Abstract
Aims:
The aim of the present study is to investigate the association between polymorphism of cytochrome P450 2C9 (CYP2C9) enzyme with head and neck squamous cell carcinoma (HNSCC) and response in patients receiving cisplatin-based radical chemoradiation (CT-RT).
Materials and Methods:
Four hundred and sixty patients suffering from locally advanced HNSCC and an equal number of healthy controls were genotyped for CYP2C9*2 and CYP2C9*013, leading to poor metabolizers (PMs) by polymerase chain reaction (PCR)-based restriction fragment length polymorphism (RFLP). Each case was assessed thoroughly for treatment response as per the World Health Organization (WHO) criteria.
Results and Analysis:
The frequency of heterozygous genotypes of both CYP2C9*2 (27.8%) and CYP2C9*3 (25%) were found to be significantly higher in the HNSCC cases as compared to the healthy controls. Tobacco intake in the form of chewing or smoking and alcohol intake resulted in several folds increase in the risk to HNSCC in the cases carrying variant genotypes of CYP2C9*2 or CYP2C9*013. Further, majority of the cases assessed for response (n = 436) carrying variant alleles of CYP2C9*2 (69.6%) or CYP2C9*3 (65.2%) were found to respond poorly to cisplatin-based radical CT-RT.
Conclusion:
The data suggests a significant association of the CYP2C9 polymorphism with HNSCC and treatment outcome underlining the importance of pretherapeutic genotyping in determining the treatment protocol.
Keywords: Chemoradiation, CYP2C9, head and neck cancer, poor metabolizers, treatment response
Introduction
Head and neck squamous cell carcinoma (HNSCC) accounts for approximately 5% of all cancers worldwide. HNSCC is often associated with heavy tobacco and alcohol use. People who use both tobacco and alcohol are at greater risk for developing these cancers.[1] Polymorphisms in genes such as cytochrome P450s (CYPs) and glutathione-S-transferases (GSTs), involved in the metabolism and detoxification of alcohol and constituents of tobacco are shown to influence an individual's susceptibility to cancer.[2] Studies including meta and pooled analysis have shown that polymorphisms involving phase I (CYP1A1, CYP2E1) and phase II (GST) enzymes are associated with tobacco-induced HNSCC.[3]
Mutations in the human genome occur spontaneously at frequencies of about 10−6 per generation due to misincorporation or damage of bases during deoxyribonucleic acid (DNA) replication. Due to the high conservation of the genetic code through generations, such a mutation can be inherited. However, if a certain mutation is seen in more than 1% of a population group, the variation is considered a polymorphism. Polymorphisms usually occur as point mutations where a single (or a few) base (s) is substituted (substitution), deleted (deletion), or an extra base is inserted (insertion). These point mutations are commonly known as single nucleotide polymorphisms (SNPs). In CYP2C9*2 polymorphism, wild type base pair CC (C = cytosine) is replaced by either CT (T = thymine) heterozygous mutant type or TT homozygous mutant type. In CYP2C9*3 polymorphism, AA (A = adenine) wild type base pair is replaced by either heterozygous AC or homozygous CC mutant base pair.
CYP2C9 is one of the major drug-metabolizing CYPs in human liver and contributes to the metabolism of a number of clinically important drugs. CYP2C9 is also known to be involved in the metabolism of some of the antineoplastic drugs such as cyclophosphamide, etoposide, tamoxifen, and ifosfamide.[4] A three-fold lower intrinsic clearance for cyclophosphamide was observed with recombinant CYP2C9*2 and CYP2C9*3 protein when compared to CYP2C9*011 protein in a yeast expression system.[5]
In addition, CYP2C9 enzyme also metabolizes several carcinogenic and mutagenic substrates including heterocyclic aromatic amines and polycyclic aromatic hydrocarbons (PAHs).[6] It has also been shown that some of the reactions catalyzed by CYP2C9 lead to detoxification of carcinogens.[7] Genetic polymorphism has also been reported for CYP2C9. Variant mutant (both homozygous and heterozygous) CYP2C9*2 and CYP2C9*3 genotypes account for ‘poor-metabolizer’ (PM) phenotype resulting in slow metabolism of drugs and other substrates metabolized by CYP2C9.[8] Significant differences are known to exist in the distribution of the variant alleles of CYP2C9 in different populations. In general, the polymorphism in CYP2C9 is more frequent in Caucasians compared to Oriental.[9] An increased frequency of CYP2C9*2 allele in the patients with lung cancer has been reported.[10] Variant alleles of CYP2C9 were reported to increase the risk of distal colorectal adenoma.[11] But, the association of CYP2C9 polymorphism has never been studied with HNSCC.
The present study, therefore, attempted to investigate the association of CYP2C9 genotypes with HNSCC risk and outcome of treatment in the patients receiving cisplatin-based radical chemoradiation (CT-RT).
Materials and Methods
A case-control study was conducted. Four hundred and sixty patients (85.2% males) suffering from locally advanced HNSCC (T3N0/T1-2 N1-2, lymph nodes ≤4 cm) and equal number of controls/healthy subjects were included in the study. The cases had biopsy-proven SCC of the oral cavity, oropharynx, larynx, or hypopharynx; and were treated with radical CT-RT with concurrent cisplatin, between November 2009 and May 2013, as per the protocol. After a complete clinical examination; they underwent biopsy, complete hemogram, liver and kidney function tests, chest radiograph, contrast-enhanced computed tomography (CT) scan of face and neck and dental prophylaxis. Those with a Karnofsky performance status of 60 or less, a second malignant neoplasm, prior radiotherapy to head and neck, or with distant metastasis were excluded. All the patients of the current study were permanent residents of Uttar Pradesh or Uttarakhand for five or more generations. Moreover, a detailed family history was taken from each of the patients regarding their race and ethnicity, before including them in the study. So, it can be safely stated that, all the subjects in this study belonged to the same ethnic group (Indo-European community) of north India. The controls/healthy subjects were selected from the permanent residents of the same locality as that of patients, by doing health camps. Moreover, the controls were frequency-matched to cases by year of birth in 5-year classes. Based on medical check-ups, controls were not found to suffer from any chronic disease.
The protocol was approved by the human ethics committee of the institution. Informed consent was obtained from each of the study subjects for inclusion in the study and before the collection of blood samples, and it was also ensured that the subject anonymity was maintained. All study subjects completed a questionnaire covering medical, residential, and occupational history, and information pertaining to dietary habits, family history of disease, smoking, tobacco-chewing, and alcohol consumption. Subjects having regular smoking habits and smoking index (cigarettes/day × 365 days) of 730 or more were classified as smokers. Likewise, smokeless tobacco dose was estimated as ‘chewing year’ (i.e., CY = frequency of tobacco chewed/day × duration of year). Those who had CY of 365 or more were considered as tobacco chewers.[12] Similarly, cumulative exposure to alcohol drinking was derived by multiplying the total yearly consumption of alcohol (in L/year) by the duration of habitual alcohol drinking (in years). Those who had cumulative exposure to alcohol about 90 L were considered as regular alcohol users in our study.[13]
DNA isolation and CYP2C9 genotyping was done by the following method: 500 μl blood samples collected in citrate-containing tubes from the study subjects were processed for the isolation of genomic DNA using the QIAamp DNA mini kit (Qiagen, CA) following the manufacturers’ protocol. For identifying the polymorphism in CYP2C9 (CYP2C9*012 and CYP2C9*013), the reaction mixture in 50 μl contained 1 × buffer (10 mM Tris-Hcl pH 8.3, 1.5-3.0 mM of MgCl 2, 25 mM KCl), 200 mM of each nucleotide, 200 nM of each of CYP2C9*2 or 2C9*013 primers, 1.5 units of Taq polymerase (MBI Fermentas, Germany), and 100 ng of genomic DNA and sterile milliQ water and was processed for polymerase chain reaction (PCR). Amplified PCR products were digested with AvaII or NsiI (MBI Fermentas, Germany) for identifying CYP2C9*2 and CYP2C9*013, respectively. The products were resolved by 3% agarose gel containing ethidium bromide as described earlier.[9]
For quality control, randomly 10% of the samples were selected and regenotyped to confirm the authenticity of the results obtained earlier and they were found to be in 100% concordance.
We determined whether genotype or allele frequencies of CYP2C9 polymorphism among the cases and controls were in Hardy-Weinberg equilibrium (HWE) using the standard Chi-square tests. The association between genetic polymorphisms and risk of HNSCC was estimated by calculating crude odds ratio (OR). A P < 0.05 was considered statistically significant. The statistical analysis was performed with the Statistical Package for Social Sciences (SPSS) software (version 17.0 for Windows; SPSS Chicago, IL).
Patients were treated with concurrent CT-RT, which included administration of 35 mg/m2 of cisplatin once every week for 7 weeks along with 70 Gy/35 fractions external beam radiation (EBRT) over 7 weeks.
One month following the completion of all treatments, treatment response was assessed by thorough clinical examination and video-laryngoscopy. On the basis of World Health Organization (WHO) criteria, the treatment outcome was divided into the following categories:
Complete response (CR): Disappearance of all known lesion (s); confirmed at 4 weeks
Partial response (PR): At least 50% decrease; confirmed at 4 weeks
Stable disease (SD): Neither PR nor PD criteria met
Progressive disease (PD): 25% increase; no CR, PR, or SD documented before increased disease or new lesion (s).
Those exhibiting CR are categorized as complete responders, while patients exhibiting PR or SD/PD are classified as non-complete responders.[14]
Results and Analysis
Nine hundred and twenty subjects consisting of 460 cases suffering from HNSCC and equal number of healthy controls, were genotyped for CYP2C9*012, CYP2C9*3 polymorphisms. The distribution of demographic variables and putative risk factors of HNSCC are summarized in Table 1. Tobacco chewing, cigarette smoking, and daily alcohol use were found to be significantly (P < 0.05) higher in patients as compared to controls [Table 1].
Table 1.
Distribution of demographic variables and putative risk factors of HNSCC cases
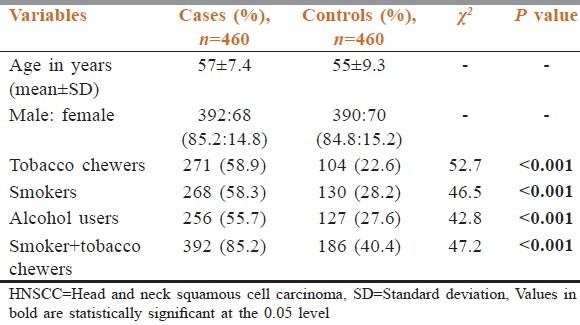
The frequency of heterozygous genotype (CT) of CYP2C9*2 polymorphism was found to be higher in the cases (27.8%) than the controls (13.5%). This increase in frequency resulted in an increased risk of HNSCC (OR: 2.3; 95% confidence interval (CI): 1.3-4.1) in cases which was found to be statistically significant [Table 2]. The frequency of homozygous genotype (TT) was slightly higher in cases (4.4%) when compared to the controls (3.2%) that resulted in an increase in risk (OR: 1.6; 95% CI: 0.5-2.4) which was not statistically significant. Adjustment of the data for age, cigarette smoking, tobacco chewing, and alcohol consumption revealed that the risk continued to be significantly increased (adjusted OR: 2.1; 95% CI: 1.2-3.9) in the cases with heterozygous genotype of CYP2C9*2 polymorphism [Table 2]. Here it should be observed that wild type genotype is more in controls than in cases which indicates that wild type genotype is less susceptible than the mutant type or reversely mutant type is more susceptible than the wild one.
Table 2.
Distribution of CYP2C9*2 and CYP2C9*3 genotypes among HNSCC cases and healthy controls
The frequency of heterozygous genotype (AC) of CYP2C9*3 was found to be increased in the cases (25%) when compared to the controls (12.2%) that increased the OR (2.1; 95% CI: 1.0-3.6) in the cases which was statistically significant [Table 2]. The frequency of homozygous mutant genotype (CC) was found to be higher in cases (4.4%) when compared to controls (2.6%), which slightly increased the risk (OR: 1.9; 95% CI: 0.5-3.2) of HNSCC [Table 2]. Adjustment of the data for age, cigarette smoking, and tobacco chewing and alcohol consumption revealed that the risk continued to be significantly increased (adjusted OR: 1.9; 95% CI: 1.0-2.9) in the cases with heterozygous genotype (AC) of CYP2C9*3 polymorphism [Table 2].
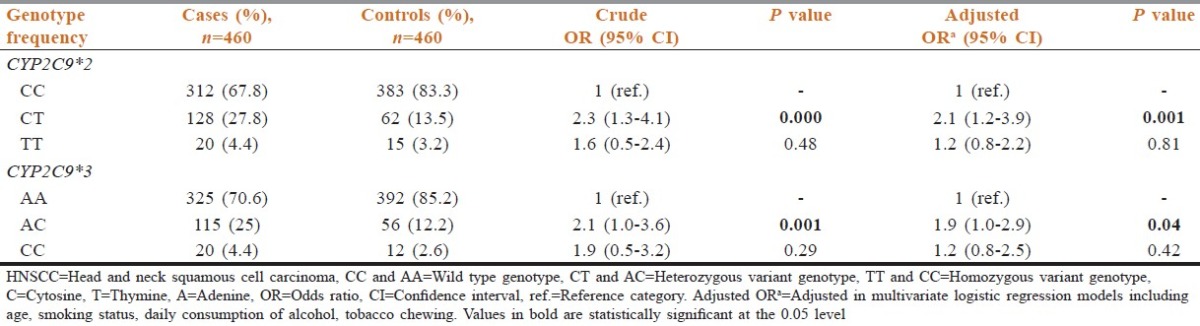
The number of individuals with variant genotypes (homozygous and heterozygous) of CYP2C9*2 was significantly increased in cases (38.7%), who were regular tobacco chewers as compared to the controls (13.5%) with similar habit of tobacco chewing [Table 3]. The increase in the frequency resulted in several fold statistically significant increase in the risk of HNSCC (OR: 4.6; 95% CI: 1.6-13.1) amongst the tobacco chewers. As observed with CYP2C9*012, the frequency of individuals who were regular tobacco chewers with variant genotypes of CYP2C9*3 was also increased significantly in cases (34.3%) when compared to the controls (16.3%). The increase in the frequency resulted in two-to three-fold statistically significant increase in the risk of HNSCC (OR: 2.6, 95% CI: 1.0-6.6) [Table 3].
Table 3.
Interaction between CYP2C9 genotypes and tobacco chewing, and risk of HNSCC
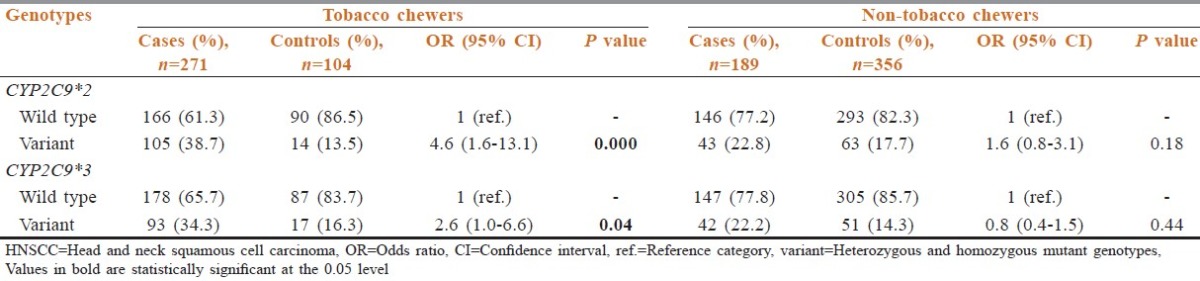
Cigarette smoking also increased the risk to HNSCC in the cases with CYP2C9 polymorphism when compared to the smokers in controls [Table 4]. The frequency of individuals who were regular smokers and carried variant mutant genotypes of CYP2C9*2 was significantly increased in the cases (39.6%) as compared to the controls (12.4%). The OR associated with cigarette smoking increased several folds in the patients with the variant genotypes of CYP2C9*2 (4.1; 95% CI: 1.6-10.9), which was found to be statistically significant (P = 0.001). As observed with CYP2C9*2 variants, the number of cases with variant (homozygous and heterozygous) genotypes of CYP2C9*3 was also increased amongst smokers (31.7%) as compared to the controls (15.4%). The increase in the frequency resulted in a statistically significant increase in the risk of HNSCC (OR: 2.2, 95% CI: 0.9-5.8) [Table 4].
Table 4.
Interaction between CYP2C9 genotypes and smoking, and risk of HNSCC
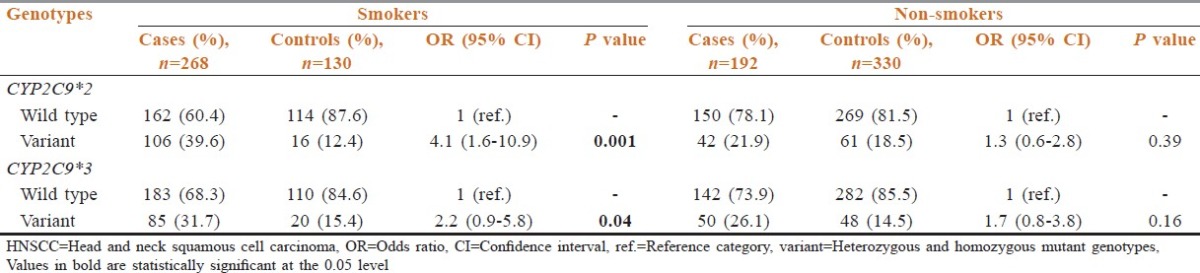
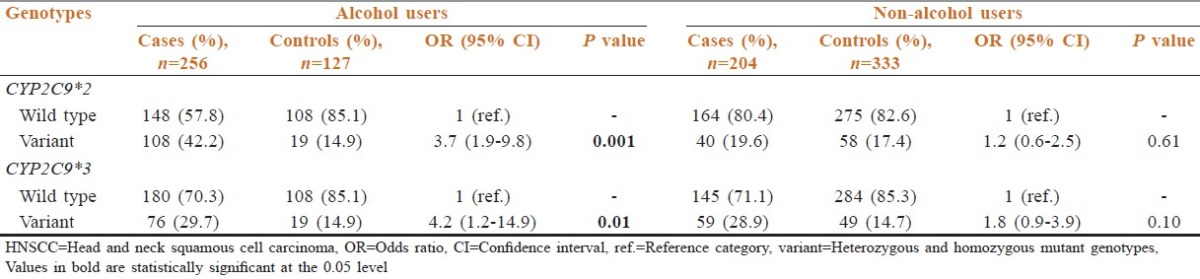
The frequency of the individuals with variant genotypes of CYP2C9*2 and who were regular alcohol users significantly increased in the cases (42.2%) when compared to the controls (14.9%) with similar habit [Table 5]. This increase is associated with almost four-fold increase in the risk (OR: 3.7; 95% CI: 1.9-9.8) to HNSCC in the cases. As observed with CYP2C9*2 variants, the number of cases with variant genotypes of CYP2C9*3 also increased amongst alcohol users (29.7%) as compared to the controls (14.9%). A statistically significant increase in risk was also observed in cases amongst the alcohol users with variant genotypes of CYP2C9*3 when compared to the alcohol users in the controls (OR: 4.2; 95% CI: 1.2-14.9) [Table 5].
Table 5.
Interaction between CYP2C9 genotypes and alcohol use, and risk of HNSCC
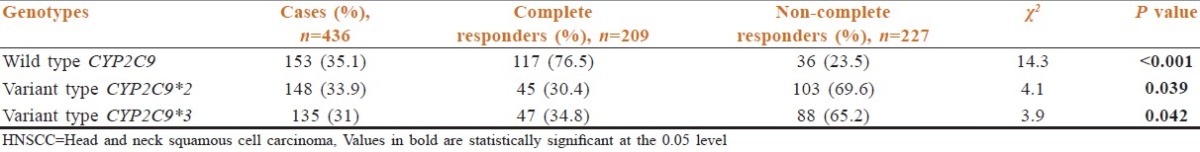
A follow-up study was also carried out in 460 patients to investigate the effects of treatment on the patients with different genotypes of CYP2C9 [Table 6]. Twenty-four patients were excluded from the final analysis, as they were either lost to follow-up (n = 9) or did not complete full coarse of CT-RT (n = 5) or at least five cycles of concurrent cisplatin (n = 10). The response to CT-RT was assessed as per the WHO criteria, in the remaining 436 patients who completed 70 Gray/35 fractions EBRT along with at least five concurrent cycles of cisplatin. Treatment-related toxicity was equally balanced between cases with wild and variant type CYP2C9 and there were no treatment breaks due to toxicity. Amongst the patients with wild type genotype of CYP2C9, 76.5% (n = 117) completely responded to CT-RT. Amongst the patients with variant genotypes of CYP2C9*012, only 30.4% (n = 45) could be categorized as complete responders, while 69.6% were found to be non-complete responders. Likewise, amongst the patients with variant CYP2C9*3 genotypes, only 34.8% (n = 47) completely responded to the treatment, while 65.2% could be categorized as non-complete responders [Table 6].
Table 6.
Treatment responses in patients of HNSCC with multiple genotypes of CYP2C9
Discussion
The data of the present study showed that functionally important polymorphism of CYP2C9 existed in north Indian population. The frequency of the variant genotypes (CYP2C9*012 and CYP2C9*013) in healthy controls was found to be higher (16.7 and 14.8%, respectively) than that reported in south Indian (7 and 1%) population.[15] This could be partly attributed to the population structure of India comprising a mixture of endogamous ethnic groups.[16] The frequency of the CYP2C9*2 genotypes in our control population was higher than that observed in other Asian populations (Chinese and Japanese), but was comparable to the Caucasians.[9] It was observed that the CYP2C9*3 polymorphism was more common in north Indian population. The frequency of the variant genotype of CYP2C9*3 was relatively higher (14.8%) when compared to the Chinese population (0.01%) and the Caucasians (7%).[9]
A relatively higher prevalence of cases with variant genotypes of CYP2C9*2 or *013 have clearly indicated that individuals inheriting PM mutant genotypes of CYP2C9 are at increased risk of developing HNSCC. The association of CYP2C9 polymorphism has not been studied with HNSCC. London et al.,[10] in 1996 have reported an increased frequency of CYP2C9*2 allele in the cases suffering from lung cancer. Since CYP2C9 is involved in the detoxification of carcinogenic substrates, the increased risk observed in cases with PM genotypes of CYP2C9 in our study could be attributed to their poor detoxifying ability.[11] Our study has indicated that the risk to HNSCC was increased in cases who were tobacco or alcohol users, suggesting that interaction of CYP2C9 genotypes with tobacco or alcohol increases the susceptibility to HNSCC. Though, tobacco chewing has been demonstrated to be the major risk factor in HNSCC as compared to cigarette smoking. PAHs are known to be generated both during cigarette smoking and tobacco chewing. The increased risk observed in cases using tobacco with PM genotypes of CYP2C9 could be explained by higher affinity of CYP2C9 for benzo (a) pyrene. CYP2C9 variant alleles have also been associated with altered metabolism of alkylating agents that are well-established mutagens.[17] A similar increase in risk in cases drinking alcohol with CYP2C9 PM genotypes have suggested that alcohol possibly interacts with CYP2C9 genotypes in increasing the risk to HNSCC. Though not much data is available on interaction of CYP2C9 genotypes and alcohol, ethanol is known to inhibit CYP2C9 activity.[18] The increase in risk could thus be possibly explained by inhibition of CYP2C9 detoxifying ability in alcohol users. Our study has further shown that chemotherapeutic response is modified in patients with PM genotypes of CYP2C9. A higher percentage of nonresponders were observed in the cases with PM genotypes of CYP2C9. Similar kind of conclusions have been given in studies with CYP2A6, CYP2D6, and CYP2C19 polymorphism and head and neck cancer.[14,19,20]
Conclusion
The present study has demonstrated a several fold increase in the risk to HNSCC in the cases with variant genotypes (PMs) of CYP2C9. The risk was even more in tobacco or alcohol users. Furthermore, it has also demonstrated that PMs of CYP2C9 modify the treatment outcome in cases receiving cisplatin-based radical CT-RT. In conclusion, genetic polymorphisms of CYP2C9 may have a role in interacting with environmental risk factors in modifying the susceptibility to HNSCC and possibly CR to CT-RT. Clearly, future prospective studies with larger sample sizes are needed to confirm these preliminary results.
Footnotes
Source of Support: Nil.
Conflict of Interest: None declared.
References
- 1.Hunter KD, Parkinson EK, Harrison PR. Profiling early head and neck cancer. Nat Rev Cancer. 2005;5:127–35. doi: 10.1038/nrc1549. [DOI] [PubMed] [Google Scholar]
- 2.Bartsch H, Nair U, Risch A, Rojas M, Wikman H, Alexandrov K. Genetic polymorphism of CYP genes, alone or in combination, as a risk modifier of tobacco-related cancers. Cancer Epidemiol Biomarkers Prev. 2000;9:3–28. [PubMed] [Google Scholar]
- 3.Hashibe M, Brennan P, Strange RC, Bhisey R, Cascorbi I, Lazarus P, et al. Meta- and pooled analyses of GSTM1, GSTT1, GSTP1, and CYP1A1 genotypes and risk of head and neck cancer. Cancer Epidemiol Biomarkers Prev. 2003;12:1509–17. [PubMed] [Google Scholar]
- 4.Van Schaik RH. Cancer treatment and pharmacogenetics of cytochrome P450 enzymes. Invest New Drugs. 2005;23:513–22. doi: 10.1007/s10637-005-4019-1. [DOI] [PubMed] [Google Scholar]
- 5.Griskevicius L, Yasar U, Sandberg M, Hidestrand M, Eliasson E, Tybring G, et al. Bioactivation of cyclophosphamide: The role of polymorphic CYP2C enzymes. Eur J Clin Pharmacol. 2003;59:103–9. doi: 10.1007/s00228-003-0590-6. [DOI] [PubMed] [Google Scholar]
- 6.Shou M, Korzekwa KR, Krausz KW, Buters JT, Grogan J, Goldfarb I, et al. Specificity of cDNA-expressed human and rodent cytochrome P450s in the oxidative metabolism of the potent carcinogen 7, 12-dimethylbenz[a] anthracene. Mol Carcinog. 1996;17:241–9. doi: 10.1002/(SICI)1098-2744(199612)17:4<241::AID-MC8>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 7.Bauer E, Guo Z, Ueng YF, Bell LC, Zeldin D, Guengerich FP. Oxidation of benzo[a] pyrene by recombinant human cytochrome P450 enzymes. Chem Res Toxicol. 1995;8:136–42. doi: 10.1021/tx00043a018. [DOI] [PubMed] [Google Scholar]
- 8.Higashi MK, Veenstra DL, Kondo LM, Wittkowsky AK, Srinouanprachanh SL, Farin FM, et al. Association between CYP2C9 genetic variants and anticoagulation-related outcomes during warfarin therapy. JAMA. 2002;287:1690–8. doi: 10.1001/jama.287.13.1690. [DOI] [PubMed] [Google Scholar]
- 9.Wang SL, Huang J, Lai MD, Tsai JJ. Detection of CYP2C9 polymorphism based on the polymerase chain reaction in Chinese. Pharmacogenetics. 1995;5:37–42. doi: 10.1097/00008571-199502000-00004. [DOI] [PubMed] [Google Scholar]
- 10.London SJ, Daly AK, Leathart JB, Navidi WC, Idle JR. Lung cancer risk in relation to the CYP2C9*011/CYP2C9*012 genetic polymorphism among African-Americans and Caucasians in Los Angeles County, California. Pharmacogenetics. 1996;6:527–33. doi: 10.1097/00008571-199612000-00006. [DOI] [PubMed] [Google Scholar]
- 11.Chan AT, Tranah GJ, Giovannucci EL, Hunter DJ, Fuchs CS. A prospective study of genetic polymorphisms in the cytochrome P-450 2C9 enzyme and the risk for distal colorectal adenoma. Clin Gastroenterol Hepatol. 2004;2:704–12. doi: 10.1016/s1542-3565(04)00294-0. [DOI] [PubMed] [Google Scholar]
- 12.Sikdar N, Mahmud SA, Paul RR, Roy B. Polymorphism in CYP1A1 and CYP2E1 genes and susceptibility to leukoplakia in Indian tobacco users. Cancer Lett. 2003;195:33–42. doi: 10.1016/s0304-3835(03)00156-3. [DOI] [PubMed] [Google Scholar]
- 13.Hung HC, Chuang J, Chien YC, Chern HD, Chiang CP, Kuo YS, et al. Genetic polymorphisms of CYP2E1, GSTM1, and GSTT1; environmental factors and risk of oral cancer. Cancer Epidemiol Biomarkers Prev. 1997;6:901–5. [PubMed] [Google Scholar]
- 14.Yadav SS, Ruwali M, Shah PP, Mathur N, Singh RL, Pant MC, et al. Association of poor metabolizers of cytochrome P450 2C19 with head and neck cancer and poor treatment response. Mutat Res. 2008;644:31–7. doi: 10.1016/j.mrfmmm.2008.06.010. [DOI] [PubMed] [Google Scholar]
- 15.Adithan C, Gerard N, Vasu S, Balakrishnan R, Shashindran CH, Krishnamoorthy R. Allele and genotype frequency of CYP2C9 in Tamilnadu population. Eur J Clin Pharmacol. 2003;59:707–9. doi: 10.1007/s00228-003-0666-3. [DOI] [PubMed] [Google Scholar]
- 16.Jose R, Chandrasekaran A, Sam SS, Gerard N, Chanolean S, Abraham BK, et al. CYP2C9 and CYP2C19 genetic polymorphisms: Frequencies in the south Indian population. Fundam Clin Pharmacol. 2005;19:101–5. doi: 10.1111/j.1472-8206.2004.00307.x. [DOI] [PubMed] [Google Scholar]
- 17.Chang TK, Yu L, Goldstein JA, Waxman DJ. Identification of the polymorphically expressed CYP2C19 and the wild-type CYP2C9-ILE359 allele as low-Km catalysts of cyclophosphamide and ifosfamide activation. Pharmacogenetics. 1997;7:211–21. doi: 10.1097/00008571-199706000-00006. [DOI] [PubMed] [Google Scholar]
- 18.Hamitouche S, Poupon J, Dreano Y, Amet Y, Lucas D. Ethanol oxidation into acetaldehyde by 16 recombinant human cytochrome P450 isoforms: Role of CYP2C isoforms in human liver microsomes. Toxicol Lett. 2006;167:221–30. doi: 10.1016/j.toxlet.2006.09.011. [DOI] [PubMed] [Google Scholar]
- 19.Ruwali M, Pant MC, Shah PP, Mishra BN, Parmar D. Polymorphism in cytochrome P450 2A6 and glutathione S-transferase P1 modifies head and neck cancer risk and treatment outcome. Mutat Res. 2009;669:36–41. doi: 10.1016/j.mrfmmm.2009.04.007. [DOI] [PubMed] [Google Scholar]
- 20.Yadav SS, Ruwali M, Pant MC, Shukla P, Singh RL, Parmar D. Interaction of drug metabolizing cytochrome P450 2D6 poor metabolizers with cytochrome P450 2C9 and 2C19 genotypes modify the susceptibility to head and neck cancer and treatment response. Mutat Res. 2010;684:49–55. doi: 10.1016/j.mrfmmm.2009.11.010. [DOI] [PubMed] [Google Scholar]


