Abstract
Background:
Contamination of water, food, and environment with antibiotic-resistant bacteria poses a serious public health issue.
Objective:
The objective was to study the bacterial pollution of the natural sources of water in east Sikkim and to determine the antimicrobial profile of the bacterial isolates.
Materials and Methods:
A total of 225 samples, 75 each during winter, summer, and monsoon season were collected from the same source in every season for bacteriological analysis by membrane filtration method. Antibiotic susceptibility test was performed using standard disc diffusion method.
Results:
A total of 19 bacterial species of the genera Escherichia, Klebsiella, Proteus, Salmonella, Shigella, Enterobacter, Citrobacter, Morganella, Pseudomonas, Acinetobacter, Flavobacterium, and Serratia were isolated and their antimicrobial sensitivity tested. Generally, most bacterial isolates except Salmonella and Shigella species were found resistant to commonly used antibiotics such as ampicillin (57.5%), trimethoprim/sulfamethoxaole (39.1%), amoxicillin/clavulanic acid (37.4%), cefixime (34.5%), tetracycline (29.1%), ceftazidime (26.3%), ofloxacin (25.9%), amikacin (8.7%), and gentamicin (2.7%) but sensitive to imipenem and piperacillin/tazobactam.
Conclusion:
Natural sources of water in east Sikkim are grossly contaminated with bacteria including enteropathogens. The consumption of untreated water from these sources might pose health risk to consumers.
Keywords: Bacterial contamination, multi drug resistance, multiple antibiotic resistance index, water sources
Introduction
Water pollution is a major health problem in the global context, there are nearly 1.7 billion cases of diarrheal disease every year.(1) Almost 90% of child deaths from diarrheal diseases are directly linked to contaminated water, and India alone accounts for 24% of all deaths under 5-years old.(2) Among the pathogens disseminated in water sources, enteric pathogens such as enterotoxigenic Escherichia coli, Shigella spp., Salmonella spp., and so forth are the ones most frequently encountered that are responsible for a variety of diseases like diarrhea, dysentery, and enteric fever. To further compound this problem, enteric bacterial pathogens have been widely reported to demonstrate resistance to several antibiotics.(3,4,5) Bacteria with highest level of resistance were isolated from environment, surface water is the main receptable reservoir of antibiotics and antibiotic-resistant bacteria in the environment.(5) The rise in antibiotic resistance has been reported in the past two decades, and antibiotic resistance still remains a global problem.(6) The determination of the efficiency of antimicrobial agents against specific pathogens, either human or animal source, is essential for proper therapy.
In rural areas of Sikkim, surface water serves as major sources of water supply for drinking and domestic purposes. The use of untreated surface water sources for drinking and for domestic purposes remains a major threat to public health, as these could serve as reservoirs for transfer of antibiotic-resistant pathogens. The present study was conducted in order to determine the level of bacterial pollution of water in rural areas and antibiogram of the bacterial isolates from the natural sources of water.
Materials and Methods
This study was conducted in rural habitations of east district of Sikkim, India [Figure 1], where sources of water are springs and streams. A total of 225 water samples, 75 each in winter, summer, and monsoon from the same source, were collected during the period from November 2011 to October 2012 from 75 villages. The samples were collected in sterile capped bottles using the standard technique described in Mackie and McCartney.(7) The bottle was labeled with full details of the source of the water and time and date of collection. The bottles were kept in an ice box and transported to microbiology laboratory within 6 h of collection.
Figure 1.
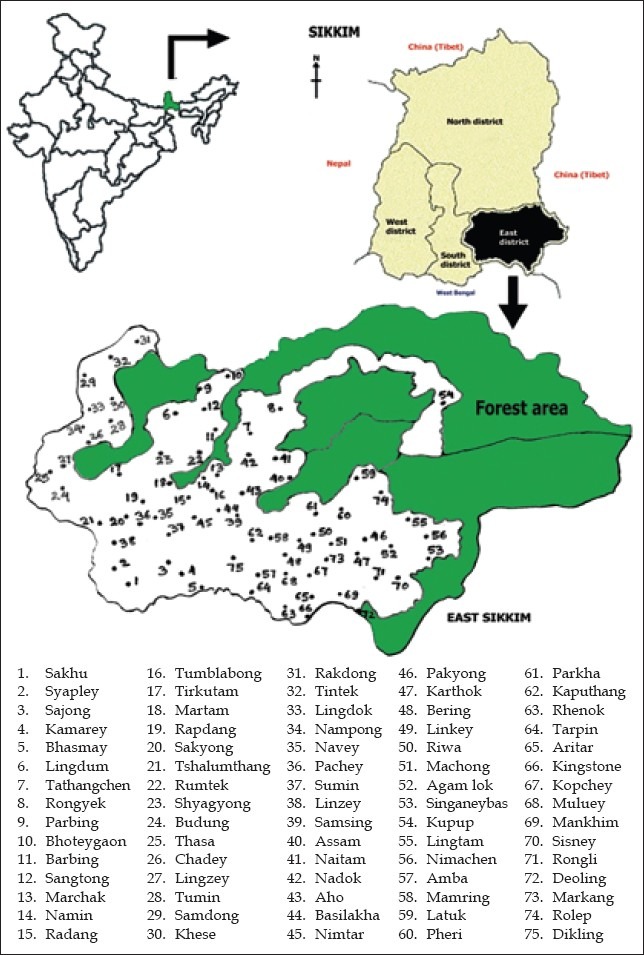
Location of study sites in the map of east district of Sikkim, India
Bacteriological analysis was done using the standard membrane filtration technique as described in Mackie and McCartney.(7) Each sample (100 ml) was filtered through a 47-mm diameter and 0.45-μm pore size cellulose acetate membrane filter grid (Millipore) held in a filtration unit. Then the membrane, with upper side upwards, with the help of sterile flat-ended forcep was transferred to the surface of MacConkey agar. Plates were incubated at 37°C. The primary identification of the bacterial isolates was done by the conventional methods of morphological and the cultural characteristics. Further confirmation was done on the basis of biochemical characteristics of the isolates.
Antimicrobial susceptibility was performed by the Kirby – Bauer disk diffusion method on Muller – Hinton agar.(8) Antibiotic discs were procured from Himedia, Mumbai, India. The antibiotics tested were ampicillin (10 μg), amoxicillin/clavulanic acid (20/10 μg), cefixime (5 μg), tetracycline (30 μg), ceftazidime (30 μg), ofloxacin (5 μg), amikacin (30 μg), gentamicin (10 μg), piperacillin/tazobactam (100/10 μg), imipenem (10 μg), chloramphenicol (30 μg), and trimethoprim/sulfamethoxaole (1.25/23.75 μg). The antimicrobials were chosen on the basis of their common use in treatment in the area. The control strain used was E. coli ATCC 25922.
Multiple antibiotic resistance (MAR) index is a useful tool for bacterial source tracking. This method helps in differentiating between human and nonhuman fecal sources.(9) MAR indices of the present isolates, belonging to Enterobacteriaceae, against the tested antibiotics were calculated based on the formula as follows.(10,11,12)
MAR index for isolates = a⁄b
a = Number of antibiotics to which the isolate is resistant
b = Number of antibiotics tested
y = Total number of resistance scored
n = Total number of isolates
x = Total number of antibiotics tested
Results
In the present study, most of the water sources were found to be contaminated in all three seasons. Out of total 541 isolates, 102 in winter, 241 in summer, and 198 in monsoon were isolated. During winter few organisms were detected as compared with that in summer and monsoon.
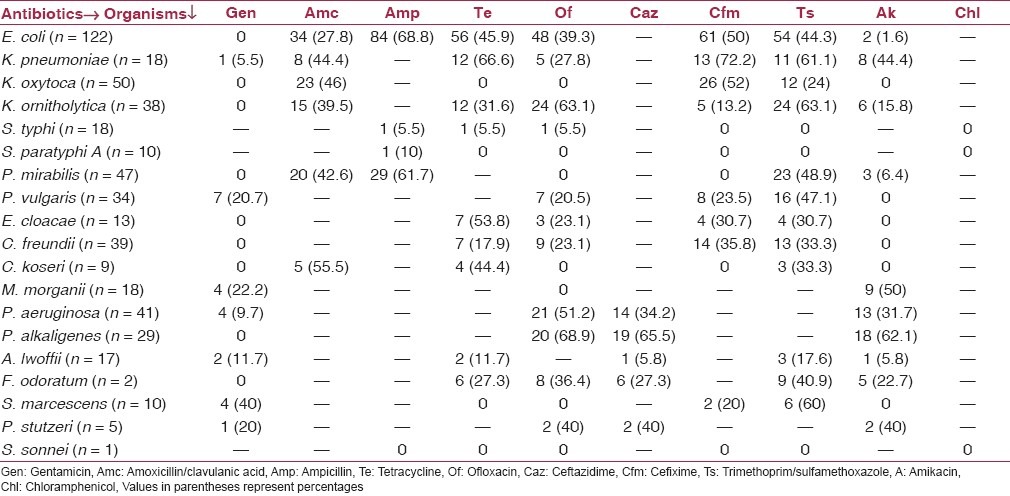
All bacterial isolates were found sensitive to imipenem and piperacillin/tazobactam but maximum resistance to ampicillin (57.5%), trimethoprim/sulfamethoxaole (39.1%), and amoxicillin/clavulanic acid (37.4%) [Figure 2]. Table 1 shows the antibiotic susceptibility pattern of the all bacterial isolates obtained in this study. E. coli showed more than 50% resistance to ampicillin and cefixime. Among Klebsiella spp. more than 50% resistance was seen to tetracycline, trimethoprim/sulfamethoxaole, ofloxacin, and cefixime. Interestingly, enteropathogens, namely, Salmonella typhi (88.8%), Salmonella paratyphi A (90%), and Shigella sonnei (100%) were found to be fairly sensitive to the antibiotics tested.
Figure 2.
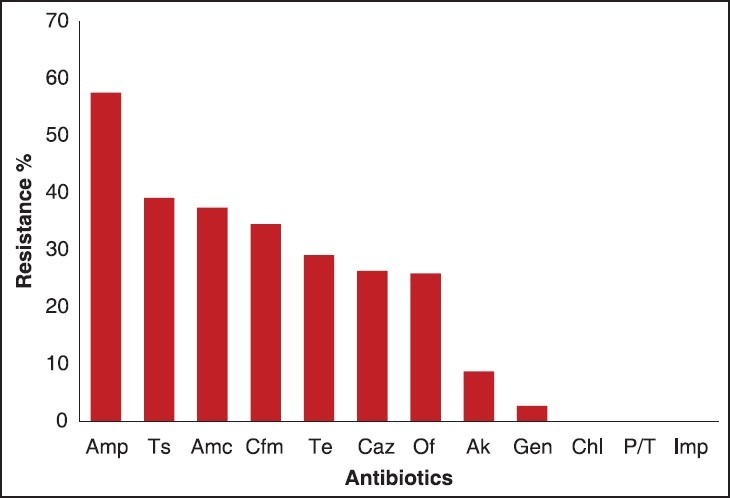
Antibiotic susceptibility test results for the isolated bacteria (n: 541) Amp: Ampicillin, Ts: Trimethoprim/sulfamethoxazole, Amc: Amoxicillin/clavulanic acid, Cfm: Cefixime,Te: Tetracycline, Caz: Ceftazidime, Of: Ofloxacin, Ak: Amikacin, Gen: Gentamicin, Chl: Chloramphenicol, P/T: Piperacillin/tazobactam, Imp: Imipenem
Table 1.
Antibiotic resistance pattern of the bacterial isolates
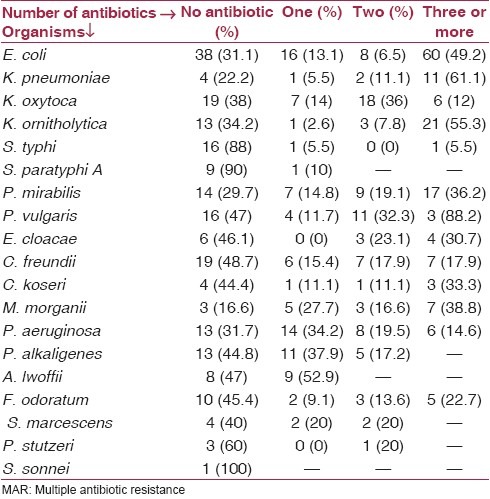
Most of the isolates showed resistance to multiple antibiotics. Of all the bacterial isolates (n = 541), 213 (39.4%) isolates were sensitive to all the antibiotics tested, 88 (16.3%) were resistant to at least one antibiotic, and 240 (44.4%) of these strains were multiresistant [Table 2].
Table 2.
Showing percentage of MAR by individual bacterial species
The overall resistance of isolates to the antibiotics is represented as MAR index. MAR index of the isolates ranged from 0 to 0.7 [Table 3], whereas MAR index of the sampling sites ranged from 0 to 0.5 [Figure 3].
Table 3.
Frequency of MAR index of the bacteria
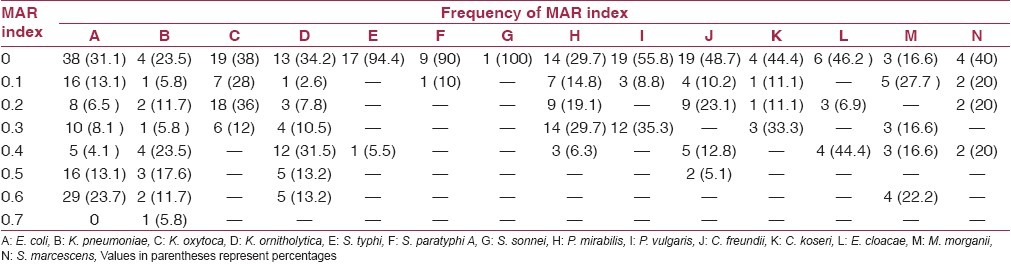
Figure 3.
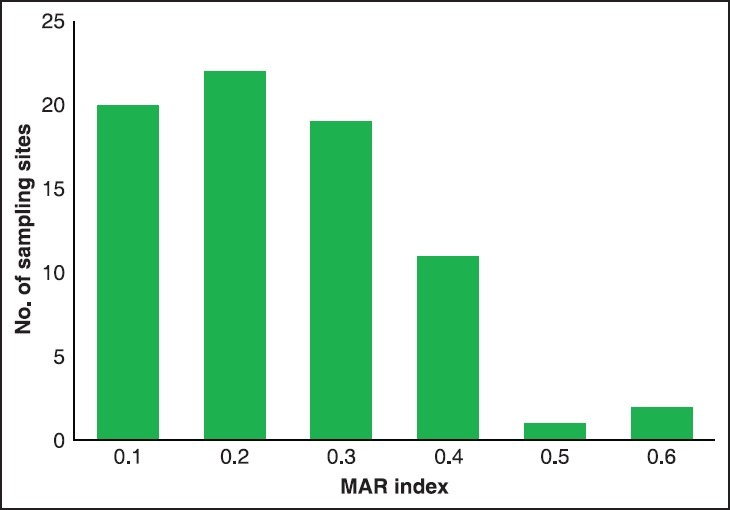
MAR index of the sampling sites, MAR: Multiple antibiotic resistance
Discussion
In the present study, the bacteria were isolated from natural sources of water and has indicated high levels of antibiotic resistance to ampicillin, whereas less resistance to amikacin and gentamicin. Similar to our study, Manisha et al., isolated multidrug-resistant E. coli, Proteus mirabilis, Proteus vulgaris, Citrobacter freundii, Morganella morganii, Pseudomonas aeruginosa, and Serratia marcescens, and the isolates showed high resistance to ampicillin and less resistance to gentamicin and amikacin.(13)
With an increase in the antibiotic load, the prevalence of acquiring resistance increases within a bacterial community. The MAR index is an excellent tool that permits us to analyze the relative prevalence of resistant bacteria in the environment. For all the sites, MAR indices were between 0.1 and 0.5. The water sources having MAR index more than 0.4 are usually from human fecal origin and MAR indices less than 0.4 are from nonhuman fecal contamination.(11,14) On the basis of this assumption, 17.3% of the samples were human fecal contaminated. When isolates are exposed to high risk sources of contamination where antibiotics are frequently used, an MAR index value >0.2 is observed. When antibiotics are seldom or never used, an MAR index value less than or equal to ≤0.2 is observed.(9,15) In this study, 36.2% of the bacterial isolates showed MAR value more than 0.2, which is a possible indication that the bacterial isolates have been exposed to several antibiotics. The high resistance seen may be due to the selective pressure exerted by the use of antibiotics in the management of bacterial infections in animals or humans in the area.
In this study, finding enteropathogens S. typhi, S. paratyphi A and S. sonnei sensitive to the commonly used antibiotics is fortunate, but the existence of other Multiple Drug Resistance (MDR) bacteria with the susceptible ones increases the chances of transfer of antibiotic resistance to the sensitive ones. This maintains a pool of resistant bacteria with a pool of resistance genes in the population that further contributes to the general increase and dissemination of bacterial resistance and can be a source of resistance genes for pathogens.(16) High resistance of E. coli to antimicrobial agents tested was observed in this study. Similar to our study, E. coli strains were isolated from water that showed high resistance for ampicillin and were resistant to more than one antibiotic.(3,17,18) In another study conducted in Agra, multidrug-resistant Klebsiella spp. was isolated from water and showed maximum resistant to gentamicin, whereas in our study Klebsiella spp. showed high sensitivity to gentamicin.(4) There can be many ways of transmission of resistant bacteria such as municipal, agricultural sewage, and human and animal excrement on the open ground surface and, these are the sources of spreading antibiotics-resistant bacteria in the aquatic environment.(19) Mode of transmission through water exposes the whole flock of population to the antibiotics. Therefore, knowledge of proper waste disposal in the environment should be acknowledged to avoid the indiscriminate spread of antibiotic-resistant bacterial strains.
The effectiveness of antibiotic treatment of diseases will decline due to the development of antibiotic resistance in the environment. It is evident from the present results that multiple antibiotic-resistant bacteria can thrive in water that can serve as an environmental reservoir and can therefore provide a route to multidrug-resistant pathogens to enter human population. The study of resistance in the environmental bacteria can help in guiding the development of strategies to counteract this resistance. From the study, it can be concluded that to detect any changing patterns, there is a need to monitor antibiotic sensitivity at regular intervals and knowledge of local presence of bacterial organisms and antibiotic sensitivities and not universal guidelines should provide the direction of antibiotic selection. Treatment of water is essential not only to kill the pathogenic bacteria but also to stop the transfer of antibiotic resistance from the resistant bacteria to the sensitive ones.
Observation of incidence of multidrug-resistant bacterial isolates in water sources of rural areas of east Sikkim in our study may serve as database for formulation of immediate and future sanitation and health-related programs in this area.
Acknowledgement
The authors are grateful to the Vice Chancellor, Sikkim Manipal University and Dean, Sikkim Manipal Institute of Medical Sciences, Tadong, Gangtok for providing facilities to conduct the valuable research and allowing us to publish this paper. The authors also thank the Departments of Forest, Rural Management and Development, Government of Sikkim for their kind support in providing various information and statistics of the study areas which led to the fruitful completion of this paper.
Footnotes
Source of Support: Nil
Conflict of Interest: None declared.
References
- 1.Diarrhoeal disease. World Health Organization (WHO) Fact sheet No. 330; 20133 [Google Scholar]
- 2.Children dying daily because of unsafe water supplies and poor sanitation and hygiene. New York: United Nation's Children Fund; 2013. Press release 2013. [Google Scholar]
- 3.Sharma BC, Rai B. Incidence of multi-drug resistance in E. coli strains isolated from three lakes of tourist attraction (Mirik lake, Jorepokhani lake and Nakhapani lake) of Darjeeling Hills, India. Indian J Fundam Appl Life Sci. 2012;2:108–14. [Google Scholar]
- 4.Verma NS, Gupta A, Dubey M, Mahajan S, Sharma R. Resistance status of some pathogenic bacteria isolated from water of Yamuna river in Agra. Asian J Exp Biol Sci. 2011;2:697–03. [Google Scholar]
- 5.Chitanand MP, Kadam TA, Gyananath G, Totewad ND, Balhal DK. Multiple antibiotic resistance indexing of coliforms to identify high risk contamination sites in aquatic environment. Indian J Microbiology. 2010;50:216–20. doi: 10.1007/s12088-010-0042-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kapil A. The challenge of antimicrobial resistance: Need to contemplate. Indian J Med Res. 2004;121:83–91. [PubMed] [Google Scholar]
- 7.Collee JG, Marmion BP, Fraser AG, Simmons A, editors. Mackie and McCartney Practical Medical Microbiology. 14th ed. London: Churchill Livingstone; 1996. pp. 889–92. [Google Scholar]
- 8.Baurer AW, Kirby WM, Sherris JC, Turuk M. Antibiotic susceptibility testing by a standardied single disk method. Am J Clin Pathol. 1966;45:493–6. [PubMed] [Google Scholar]
- 9.Scott TM, Rose JB, Jenkins TM, Farrah SR, Lukasik J. Microbial source tracking: Current methodology and future directions. Appl Environ Microbiol. 2002;68:5796–803. doi: 10.1128/AEM.68.12.5796-5803.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Krumperman PH. Multiple antibiotic resistance indexing Escherichia coli to identify risk sources of faecal contamination of foods. Appl Environ Microbiol. 1983;46:165–70. doi: 10.1128/aem.46.1.165-170.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tambekar DH, Hirulkar NB, Waghmare AS. MAR indexing to discriminate the source of faecal contamination in drinking water. Nat Environ Poll Tech. 2005;4:525–8. [Google Scholar]
- 12.Hinton M, Hedges AJ, Linton AH. The ecology of Escherichia coli in market calves fed a milk-substitute diet. J Appl Bacteriol. 1985;58:27–35. doi: 10.1111/j.1365-2672.1985.tb01426.x. [DOI] [PubMed] [Google Scholar]
- 13.Mandal MD, Mandal S, Pal NK. Antibiotic resistance prevalence and pattern in environmental bacterial isolates. Open Antimicrob Agents J. 2011;3:45–52. [Google Scholar]
- 14.Kaneene BJ, Miller R, Sayah R, Johnson YJ, Gilliland D, Gardiner JC. Considerations when using discrimination function analysis of antimicrobial resistance profiles to identify sources of faecal contamination of surface water in Michigan. Appl Environ Microbial. 2007;73:2878–90. doi: 10.1128/AEM.02376-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vergidis PI, Falagas ME. Multidrug-resistant gram-negative bacterial infections: The emerging threat and potential novel treatment options. Curr Opin Investig Drugs. 2008;9:176–83. [PubMed] [Google Scholar]
- 16.Pontes DS, Pinheiro FA, Lima-Bittencourt CI, Guedes RL, Cursino L, Barbosa F, et al. Multiple antimicrobial resistance of Gram negative bacteria from natural oligotrophic lakes under distinct anthropogenic influence in a tropical region. Microb Ecol. 2009;58:762–72. doi: 10.1007/s00248-009-9539-3. [DOI] [PubMed] [Google Scholar]
- 17.Cardonha AM, Vieira RS, Rodrigues DP, Macrae A, Peiramo G, Teophilo GN. Faecal pollution in water from storm sewers and adjacent seashore in Natal, Rio Grande to Norte, Brazil. Int Microbiol. 2004;7:213–8. [PubMed] [Google Scholar]
- 18.Rasheed F, Khan A, Kazmi SU. Bacteriological analysis, antimicrobial susceptibility and detection of 16S rRNA gene of Helicobacter pylori by PCR in drinking water samples of earthquake affected areas and other parts of Pakistan. Malays J Microbiol. 2009;5:123–7. [Google Scholar]
- 19.Segura PA, François M, Gagnon C, Sauvé S. Review of the occurrence of anti-infectives in contaminated wastewaters and natural and drinking waters. Environ Health Perspect. 2009;117:675–84. doi: 10.1289/ehp.11776. [DOI] [PMC free article] [PubMed] [Google Scholar]


