11:572–580. doi: 10.1038/sj.leu.2400598. [DOI] [PubMed] [Google Scholar]
- Buhmann R, Nolte A, Westhaus D, Emmerich B, Hallek M. CD40-activated B-cell chronic lymphocytic leukemia cells for tumor immunotherapy: stimulation of allogeneic versus autologous T cells generates different types of effector cells. Blood. 1999;93:1992–2002. [PubMed] [Google Scholar]
- Pascutti MF, Jak M, Tromp JM, Derks IA, Remmerswaal EB, Thijssen R, et al. IL-21 and CD40L signals from autologous T cells can induce antigen-independent proliferation of CLL cells. Blood. 2013;122:3010–3019. doi: 10.1182/blood-2012-11-467670. [DOI] [PubMed] [Google Scholar]
- Gold MR. To make antibodies or not: signaling by the B-cell antigen receptor. Trends Pharmacol Sci. 2002;23:316–324. doi: 10.1016/s0165-6147(02)02045-x. [DOI] [PubMed] [Google Scholar]
- Kazi JU, Kabir NN, Ronnstrand L. Protein kinase C (PKC) as a drug target in chronic lymphocytic leukemia. Med Oncol. 2013;30:757. doi: 10.1007/s12032-013-0757-7. [DOI] [PubMed] [Google Scholar]
- Apollonio B, Scielzo C, Bertilaccio MT, Ten Hacken E, Scarfo L, Ranghetti P, et al. Targeting B-cell anergy in chronic lymphocytic leukemia. Blood. 2013;121:3879–3888. doi: 10.1182/blood-2012-12-474718. [DOI] [PubMed] [Google Scholar]
- Zupo S, Isnardi L, Megna M, Massara R, Malavasi F, Dono M, et al. CD38 expression distinguishes two groups of B-cell chronic lymphocytic leukemias with different responses to anti-IgM antibodies and propensity to apoptosis. Blood. 1996;88:1365–1374. [PubMed] [Google Scholar]
- Nishizuka Y. The molecular heterogeneity of protein kinase C and its implications for cellular regulation. Nature. 1988;334:661–665. doi: 10.1038/334661a0. [DOI] [PubMed] [Google Scholar]
- Mond JJ, Feuerstein N, June CH, Balapure AK, Glazer RI, Witherspoon K, et al. Bimodal effect of phorbol ester on B-cell activation: Implication for the role of protein kinase C. J Biol Chem. 1991;266:4458–4463. [PubMed] [Google Scholar]
- Tangye SG, Weston KM, Raison RL. Phorbol ester activates CD5+ leukaemic B cells via a T cell-independent mechanism. Immunol Cell Biol. 1995;73:44–51. doi: 10.1038/icb.1995.7. [DOI] [PubMed] [Google Scholar]
- van Kooten C, Rensink I, Aarden L, van Oers R. Effect of IL-4 and IL-6 on the proliferation and differentiation of B-chronic lymphocytic leukemia cells. Leukemia. 1993;7:618–624. [PubMed] [Google Scholar]
- Manocha S, Matrai Z, Osthoff M, Carter A, Pettitt AR. Correlation between cell size and CD38 expression in chronic lymphocytic leukaemia. Leuk Lymphoma. 2003;44:797–800. doi: 10.1080/1042819031000068034. [DOI] [PubMed] [Google Scholar]
- Bohnhorst JO, Bjorgan MB, Thoen JE, Natvig JB, Thompson KM. Bm1–Bm5 classification of peripheral blood B cells reveals circulating germinal center founder cells in healthy individuals and disturbance in the B cell subpopulations in patients with primary Sjogren's syndrome. J Immunol. 2001;167:3610–3618. doi: 10.4049/jimmunol.167.7.3610. [DOI] [PubMed] [Google Scholar]
- Chiorazzi N, Ferrarini M. Cellular origin(s) of chronic lymphocytic leukemia: cautionary notes and additional considerations and possibilities. Blood. 2011;117:1781–1791. doi: 10.1182/blood-2010-07-155663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein U, Casola S, Cattoretti G, Shen Q, Lia M, Mo T, et al. Transcription factor IRF4 controls plasma cell differentiation and class-switch recombination. Nat Immunol. 2006;7:773–782. doi: 10.1038/ni1357. [DOI] [PubMed] [Google Scholar]
- Dong HY, Browne P, Liu Z, Gangi M. PAX-5 is invariably expressed in B-cell lymphomas without plasma cell differentiation. Histopathology. 2008;53:278–287. doi: 10.1111/j.1365-2559.2008.03091.x. [DOI] [PubMed] [Google Scholar]
- Jourdan M, Caraux A, Caron G, Robert N, Fiol G, Reme T, et al. Characterization of a transitional preplasmablast population in the process of human B cell to plasma cell differentiation. J Immunol. 2011;187:3931–3941. doi: 10.4049/jimmunol.1101230. [DOI] [PubMed] [Google Scholar]
- Vuillier F, Dumas G, Magnac C, Prevost MC, Lalanne AI, Oppezzo P, et al. Lower levels of surface B-cell-receptor expression in chronic lymphocytic leukemia are associated with glycosylation and folding defects of the mu and CD79a chains. Blood. 2005;105:2933–2940. doi: 10.1182/blood-2004-09-3643. [DOI] [PubMed] [Google Scholar]
- Minuzzo S, Indraccolo S, Tosello V, Piovan E, Cabrelle A, Trentin L, et al. CD40 activation of B-CLL cells is associated with augmented intracellular levels of CD79b and increased BCR expression in a subset of patients. Leukemia. 2005;19:1099–1101. doi: 10.1038/sj.leu.2403772. [DOI] [PubMed] [Google Scholar]
- Lanemo Myhrinder A, Hellqvist E, Sidorova E, Soderberg A, Baxendale H, Dahle C, et al. A new perspective: molecular motifs on oxidized LDL, apoptotic cells, and bacteria are targets for chronic lymphocytic leukemia antibodies. Blood. 2008;111:3838–3848. doi: 10.1182/blood-2007-11-125450. [DOI] [PubMed] [Google Scholar]
- Zwick C, Fadle N, Regitz E, Kemele M, Stilgenbauer S, Buhler A, et al. Autoantigenic targets of B-cell receptors derived from chronic lymphocytic leukemias bind to and induce proliferation of leukemic cells. Blood. 2013;121:4708–4717. doi: 10.1182/blood-2012-08-447904. [DOI] [PubMed] [Google Scholar]
- Ma Y, Hendershot LM. The unfolding tale of the unfolded protein response. Cell. 2001;107:827–830. doi: 10.1016/s0092-8674(01)00623-7. [DOI] [PubMed] [Google Scholar]
- Kaufman RJ. Orchestrating the unfolded protein response in health and disease. J Clin Invest. 2002;110:1389–1398. doi: 10.1172/JCI16886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwakoshi NN, Lee AH, Glimcher LH. The X-box binding protein-1 transcription factor is required for plasma cell differentiation and the unfolded protein response. Immunol Rev. 2003;194:29–38. doi: 10.1034/j.1600-065x.2003.00057.x. [DOI] [PubMed] [Google Scholar]
- Yoshida H, Matsui T, Yamamoto A, Okada T, Mori K. XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell. 2001;107:881–891. doi: 10.1016/s0092-8674(01)00611-0. [DOI] [PubMed] [Google Scholar]
- Hebert DN, Simons JF, Peterson JR, Helenius A. Calnexin, calreticulin, and Bip/Kar2p in protein folding. Cold Spring Harb Symp Quant Biol. 1995;60:405–415. doi: 10.1101/sqb.1995.060.01.045. [DOI] [PubMed] [Google Scholar]
- Ochiai K, Katoh Y, Ikura T, Hoshikawa Y, Noda T, Karasuyama H, et al. Plasmacytic transcription factor Blimp-1 is repressed by Bach2 in B cells. J Biol Chem. 2006;281:38226–38234. doi: 10.1074/jbc.M607592200. [DOI] [PubMed] [Google Scholar]
- Jourdan M, Caraux A, De Vos J, Fiol G, Larroque M, Cognot C, et al. An in vitro model of differentiation of memory B cells into plasmablasts and plasma cells including detailed phenotypic and molecular characterization. Blood. 2009;114:5173–5181. doi: 10.1182/blood-2009-07-235960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tunyaplin C, Shaffer AL, Angelin-Duclos CD, Yu X, Staudt LM, Calame KL. Direct repression of prdm1 by Bcl-6 inhibits plasmacytic differentiation. J Immunol. 2004;173:1158–1165. doi: 10.4049/jimmunol.173.2.1158. [DOI] [PubMed] [Google Scholar]
- Sciammas R, Shaffer AL, Schatz JH, Zhao H, Staudt LM, Singh H. Graded expression of interferon regulatory factor-4 coordinates isotype switching with plasma cell differentiation. Immunity. 2006;25:225–236. doi: 10.1016/j.immuni.2006.07.009. [DOI] [PubMed] [Google Scholar]
- De Vos J, Couderc G, Tarte K, Jourdan M, Requirand G, Delteil MC, et al. Identifying intercellular signaling genes expressed in malignant plasma cells by using complementary DNA arrays. Blood. 2001;98:771–780. doi: 10.1182/blood.v98.3.771. [DOI] [PubMed] [Google Scholar]
- Dirks W, Rome D, Ringel F, Jager K, MacLeod RA, Drexler HG. Expression of the growth arrest-specific gene 6 (GAS6) in leukemia and lymphoma cell lines. Leuk Res. 1999;23:643–651. doi: 10.1016/s0145-2126(99)00075-2. [DOI] [PubMed] [Google Scholar]
- Matutes E, Polliack A. Morphological and immunophenotypic features of chronic lymphocytic leukemia. Rev Clin Exp Hematol. 2000;4:22–47. doi: 10.1046/j.1468-0734.2000.00002.x. [DOI] [PubMed] [Google Scholar]
- Inamdar KV, Bueso-Ramos CE. Pathology of chronic lymphocytic leukemia: an update. Ann Diagn Pathol. 2007;11:363–389. doi: 10.1016/j.anndiagpath.2007.08.002. [DOI] [PubMed] [Google Scholar]
- Medina F, Segundo C, Jimenez-Gomez G, Gonzalez-Garcia I, Campos-Caro A, Brieva JA. Higher maturity and connective tissue association distinguish resident from recently generated human tonsil plasma cells. J Leukoc Biol. 2007;82:1430–1436. doi: 10.1189/0507279. [DOI] [PubMed] [Google Scholar]
- Medina F, Segundo C, Campos-Caro A, Gonzalez-Garcia I, Brieva JA. The heterogeneity shown by human plasma cells from tonsil, blood, and bone marrow reveals graded stages of increasing maturity, but local profiles of adhesion molecule expression. Blood. 2002;99:2154–2161. doi: 10.1182/blood.v99.6.2154. [DOI] [PubMed] [Google Scholar]
- Gutierrez NC, Ocio EM, de Las Rivas J, Maiso P, Delgado M, Ferminan E, et al. Gene expression profiling of B lymphocytes and plasma cells from Waldenstrom's macroglobulinemia: comparison with expression patterns of the same cell counterparts from chronic lymphocytic leukemia, multiple myeloma and normal individuals. Leukemia. 2007;21:541–549. doi: 10.1038/sj.leu.2404520. [DOI] [PubMed] [Google Scholar]
- Feyler S, O'Connor SJ, Rawstron AC, Subash C, Ross FM, Pratt G, et al. IgM myeloma: a rare entity characterized by a CD20–CD56–CD117- immunophenotype and the t(11;14) Br J Haematol. 2008;140:547–551. doi: 10.1111/j.1365-2141.2007.06969.x. [DOI] [PubMed] [Google Scholar]
- Deaglio S, Vaisitti T, Aydin S, Ferrero E, Malavasi F. In-tandem insight from basic science combined with clinical research: CD38 as both marker and key component of the pathogenetic network underlying chronic lymphocytic leukemia. Blood. 2006;108:1135–1144. doi: 10.1182/blood-2006-01-013003. [DOI] [PubMed] [Google Scholar]
- Deaglio S, Capobianco A, Bergui L, Durig J, Morabito F, Duhrsen U, et al. CD38 is a signaling molecule in B-cell chronic lymphocytic leukemia cells. Blood. 2003;102:2146–2155. doi: 10.1182/blood-2003-03-0989. [DOI] [PubMed] [Google Scholar]
- Pittner BT, Shanafelt TD, Kay NE, Jelinek DF. CD38 expression levels in chronic lymphocytic leukemia B cells are associated with activation marker expression and differential responses to interferon stimulation. Leukemia. 2005;19:2264–2272. doi: 10.1038/sj.leu.2403975. [DOI] [PubMed] [Google Scholar]
- Pearce L, Morgan L, Lin TT, Hewamana S, Matthews RJ, Deaglio S, et al. Genetic modification of primary chronic lymphocytic leukemia cells with a lentivirus expressing CD38. Haematologica. 2010;95:514–517. doi: 10.3324/haematol.2009.014381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh YO, Keating MJ, Saffer HL, Jilani I, Lerner S, Albitar M. Higher levels of surface CD20 expression on circulating lymphocytes compared with bone marrow and lymph nodes in B-cell chronic lymphocytic leukemia. Am J Clin Pathol. 2001;116:437–443. doi: 10.1309/438N-E0FH-A5PR-XCAC. [DOI] [PubMed] [Google Scholar]
- Arce S, Luger E, Muehlinghaus G, Cassese G, Hauser A, Horst A, et al. CD38 low IgG-secreting cells are precursors of various CD38 high-expressing plasma cell populations. J Leukoc Biol. 2004;75:1022–1028. doi: 10.1189/jlb.0603279. [DOI] [PubMed] [Google Scholar]
- Guikema JE, Hovenga S, Vellenga E, Conradie JJ, Abdulahad WH, Bekkema R, et al. CD27 is heterogeneously expressed in multiple myeloma: low CD27 expression in patients with high-risk disease. Br J Haematol. 2003;121:36–43. doi: 10.1046/j.1365-2141.2003.04260.x. [DOI] [PubMed] [Google Scholar]
- Kaminski DA, Wei C, Qian Y, Rosenberg AF, Sanz I. Advances in human B cell phenotypic profiling. Front Immunol. 2012;3:302. doi: 10.3389/fimmu.2012.00302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henn AD, Laski M, Yang H, Welle S, Qiu X, Miao H, et al. Functionally distinct subpopulations of CpG-activated memory B Cells. Sci Rep. 2012;2:345. doi: 10.1038/srep00345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bataille R, Jego G, Robillard N, Barille-Nion S, Harousseau JL, Moreau P, et al. The phenotype of normal, reactive and malignant plasma cells: identification of ‘many and multiple myelomas' and of new targets for myeloma therapy. Haematologica. 2006;91:1234–1240. [PubMed] [Google Scholar]
- Jego G, Robillard N, Puthier D, Amiot M, Accard F, Pineau D, et al. Reactive plasmacytoses are expansions of plasmablasts retaining the capacity to differentiate into plasma cells. Blood. 1999;94:701–712. [PubMed] [Google Scholar]
- Brisslert M, Bokarewa M, Larsson P, Wing K, Collins LV, Tarkowski A. Phenotypic and functional characterization of human CD25+ B cells. Immunology. 2006;117:548–557. doi: 10.1111/j.1365-2567.2006.02331.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soma LA, Craig FE, Swerdlow SH. The proliferation center microenvironment and prognostic markers in chronic lymphocytic leukemia/small lymphocytic lymphoma. Hum Pathol. 2006;37:152–159. doi: 10.1016/j.humpath.2005.09.029. [DOI] [PubMed] [Google Scholar]
- Hartmann TN, Grabovsky V, Wang W, Desch P, Rubenzer G, Wollner S, et al. Circulating B-cell chronic lymphocytic leukemia cells display impaired migration to lymph nodes and bone marrow. Cancer Res. 2009;69:3121–3130. doi: 10.1158/0008-5472.CAN-08-4136. [DOI] [PubMed] [Google Scholar]
- Kindler V, Zubler RH. Memory, but not naive, peripheral blood B lymphocytes differentiate into Ig-secreting cells after CD40 ligation and costimulation with IL-4 and the differentiation factors IL-2, IL-10, and IL-3. J Immunol. 1997;159:2085–2090. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.