Abstract
Relatively little progress has been made in determining the in vivo regulation of glutathione S-transferase P (GSTP), particularly the human enzyme hGSTP1, despite being identified as a significant factor in carcinogenesis and development of drug resistance in tumor cell lines. Here we report the characterization of a transgenic reporter mouse that reveals how hGSTP1 is regulated in vivo by chemopreventive agents. Basal expression was found in crypts and villi of the small and large intestine, bronchiolar epithelial cells, the epidermis and hair follicles, gall bladder epithelium, choroid plexus and biliary epithelium. Expression was induced in different tissues by the antioxidant chemopreventive agents ethoxyquin (EQ) and butylated hydroxyanisole (BHA). However, genetic deletion of the Nrf2 transcription factor, which directs central genetic programs of detoxification and protection against oxidative stress, increased rather than attenuated GSTP1 expression. In vitro investigations with mouse embryonic fibroblasts revealed factor(s) in addition to Nrf2 that control the expression of GSTP1, offering further insights into regulation. The new reporter mouse described here provides a useful tool to gain deeper insights into the mechanisms of action of chemopreventive compounds and other environmental agents.
Keywords: human GSTP1, regulation, reporter
Introduction
The glutathione S-transferases (GSTs) comprise a family of dimeric Phase II drug metabolizing enzymes (EC 2.5.1.18) whose principal function is to catalyse the conjugation of reduced glutathione to reactive electrophiles as part of a cellular adaptive response to chemical toxins ands oxidative stress (1). Glutathione S-transferase P (GSTP) has been reported to be significantly elevated in a range of animal and human tumour types, although the role of this enzyme in the tumorigenic process is still unclear; indeed, the endogenous role(s) of GSTP have yet to be fully elucidated (2, 3). Cell lines made resistant to toxic chemicals, including anti-cancer drugs, express high levels of GSTP (4); likewise, cell lines in which GSTP is over-expressed are found to be resistant to a range of drugs and chemicals (5), even though many of these compounds are not known to be substrates of the enzyme. It appears, therefore, that GSTP is likely to have actions which are not mediated by its primary catalytic function (6). This is supported by a growing body of information which suggests that GSTP can associate with kinases involved in signalling cellular responses to stress (e.g. Jun N-terminal kinase (JNK), apoptosis signal-regulating kinase (ASK1)), inhibiting kinase function and suppressing apoptosis (7, 8). Further, GSTP has been shown to play a role in protein glutathionylation and to be essential for the regeneration of the active site cysteine in the 1-Cys peroxiredoxin Prdx6 (9).
Knockout mice lacking GSTP display no overt phenotype due to the absence of the enzyme but have increased susceptibility to chemically-induced skin and lung tumours (10, 11) and are almost completely resistant to the hepatotoxic effects of acetaminophen (12). Furthermore, when GstP null mice are crossed to the APCMin line, a marked increased incidence of colonic adenomas is observed. These, like those in humans, are located in the rectal area of the large intestine, identifying the GstP null × APCMin hybrid as a model which closely recapitulates human colorectal cancer (13).
The regulation of GSTP expression has been studied extensively in mouse and rat, but less so in humans (14, 15). We and others have previously characterised the human GSTP1 promotor in vitro, identifying a number of elements through which transcription of the gene is induced or repressed (16, 17). In particular, it contains an AP-1 site (position −73 to −54) which has been extensively characterised in a multi-drug resistant derivative (VCREMS) of the breast cancer cell line, MCF-7 (18). The identification and characterisation of an antioxidant response element (ARE) (19), which potentially overlaps with AP-1 regulatory sequences, raises the possibility that the hGSTP1 gene may also be regulated by the Nrf2/KEAP1 system (20).
In order to gain further insights into the in vivo regulation of GSTP, we have generated a transgenic mouse model containing the entire human GSTP1 gene, plus 2.5kb of promotor region, fused in-frame to lacZ and cloned into the ROSA26 locus (21). We now report that this mouse line recapitulates the known pattern of GSTP expression in man and demonstrate for the first time that the expression level of the human gene can be profoundly induced by exogenous factors such as cancer chemopreventive agents. These data provide detail on how environmental factors influence cancer susceptibility.
Materials & Methods
Chemicals
All chemicals were obtained from Sigma Fine Chemicals (Poole, Dorset, UK) unless otherwise stated. Mirsky’s solution was purchased from National Diagnostics, AGTC Bioproducts, East Yorks, UK.
Generation of [hGSTP1LacZ]ROSA26 mice
hGSTP1::LacZROSA26 mice were generated by Taconic Artemis, Cologne, Germany, using approximately 500bp of the 5′-UTR and the entire hGSTP1 gene fused to LacZ (Figure 1). This genomic sequence was fused in-frame to β-galactosidase, and the whole construct cloned into the ubiquitously transcriptionally active ROSA26 region in order to optimise the chances of generating a mouse line in which the expression of the human GSTP gene is faithfully re-capitulated. This construct was targeted to the ROSA26 region in C57BL/6 embryonic stem (ES) cells by homologous recombination. The line was maintained in the hemizygous state by crossing to C57BL/6 (wild-type) mice and the presence of the genetic alteration was verified by PCR. hGSTP1::LacZROSA26 Nrf2−/− mice were generated by crossing hGSTP1::LacZROSA26 mice with Nrf2−/− mice (22), obtained originally from the laboratory of Professor Masayuki Yamamoto and subsequently back-crossing to C57BL/6 for 6 generations. It is important to note that the strategy used to generate the Nrf2 null mouse replaced the C-terminal half of Nrf2 with LacZ, potentially confounding data generated in crosses with the hGSTP1::LacZROSA26 line. However, neither basal nor induced reporter activity was detected in any of the tissues examined during prior testing of the Nrf2 null mice for LacZ expression in our laboratory (data not shown), so we are confident that the LacZ activity detected in hGSTP1::LacZROSA26 (Nrf2−/−) mice is derived from the reporter driven by hGSTP1 regulatory elements.
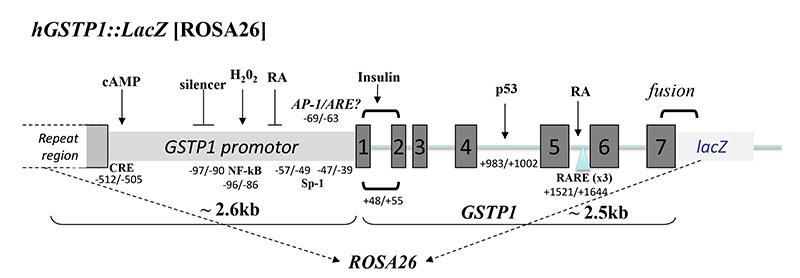
Figure 1. Generation of hGSTP1::LacZ mice.
The construct used to generate the of hGSTP1::LacZROSA26 mouse. Approximately 2.6 kb of the human GSTP1 promotor and the entire GSTP1 gene (~2.5kb) were cloned, fused in-frame with lacZ, into the ROSA26 gene locus by homologous recombination in C57BL/6 embryonic stem cells. Positions of known transcription factor binding sites, and previously reported regulators of gene induction or repression, are shown: cAMP/CRE; NF-kB; RA(32); AP-1/Sp1(18, 48, 49); Insulin(32, 50); p53(31); RA/RARE(50).
Mice were kept in open-top cages with access to RM1 rodent food (Special Diet Services) and water ad libitum and a 12h light/dark cycle. All animal procedures were carried out under Home Office Project licence according to the Animal (Scientific) Procedures Act (1986) and after local ethical review.
Treatment of mice
We have previously encountered difficulties, including reluctance of the animals to consume adulterated food and associated toxicities, when administering ethoxyquin (EQ) and butylated hydroxyanisole (BHA) via the diet. In this study, therefore, these compounds were administered by gavage over a shorter period. Adult male hGSTP1::LacZROSA26 mice (8-12 weeks) on an Nrf2+/+ or Nrf2−/− background were therefore treated with EQ or BHA by gavage (350mg/kg body weight in corn oil daily for two days). Control groups received vehicle alone. For experiments with 1-aminobenzotriazole (ABT) mice were treated as above but received ABT (50mg/kg body weight p.o.) 2 hours before treatment with EQ or corn oil. Mice were sacrificed by a rising concentration of CO2 24h after the final dose. Organs were removed rapidly, rinsed in ice-cold sterile phosphate-buffered saline (PBS) and either snap-frozen in liquid nitrogen for microsome preparation or fixed for LacZ staining. Blood was also collected by cardiac puncture into heparinized tubes. Plasma for biochemical analysis (undertaken by the Clinical Pathology Service, MRC Harwell) was prepared and stored at −20°C until required. hGSTP1::LacZROSA26 Nrf2+/+ and hGSTP1::LacZROSA26 Nrf2−/− mice treated with vehicle, EQ or BHA showed no evidence of alterations in ALT, LDH, creatinine or glucose levels compared with historical wild-type controls, although they did exhibit a small (non-significant) increase in serum bilirubin (data not shown).
Preparation of subcellular fractions and immunoblotting
Microsomal and cytosolic fractions were prepared from mouse tissues by differential centrifugation according to standard procedures and protein concentrations were determined using the Bio-Rad Protein Assay Reagent (Bio-Rad, Herts., UK). Western blot analysis was carried out loading 5μg microsomal protein per lane and using polyclonal antisera raised against β-galactosidase (Sigma, Dorset, UK), GSTP1 (BD Biosciences), NQO1 (Abcam), GSTA1 & GSTM1 (generous gifts from Professor John Hayes), and rat P450s, as described previously (23). GAPDH was used as a loading control (Sigma, Dorset, UK). Immunoreactive proteins were detected using donkey anti-rabbit horseradish peroxidase IgG (DAKO, Ely, UK) as a secondary antibody and visualized by chemiluminescence (ECL+, GE Healthcare, Bucks, UK) using a Fujifilm LAS-3000 mini imaging system (Fujifilm UK Ltd, UK). Densitometric analysis was performed using Multi Gauge V2.2 software (Fujifilm UK Ltd, UK).
Tissue sections and LacZ staining
Liver samples were fixed in PBS containing 2mM MgCl2 and 1% para-formaldehyde for 3 hours. All other tissues were fixed in Mirsky’s solution overnight. After fixation, samples were washed 3 times in PBS + 2mM MgCl2, dehydrated overnight in the same solution containing 30% sucrose, embedded in Cryo-M-Bed embedding compound (Bright, Huntingdon, Cambs, UK) and frozen in a dry ice/iso-pentane bath. Cryo-sectioning was performed using a Model OTF Cryostat (Bright, Huntingdon, Cambs, UK) set to −20°C. The samples were equilibrated to chamber temperature for 30 minutes before sectioning. Sections (10 μM) were cut and placed on adhesion slides, rehydrated in PBS + 2mM MgCl2 for 5 minutes and incubated overnight at 37°C in X-gal staining solution (PBS + 2mM MgCl2, 0.01% sodium deoxycholate, 0.02% Igepal CA630, 5mM potassium ferricyanide, 5mM potassium ferrocyanide and 1mg/ml 5-bromo-4-chloro-3-indolyl b-D-galactopyranosidase). The next day, they were washed in PBS + 2mM MgCl2 and counterstained in Nuclear Fast Red for 10 minutes. Following counterstaining the slides washed in distilled water and dehydrated through 70% and 95% ethanol before incubating in Histoclear (VWR, Leicestershire, UK) for 3 minutes, air-drying and coverslipping using DPX mountant (VWR, Leicestershire, UK). Images were captured using a Zeiss 12MP digital camera and AxioVision software (v4.5) (Carl Zeiss, Welwyn Garden City, UK) attached to an Olympus IX50 inverted light microscope.
Preparation of mouse embryo fibroblasts
Embryos were harvested under sterile conditions and skeletal/connective tissue was scissor-minced then trypsinized in PBS for 5 min at 37°C. Mouse embryo fibroblast (MEF) medium (Dulbecco’s Modified Eagle Medium (DMEM) containing 10% foetal calf serum, 1% L-glutamine and 1% penicillin/streptomycin) was added to inactivate the trypsin and the tissue was disaggregated by pipetting before being centrifuged at room temperature for 5 min. The pellet was disaggregated in trypsin and incubated for 10 min at 37°C, then MEF media was added to inhibit the trypsin before allowing the debris to settle, plating out the supernatant and incubating at 37°C in an atmosphere containing 5% CO2. Following attachment, the cells were washed with sterile PBS, re-fed with fresh MEF media and grown to confluency, at which point they were harvested by trypinisation and passaged onto fresh plates. Cells were not passaged beyond passage 4. Backup stocks were frozen at each passage and stored in 90% foetal calf serum/10% DMSO at −80°C.
Induction experiments using mouse embryo fibroblasts
MEFs (5 × 105 cells/10cm2 dish) were plated in DMEM and allowed to grow for 24h before being exposed to fresh medium containing either DMSO (0.1% (v/v)) alone or tBHQ (final concentration 20μM). After up to 24 h exposure they were harvested and used for preparation of lysates. Briefly, the cells were washed 3 times in ice-cold PBS and each 10 cm2 dish was covered with 300μl buffer (25 mM Tris HCl pH7.4 containing 150 mM NaCl, 5 mM EDTA, 1% (v/v) Nonidet 40, 0.5% (w/v) sodium deoxycholate, 0.1% (w/v) sodium dodecyl sulphate and cOmplete ULTRA protease inhibitor cocktail (Roche Diagnostics, Mannheim, Germany)). The cells were scraped into an Eppendorf tube and vortexed for 10 s, kept on ice for 30 min, and centrifuged (16,000 g for 5 minutes at 4°C). The supernatant was transferred to a clean tube and stored at −80°C. After protein estimation, samples were separated by SDS-PAGE and immunoblotted as described.
Results
Hemizygous hGSTP1::LacZROSA26 mice generated and maintained on a C57BL/6 background were fertile, with normal litter sizes and sex ratios. The transgene was transmitted in a Mendelian fashion and the mice displayed no overt phenotype or welfare issues.
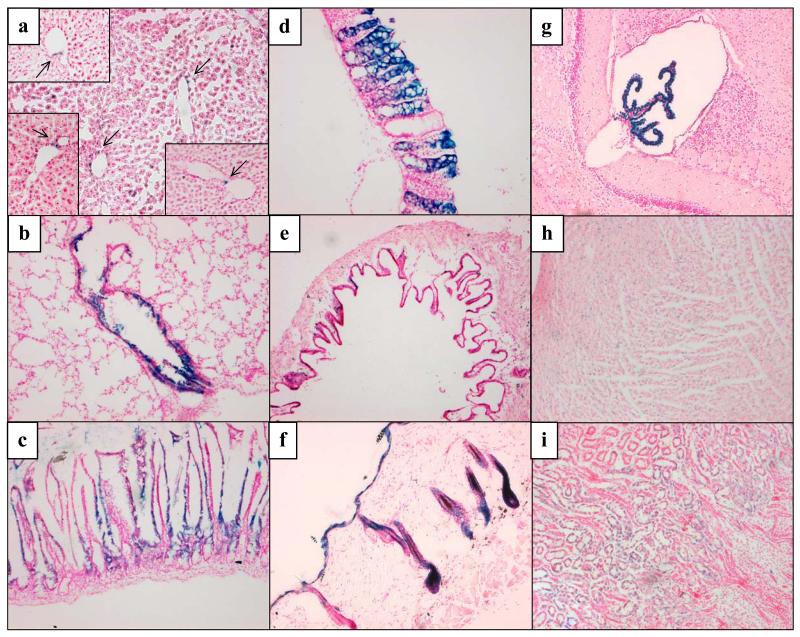
An initial tissue screen characterizing the constitutive expression of the LacZ reporter (and thus hGSTP1) in hGSTP1::LacZROSA26 mice (Figure 2) revealed that the main sites of reporter expression were the liver, lung and intestine. In the liver, LacZ staining was only visible in cells in the periportal area (Zone 1), consistent with reporter expression in the biliary epithelium. The LacZ staining observed in the lung was more intense, but this was located almost exclusively in bronchiolar epithelial cells, only occasional cells being stained in the alveolar parenchyma. In the small intestine, constitutive expression was noted in the mid-region of the villus mucosa, but not in every villus. In the large intestine, reporter expression was more uniform from the crypt to top of the villus, but again the pattern of expression was mosaic along the length of the colon. This may reflect the role of specific crypt stem cell populations in generating the mucosal population of the intestinal epithelium or the absence of essential cis-acting elements in certain cell populations, and has been observed in other transgenic reporter models (24, 25).
Figure 2. Constitutive expression of the hGSTP1::LacZ ROSA26 reporter in mouse tissues.
Adult male hGSTP1::LacZROSA26 mice (n=3) were sacrificed, tissues removed and processed for cryo-sectioning and β-galactosidase staining as described in Materials & Methods. Representative sections are shown from the following tissues: a – liver (insets show additional hepatic staining, indicated by arrows); b – lung; c – small intestine; d – large intestine; e – gall bladder; f – skin; g – choroid plexus (brain); h – heart; i – kidney. Photomicrographs acquired as detailed in Materials & Methods; bright field, magnification x10.
Low levels of reporter expression were also noted in the gall bladder epithelium, hair follicle and epidermis and across the whole of the tissue in heart and kidney, where it was punctate in nature. The brain, however, did not exhibit LacZ staining except in the choroid plexus where expression of the reporter was readily detectable, consistent with literature reports on expression of GSTP in the human brain (26).
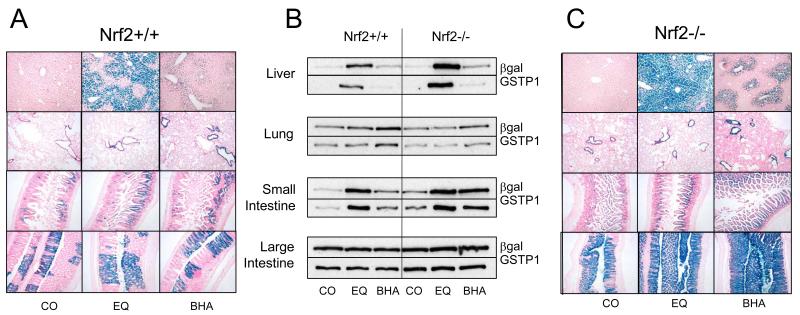
In the light of the above results, and in order to investigate whether exogenous chemopreventive agents were able to regulate GSTP1 expression in vivo, hGSTP1::LacZROSA26 mice were treated with EQ or BHA and marked changes in reporter expression were observed in the liver, lung, and small and large intestine (Figure 3A). In the liver, EQ treatment elicited significant LacZ staining in hepatocytes, predominantly in the pericentral (Zone 3) region, but also spreading into Zone 2. Following BHA administration, although significant induction was observed the effects were less marked than with ethoxyquin and were restricted to the pericentral (Zone 3) region. Conversely, in the lung, BHA significantly induced expression of the LacZ reporter whereas EQ did not. Pulmonary LacZ staining was almost exclusively restricted to the bronchiolar epithelial cells under both basal and induced conditions, although a small number of cells in the alveolar parenchyma were also positive for reporter gene expression in untreated tissue and this expression increased marginally in response to EQ and BHA treatment. Reporter expression in the small intestine sections was marginally induced by both EQ and BHA (more so by the latter), but remained patchy throughout the tissue and was absent from some regions even after induction. Reporter expression was confined to the mid-region of the villus, punctate staining being observed at the base of the crypt and in the muscularis propria. In the large intestine, patchy reporter expression was detected throughout the length of the tissue (but was uniform along the length of those crypts which were positive) and remained essentially unchanged following EQ and BHA treatment. Western blotting demonstrated that the detection of the reporter, as evidenced by LacZ staining, (Figure 3B) generally reflected the level of expression of both β-galactosidase and hGSTP1 protein in each tissue (Supplemental Figure 1). However, it should be noted that whereas Western blots involve analysis of an entire tissue, β-galactosidase staining shows the effects in specific cell types, often resulting in different changes being observed between these two methods.
Figure 3. hGSTP1::LacZROSA26 and hGSTP1::LacZROSA26::Nrf2−/− – basal and induced expression.
Adult male hGSTP1::LacZROSA26 and hGSTP1::LacZROSA26 (Nrf2−/−) mice (n=3) were treated with EQ or BHA (350mg/kg, p.o.) or corn-oil vehicle and then sacrificed, tissues removed and processed for β-galactosidase staining and Western blotting as described in Materials & Methods. Representative sections from liver, lung, small intestine and large intestine are shown in panel A (hGSTP1::LacZROSA26) and panel C (hGSTP1::LacZROSA26 (Nrf2−/−)). Western blots of pooled microsomal samples against β-galactosidase and hGSTP1 are shown in panel B. Photomicrographs acquired as detailed in Materials & Methods; bright field, magnification x10. CO = corn oil, EQ = ethoxyquin, BHA = butylated hydroxyanisole.
Chemopreventive agents such as BHA and EQ are thought to exert their protective properties through the activation of the transcription factor Nrf2, which in turn induces expression of genes which contain the antioxidant response element (ARE) in their promotors. In order to clarify the role of the ARE in the induction of the hGSTP1::LacZROSA26 reporter, we crossed the reporter mice onto a Nrf2−/− background and repeated the induction protocols with EQ and BHA (Figure 3C). Deletion of Nrf2 had no effect on basal reporter expression in the liver, but resulted in a more marked response to EQ treatment, leading to pan-acinar staining in hepatocytes (Figures 3B,C) although Kupffer cells, stromal cells and vascular and biliary epithelium remained unstained. Similarly, expression was higher following BHA treatment on the Nrf2−/− background than in the presence of Nrf2, but in this case it remained confined to pericentral (Zone 3) hepatocytes (Figure 3A vs Figure 3C). In the lung, deletion of Nrf2 made no difference to basal or induced level or location of reporter expression in the case of EQ, which remained almost exclusively confined to the bronchiolar epithelium, but LacZ staining was more intense following BHA treatment, independent of Nrf2 status. LacZ staining in the small intestine was unchanged after corn oil or EQ treatment but was elevated and more widely distributed after BHA administration (Figure 3B,C). Both basal and induced (by EQ or BHA) reporter gene expression were elevated in the large intestine; furthermore, rather than being patchy or mosaic in nature (as in Nrf2+/+ (Figure 3A)), LacZ staining was found throughout the large intestine (Figure 3C), although the lamina propria, muscularis propria and lymphoid aggregates remained negative. For liver, lung and small intestine, hGSTP1 expression mirrored that of β-galactosidase, as evidenced by Western blotting, although it should be noted that expression of β-galactosidase and induction of hGSTP1 in liver after EQ and BHA treatment are difficult to estimate accurately due to the extremely low level of expression of these proteins in control mice (Figure 3B; Supplemental Figure 1). Furthermore, in Supplemental Figure 1, the values for EQ and BHA are shown with respect to their equivalent control; however, the fold-change in absolute expression values between Nrf2 wild-type and null genotypes for β-galactosidase and hGSTP1 in liver after EQ treatment (1.9- and 2.3-fold, respectively) are consistent with the increased expression observed on the Western blot (Figure 3B).
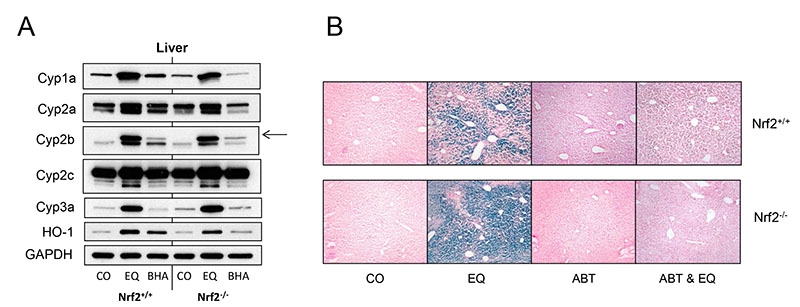
In order to establish why deletion of Nrf2 resulted in an increased expression of hGSTP1, we investigated the role of metabolism in EQ-mediated regulation of hGSTP1. Western blot analysis of the hepatic expression of several P450 enzymes following induction by EQ and BHA indicated that treatment with EQ increased the expression of Cyp1a, Cyp2b and Cyp3a at the protein level but only had a marginal effect, if any, on Cyp2a and Cyp2c expression, while BHA only induced the expression of Cyp2b, and to a much lesser extent than EQ. Nrf2 status did not affect this pattern of expression (Figure 4A; Supplemental Figure 2). These results suggest that differences between Nrf2+/+ and Nrf2−/− mice in the effects of EQ are not a function of different responsiveness to P450 induction by this antioxidant. However, this does not preclude the possibility that P450-mediated metabolism plays a role in these effects. In order to investigate this further, we used the cytochrome P450 inhibitor ABT to interfere with the metabolism of EQ and determined the consequences for reporter gene expression. As expected, EQ upregulated reporter expression in hGSTP1::LacZROSA26 Nrf2+/+ mice, and reporter induction was more marked on the Nrf2−/− background. In mice treated with ABT or vehicle, hepatic LacZ staining was completely absent, regardless of the presence of Nrf2 (Figure 4B). However, when EQ treatment was preceded by dosing with ABT, reporter gene induction was severely attenuated, being completely abolished in the absence of Nrf2, while only a very few LacZ positive hepatocytes were found in mice with functional Nrf2 (Figure 4B). These results indicate that P450-mediated metabolism of EQ is an essential step in the induction of hGSTP1 expression, and also for the activation of Nrf2 by EQ. Biochemical analysis of serum from hGSTP1::LacZROSA26 Nrf2+/+ and hGSTP1::LacZROSA26 Nrf2−/− mice treated with ABT + EQ showed no changes in ALT or glucose levels, but approximately 5-fold increases for LDH, bilirubin and creatinine following administration of both ABT and EQ (data not shown).
Figure 4. P450 activity is essential for EQ induction of reporter expression in hGSTP1::LacZROSA26 mice.
Adult male hGSTP1::LacZROSA26 mice, wild-type or nulled for Nrf2 (n=2 or 3), were treated with EQ (350mg/kg, p.o.) with or without pre-treatment with ABT (50mg/kg, p.o.) or corn-oil vehicle and then sacrificed, tissues removed and processed for Western blotting using HO-1 and P450 antisera (A) and β-galactosidase staining (B) as described inMaterials & Methods. Representative liver sections are shown in panel A; in panel B, arrow indicates the band for Cyp2b10. Photomicrographs acquired as detailed in Materials & Methods; bright field, magnification x10. CO = corn oil, EQ = ethoxyquin, ABT = 1-aminobenzotriazole.
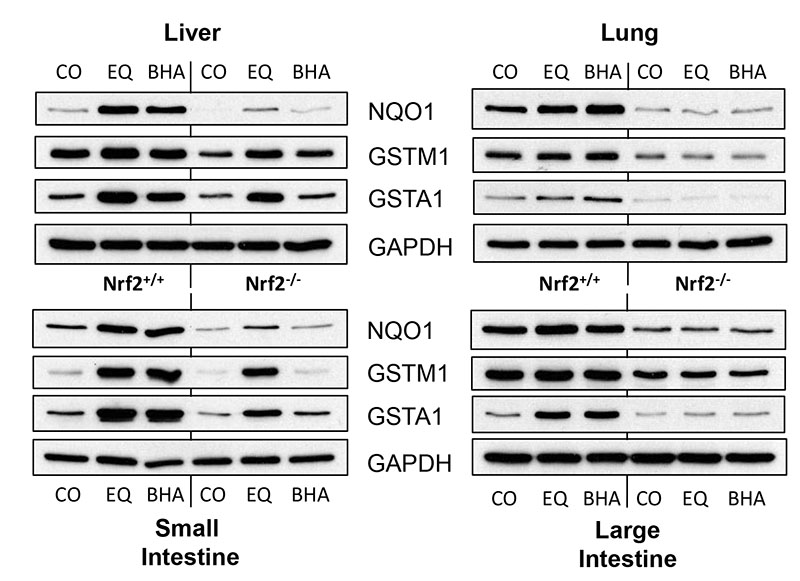
The effects of Nrf2 deletion on the expression of the hGSTP1::LacZROSA26 reporter contrast with the way this deletion affects the expression of other genes (such as haem oxygenase 1 (HO-1)) which are known to be regulated by Nrf2 via the ARE. In our model, induction of HO-1 by EQ and BHA was attenuated and essentially abolished, respectively, in the absence of Nrf2 (Figure 4A). We went on to investigate the effects of EQ and BHA on expression of other ARE-regulated genes in hGSTP1::LacZROSA26 mice (with or without functional Nrf2) by means of Western blot analysis of NQO1, GSTA1 and GSTM1 in cytosolic fractions from liver, lung, small and large intestine (Figure 5; Supplemental Figure 3A). Hepatic expression of NQO1 was significantly upregulated by both EQ and BHA in the presence of functional Nrf2 but was not changed above basal values in Nrf2−/− mice; a similar pattern was observed in the other tissues studied (Supplemental Figure 3B). In the case of GSTM1, basal and induced expression levels in hGSTP1::LacZROSA26 Nrf2+/+ mice were reduced to varying degrees (but not abolished) in liver, lung and large intestine in the absence of Nrf2. Interestingly, this was not the case in the small intestine. In this tissue GSTM1 expression was not increased in hGSTP1::LacZROSA26 Nrf2−/− mice following BHA treatment, but induction by EQ was essentially undiminished and thus appeared to be independent of Nrf2 status. The induction of GSTA1 observed in lung and large intestine of hGSTP1::LacZROSA26 Nrf2+/+ mice following EQ or BHA treatment was completely abolished on the Nrf2−/− genetic background, although some induction was observed in the liver and small intestine of this genotype after both treatments.
Figure 5. Enzyme induction in tissues from hGSTP1::LacZROSA26 + Nrf2 following treatment with EQ and BHA.
Adult male hGSTP1::LacZROSA26 mice, wild-type or nulled for Nrf2 (n=3), were treated with EQ or BHA (350mg/kg, p.o.) or corn-oil vehicle and then sacrificed, tissues (liver, lung, small and large intestine) removed and processed for Western blotting using antisera against the proteins shown as described in Materials & Methods. Cytosolic fractions were pooled within a treatment group. CO = corn oil, EQ = ethoxyquin, BHA = butylated hydroxyanisole.
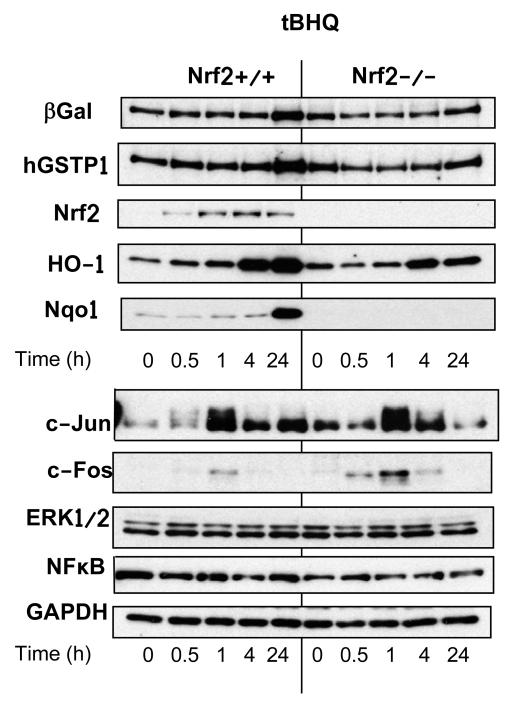
In order to model the regulation of hGSTP1 and the LacZ reporter by signalling through the ARE, we used tBHQ (the active metabolite of BHA) as an example of a compound which has been shown unequivocally to act via an antioxidant mechanism. The regulation of β-galactosidase (representing the hGSTP1::LacZ reporter), hGSTP1 and various antioxidant response proteins was investigated by Western blotting in MEFs prepared from both hGSTP1::LacZROSA26 Nrf2+/+ and hGSTP1::LacZROSA26 Nrf2−/− mice. Treatment of hGSTP1::LacZROSA26 Nrf2+/+ MEFs with tBHQ led to an increased expression of hGSTP1 at 24h; this increase was attenuated in fibroblasts derived from Nrf2−/− mice, indicating a role for the ARE in hGSTP regulation (Figure 6; Supplemental Figure 4). The expression of Nrf2 in Nrf2+/+ fibroblasts began to be upregulated within 30-50 min of exposure, peaked after 4 hours and was beginning to decline by 24 h. There was also a time-dependent increase in the expression of HO-1, which increased steadily over the first 24 h of exposure, while there was little or no change in NQO-1 expression at the earlier timepoints, increased expression only being detected after 24h. In contrast, the expression of NFκB and ERK1/2 did not change over the 24h period of exposure to tBHQ, and while there was increased expression of both c-Jun and c-Fos peaking after 1h, the increase was barely detectable in the latter case. When Nrf2−/− fibroblasts were treated in the same way, the pattern of β-galactosidase and hGSTP1 expression was the same as on the Nrf2+/+ background, while the expression of Nrf2 was, as expected, below the limit of detection (Supplemental Figure 4). In contrast with the observations in Nrf2+/+ cells, the expression of NQO-1 was completely abolished in Nrf2−/− fibroblasts, and HO-1 expression was somewhat lower although it exhibited the same general time course as in Nrf2+/+ cells. Protein quantitation shows the expression profile for c-Fos to be virtually identical in mice with and without functional Nrf2, while for c-Jun induction was observed in both genotypes, albeit lower and less sustained in the absence of Nrf2 (Supplemental Figure 4). The expression of ERK1/2 and NFκB did not alter from basal levels over the 24h period, regardless of the Nrf2 status of the cells. There was no change in expression of β-galactosidase protein in untreated MEFs, or MEFs treated only with DMSO, over the 24h period (data not shown).
Figure 6. Induction of gene expression by tBHQ in mouse embryonic fibroblasts from hGSTP1::LacZROSA26 and hGSTP1::LacZROSA26 (Nrf2−/−) mice.
Mouse embryonic fibroblasts were made from hGSTP1::LacZROSA26 mice, wild-type or nulled for Nrf2, and treated with tBHQ as described in Materials & Methods. Cells were harvested at the timepoints indicated, cell lysates prepared and immunoblotted for various proteins as shown. Results are representative of more than one independent experiment.
Discussion
We and others have demonstrated that GSTP can play a critical role in both chemically-induced and genetically-initiated carcinogenesis (11, 13, 27). GSTP is also often over-expressed in human tumours such as lung, colon and ovary (28). Understanding the endogenous and exogenous factors which control GSTP expression is therefore important in defining those variables which may influence cancer susceptibility and drug response in cancer patients. (2, 29). To that end, we have generated a transgenic mouse model containing the complete sequence of hGSTP together with a LacZ reporter gene to permit histochemical localisation of the expressed products of the transgene. The reporter construct contained 2.6kb of flanking sequence upstream of the GSTP transcription start site, encompassing the reported regulatory elements in the promotor (18, 30) and the entire coding sequence, since regulatory motifs have also been reported in both intronic and exonic regions of the GSTP gene (31, 32). We demonstrate that this model recapitulates the expression profile of hGSTP in vivo, allowing further investigation concerning the regulation and expression profile of this gene, and providing clues to the endogenous role(s) of GSTP in cellular homeostasis.
Expression of hGSTP1 did not cause any overt phenotypic changes in the hGSTP1::LacZROSA26 mice, which bred normally and exhibited Mendelian inheritance of the transgene. A tissue screen for β-galactosidase activity found that the reporter gene was expressed to varying degrees throughout the body, in a manner which generally reflected what had been previously reported for expression of hGSTP1 (33). In the livers of hGSTP1::LacZROSA26 mice, reporter expression was evident around the portal area and in the epithelium of the gall bladder. This corresponds with the restricted expression of hGSTP1 in human biliary epithelium and gall bladder (34, 35), whereas mGstp1/p2 has been reported to be a major isoenzyme in male mouse liver (36). GSTP1 has previously been reported to be the major GST isoform in human lung (37), being expressed predominantly in the bronchiolar epithelium (38) as observed in the current study. Similarly, consistent with our findings, hGSTP1 expression has been reported in both villi and crypts of the small intestine, while in the colonic mucosa it was the only GST isoform identifiable (34). LacZ staining was also seen in the epidermis and sebaceous glands of the skin, heart muscle, choroid plexus and kidney tubules, consistent with the pattern of GSTP1 expression in human tissues (33). In particular, investigation of the expression of GST in human brain has indicated localisation to interfaces between blood, brain and cerebrospinal fluid (CSF), including the choroid plexus, leading to speculation that GSTP might function to protect the brain from blood- and CSF-borne toxins (26). These data are also consistent with the expression of hGSTP1 measured by quantitation of RNA across tissues as reported in the BioGPS database (http://biogps.org) (39).
The pattern of reporter expression in hGSTP1::LacZROSA26 mice demonstrates the dominant effect of cis-regulatory sequences over the prevailing cellular environment in the regulation of hGSTP1 expression, the distribution observed being more reflective of the human pattern (reflecting the origin of the construct used) than the mouse pattern (determined by trans-acting factors within the mouse cellular environment). Thus, cis-acting elements in the 5′ region of the human GSTP1 gene are sufficient to establish the human pattern of expression. The utility of humanised mice for studying the biological role of hGSTP1 is illustrated by studies in which a different humanised mouse line was used to address species differences in the toxicity of acetaminophen. When hepatotoxicity due to acetaminophen (300 mg/kg, administered either i.p. or p.o.) was compared in wild-type, mGstp1/2 null and GSTP1 humanised mice, the humanised mice exhibited a response which was intermediate between those of the wild-type and null mice, consistent with the expression of hGSTP1 in non-parenchymal cell types (Kupffer cells, endothelial cells and bile ducts) but not hepatocytes in the humanised line compared with expression in all (or the majority of) hepatocytes in wild type mice and a complete absence of GSTP in null mice (35). The reporter line we describe will be of utility in the further pursuit of these issues because the construct it contains includes more of the 5′ regulatory region of hGSTP1 and it carries a reporter gene for localisation of hGSTP1 expression as well as the complete coding sequence of hGSTP1 for functional studies.
Having established that basal expression of the LacZ reporter in the hGSTP1::LacZROSA26 mice reflected the expression profile of hGSTP1, we looked at how this expression could be altered in four key tissues (liver, lung, small and large intestine) following treatment with the inducing agents EQ and BHA, both of which have been reported to induce gene expression via the Nrf2/Keap1 system and the ARE (40). Interestingly, however, the induction of reporter expression in both liver and intestine following treatment with EQ or BHA was potentiated by the removal of Nrf2, a phenomenon that contrasted with the attenuated expression and induction of NQO1, and to a lesser extent GSTM1 and GSTA1, in the absence of Nrf2. It is also of note that the induction of oxidative stress in the liver, as measured HO-1 expression, is mediated at least in part by Nrf2, since EQ exhibits both Nrf2-dependent and independent action. Conversely, pulmonary expression of hGSTP1, as determined by both LacZ staining and immunoblotting for the hGSTP1/βGal fusion protein, was unchanged regardless of Nrf2 genotype. It is interesting to note that both basal and induced expression of NQO1, GSTM1, GSTA1 in the lung was severely attenuated in the absence of Nrf2 (Supplemental Figures 3A and 3B). This may indicate, either that the ARE does not play a significant role in regulating the expression of hGSTP1, and/or that pulmonary expression is already at maximal levels, possibly due to a critical role played by the enzyme in this tissue (11, 41).
There are several possible explanations for these findings; for example, Nrf2 may not regulate expression of hGSTP directly; instead it may affect levels of an enzyme(s) involved in the metabolism of EQ and BHA or their derivatives. In the absence of Nrf2, altered metabolism could lead to the accumulation of one or more metabolites that, in turn, result in increased hGSTP induction through other regulatory elements such as AP-1 or NFκB. These data are interesting as they illustrate not only that chemopreventive agents exert their effects by multiple pathways in addition to their effects on Nrf2 signalling, but also that GSTP is regulated in vivo by cellular stress pathways such as AP-1, independent of Nrf2. Our experiments with ABT demonstrate that at least two mechanisms are involved in transgene (and hence hGSTP1) induction by EQ and that one of them requires P450-mediated metabolic activation. Although the identity of the P450(s) involved is unclear, it is likely to involve members of the Cyp2b family, since EQ upregulated the expression of Cyp2b10 in our experiments and has previously been shown to strongly induce the de-ethylation of benzyloxyresorufin (42). Furthermore, microarray analysis of mouse liver following EQ administration revealed a gene expression profile similar to that observed following phenobarbital treatment: the expression of 70 genes was increased at least 2-fold in response to PB and 23 of these were also upregulated to a similar extent following treatment with EQ (unpublished). The most likely explanation for the potentiation of GSTP expression in Nrf2−/− mice is that both EQ and BHA activate other signalling cascades, such as AP-1 signalling, and that as a consequence of the attenuation of the antioxidant Nrf2 response these stress response pathways are further induced.
It is clear from our in vitro data that tBHQ is capable of inducing the expression of hGSTP and that this is compromised by the absence of Nrf2, at least in vitro, suggesting that Nrf2 can regulate expression of hGSTP via the ARE (Figure 6, Supplemental Figure 4). Loss of Nrf2 may allow other transcription factors (e.g. Fos and Jun) to access the embedded AP-1 site within the ARE (43), thus increasing gene expression. The contrast between in vitro and in vivo findings with regard to increased expression in the absence of Nrf2 may be due to a lack of the relevant transcription factor(s) in the MEF system. These data differ from those in a previous report stating that tBHQ and sulforaphane fail to induce hGSTP expression in HaCaT cells (44). These differences may simply reflect the different models employed: HaCaT is a transformed human keratinocyte cell line and may lack essential metabolic and/or regulatory factors required for induction by tBHQ and sulforaphane.
There has been intense debate regarding the consensus sequence of the ARE, which apparently varies in terms of sequence, numbers of elements and genomic organisation not only between species but also between genes within the same organism (19, 45, 46). While a simple definition of the ARE is not possible, Hayes et al., (20) have argued that the variable context and content of the ARE lends itself to recognizing four classes of enhancer element:
Classes 1 and 2 contain the 16bp extended ARE (5′-TMAnnRTGAYnnnGCR-3′, where M = A/C, R = A/G and Y = C/T and the 9bp core ARE is in bold), with or without an embedded AP-1 site (underlined), respectively.
Classes 3 and 4 contain a minimal ARE (5′-RTGAYnnnGCR-3′), with or without an embedded AP-1 site (underlined), respectively.
Examination of GSTP promotors in mouse, rat and human reveals an ARE within the first 150bp upstream of the transcription start site in each case; the rodent sequences consist of a 9bp core with an embedded AP-1 site, while the corresponding element in the human gene is, under the classification outlined above, a Class 3 ARE. It is also noteworthy that the rat GSTP promotor contains a further ARE approximately 2.5kb upstream of the ATG. This has been termed the GPEI (47) and comprises a Class 1 ARE. The human GSTP gene has another ARE at approximately −2166bp (44).
In summary, we have generated a transgenic mouse model which recapitulates the expression profile of hGSTP in vivo, and shown that cellular levels of human GSTP can be profoundly affected in vivo by environmental factors in a predominantly Nrf2-independent manner. These data demonstrate how dietary components and other exogenous factors may, through modulation of GSTP levels, affect cancer susceptibility and thus potentially the outcome of chemotherapy. The hGSTP1::LacZROSA26 model will also be a valuable tool with which to investigate the endogenous role(s) of GSTP in cellular homeostasis, response to toxic insults and mechanisms of carcinogenesis.
Supplementary Material
Acknowledgements
Catherine Hughes, Susanne van Schelven and Jennifer Kennedy are thanked for excellent technical assistance. Probit Chakravarty of Cancer Research UK Bioinformatics & Biostatistics Group, London Research Institute, is also thanked for invaluable bioinformatics assistance.
CRW is funded by a Cancer Research UK Programme Grant C4639/A10822.
Abbreviations
- ABT
1-aminobenzotriazole
- ALT
alanine aminotransferase
- ARE
antioxidant response element
- ASK1
apoptosis signal-regulating kinase
- BHA
butylated hydroxyanisole
- CSF
cerebrospinal fluid
- DMEM
Dulbecco’s modified Eagle medium
- EQ
ethoxyquin
- ES cells
embryonic stem cells
- GST
glutathione transferase
- HO-1
haem oxygenase-1
- I3C
indole-3-carbinole
- JNK
jun N-terminal kinase
- LDH
lactate dehydrogenase
- MEF
mouse embryo fibroblast
- NQO-1
NAD(P)H:quinone oxidoreductase
- PBS
phosphate-buffered saline
- PCR
polymerase chain reaction
- Prdx6
peroxiredoxin 6
- UTR
untranslated region
References
- 1.Hayes JD, Flanagan JU, Jowsey IR. Glutathione transferases. Annu Rev Pharmacol Toxicol. 2005;45:51–88. doi: 10.1146/annurev.pharmtox.45.120403.095857. [DOI] [PubMed] [Google Scholar]
- 2.Henderson CJ, McLaren AW, Moffat GJ, Bacon EJ, Wolf CR. Pi-class glutathione S-transferase: regulation and function. Chemico-Biological Interactions. 1998;111-112:69–82. doi: 10.1016/s0009-2797(97)00176-2. [DOI] [PubMed] [Google Scholar]
- 3.Henderson CJ, Wolf CR. Knockout and transgenic mice in glutathione transferase research. Drug Metab Rev. 2011;43:152–64. doi: 10.3109/03602532.2011.562900. [DOI] [PubMed] [Google Scholar]
- 4.Wareing CJ, Black SM, Hayes JD, Wolf CR. Increased levels of alpha-class and pi-class glutathione S-transferases in cell lines resistant to 1-chloro-2,4-dinitrobenzene. European Journal of Biochemistry. 1993;217:671–6. doi: 10.1111/j.1432-1033.1993.tb18292.x. [DOI] [PubMed] [Google Scholar]
- 5.McLellan LI, Wolf CR. Glutathione and glutathione-dependent enzymes in cancer drug resistance. Drug Resist Updat. 1999;2:153–64. doi: 10.1054/drup.1999.0083. [DOI] [PubMed] [Google Scholar]
- 6.Tew KD. Redox in redux: Emergent roles for glutathione S-transferase P (GSTP) in regulation of cell signaling and S-glutathionylation. Biochem Pharmacol. 2007;73:1257–69. doi: 10.1016/j.bcp.2006.09.027. [DOI] [PubMed] [Google Scholar]
- 7.Elsby R, Kitteringham NR, Goldring CE, Lovatt CA, Chamberlain M, Henderson CJ, et al. Increased constitutive c-Jun N-terminal kinase signaling in mice lacking glutathione S-transferase Pi. Journal of Biological Chemistry. 2003;278:22243–9. doi: 10.1074/jbc.M301211200. [DOI] [PubMed] [Google Scholar]
- 8.Wu Y, Fan Y, Xue B, Luo L, Shen J, Zhang S, et al. Human glutathione S-transferase P1-1 interacts with TRAF2 and regulates TRAF2-ASK1 signals. Oncogene. 2006;25:5787–800. doi: 10.1038/sj.onc.1209576. [DOI] [PubMed] [Google Scholar]
- 9.Manevich Y, Hutchens S, Tew KD, Townsend DM. Allelic variants of glutathione S-transferase P1-1 differentially mediate the peroxidase function of peroxiredoxin VI and alter membrane lipid peroxidation. Free radical biology and medicine. 2013;54:62–70. doi: 10.1016/j.freeradbiomed.2012.10.556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Henderson CJ, Smith AG, Ure J, Brown K, Bacon EJ, Wolf CR. Increased skin tumorigenesis in mice lacking pi class glutathione S-transferases. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:5275–80. doi: 10.1073/pnas.95.9.5275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ritchie KJ, Henderson CJ, Wang XJ, Vassieva O, Carrie D, Farmer PB, et al. Glutathione transferase pi plays a critical role in the development of lung carcinogenesis following exposure to tobacco-related carcinogens and urethane. Cancer Research. 2007;67:9248–57. doi: 10.1158/0008-5472.CAN-07-1764. [DOI] [PubMed] [Google Scholar]
- 12.Henderson CJ, Wolf CR, Kitteringham N, Powell H, Otto D, Park BK. Increased resistance to acetaminophen hepatotoxicity in mice lacking glutathione S-transferase Pi. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:12741–5. doi: 10.1073/pnas.220176997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ritchie KJ, Walsh S, Sansom OJ, Henderson CJ, Wolf CR. Markedly enhanced colon tumorigenesis in Apc(Min) mice lacking glutathione S-transferase Pi. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:20859–64. doi: 10.1073/pnas.0911351106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ikeda H, Serria MS, Kakizaki I, Hatayama I, Satoh K, Tsuchida S, et al. Activation of mouse Pi-class glutathione S-transferase gene by Nrf2(NF-E2-related factor 2) and androgen. The Biochemical journal. 2002;364:563–70. doi: 10.1042/BJ20011756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sakai M, Muramatsu M. Regulation of glutathione transferase P: a tumor marker of hepatocarcinogenesis. Biochemical and biophysical research communications. 2007;357:575–8. doi: 10.1016/j.bbrc.2007.03.174. [DOI] [PubMed] [Google Scholar]
- 16.Moffat GJ, McLaren AW, Wolf CR. Involvement of Jun and Fos proteins in regulating transcriptional activation of the human pi class glutathione S-transferase gene in multidrug-resistant MCF7 breast cancer cells. The Journal of biological chemistry. 1994;269:16397–402. [PubMed] [Google Scholar]
- 17.Morrow CS, Cowan KH, Goldsmith ME. Structure of the human genomic glutathione S-transferase-pi gene. Gene. 1989;75:3–11. doi: 10.1016/0378-1119(89)90377-6. [DOI] [PubMed] [Google Scholar]
- 18.Moffat GJ, McLaren AW, Wolf CR. Transcriptional and post-transcriptional mechanisms can regulate cell-specific expression of the human Pi-class glutathione S-transferase gene. Biochemical Journal. 1997;324:91–5. doi: 10.1042/bj3240091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nioi P, McMahon M, Itoh K, Yamamoto M, Hayes JD. Identification of a novel Nrf2-regulated antioxidant response element (ARE) in the mouse NAD(P)H:quinone oxidoreductase 1 gene: reassessment of the ARE consensus sequence. The Biochemical journal. 2003;374:337–48. doi: 10.1042/BJ20030754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hayes JD, McMahon M, Chowdhry S, Dinkova-Kostova AT. Cancer chemoprevention mechanisms mediated through the Keap1-Nrf2 pathway. Antioxid Redox Signal. 2010;13:1713–48. doi: 10.1089/ars.2010.3221. [DOI] [PubMed] [Google Scholar]
- 21.Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat Genet. 1999;21:70–1. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- 22.Itoh K, Chiba T, Takahashi S, Ishii T, Igarashi K, Katoh Y, et al. An Nrf2/small Maf heterodimer mediates the induction of phase II detoxifying enzyme genes through antioxidant response elements. Biochemical and biophysical research communications. 1997;236:313–22. doi: 10.1006/bbrc.1997.6943. [DOI] [PubMed] [Google Scholar]
- 23.Forrester LM, Henderson CJ, Glancey MJ, Back DJ, Park BK, Ball SE, et al. Relative expression of cytochrome P450 isoenzymes in human liver and association with the metabolism of drugs and xenobiotics. Biochem J. 1992;281(Pt 2):359–68. doi: 10.1042/bj2810359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hirsch MR, d’Autreaux F, Dymecki SM, Brunet JF, Goridis C. A Phox2b::FLPo transgenic mouse line suitable for intersectional genetics. Genesis. 2013;51:506–14. doi: 10.1002/dvg.22393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sweetser DA, Hauft SM, Hoppe PC, Birkenmeier EH, Gordon JI. Transgenic mice containing intestinal fatty acid-binding protein-human growth hormone fusion genes exhibit correct regional and cell-specific expression of the reporter gene in their small intestine. Proceedings of the National Academy of Sciences of the United States of America. 1988;85:9611–5. doi: 10.1073/pnas.85.24.9611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Carder PJ, Hume R, Fryer AA, Strange RC, Lauder J, Bell JE. Glutathione S-transferase in the human brain. Neuropathology and Applied Neurobiology. 1990;16:293–303. doi: 10.1111/j.1365-2990.1990.tb01264.x. [DOI] [PubMed] [Google Scholar]
- 27.Satoh K, Kitahara A, Soma Y, Inaba Y, Hatayama I, Sato K. Purification, induction, and distribution of placental glutathione transferase: a new marker enzyme for preneoplastic cells in the rat chemical hepatocarcinogenesis. Proceedings of the National Academy of Sciences of the United States of America. 1985;82:3964–8. doi: 10.1073/pnas.82.12.3964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McIlwain CC, Townsend DM, Tew KD. Glutathione S-transferase polymorphisms: cancer incidence and therapy. Oncogene. 2006;25:1639–48. doi: 10.1038/sj.onc.1209373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Henderson CJ, Wolf CR. Disruption of the glutathione transferase pi class genes. Methods in Enzymology. 2005;401:116–35. doi: 10.1016/S0076-6879(05)01007-4. [DOI] [PubMed] [Google Scholar]
- 30.Morrow CS, Goldsmith ME, Cowan KH. Regulation of human glutathione S-transferase pi gene transcription: influence of 5′-flanking sequences and trans-activating factors which recognize AP-1-binding sites. Gene. 1990;88:215–25. doi: 10.1016/0378-1119(90)90034-o. [DOI] [PubMed] [Google Scholar]
- 31.Lo HW, Stephenson L, Cao X, Milas M, Pollock R, Ali-Osman F. Identification and functional characterization of the human glutathione S-transferase P1 gene as a novel transcriptional target of the p53 tumor suppressor gene. Molecular cancer research: MCR. 2008;6:843–50. doi: 10.1158/1541-7786.MCR-07-2105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xia C, Taylor JB, Spencer SR, Ketterer B. The human glutathione S-transferase P1-1 gene: modulation of expression by retinoic acid and insulin. Biochem J. 1993;292(Pt 3):845–50. doi: 10.1042/bj2920845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Terrier P, Townsend AJ, Coindre JM, Triche TJ, Cowan KH. An immunohistochemical study of pi class glutathione S-transferase expression in normal human tissue. Am J Pathol. 1990;137:845–53. [PMC free article] [PubMed] [Google Scholar]
- 34.Hayes PC, Harrison DJ, Bouchier IA, McLellan LI, Hayes JD. Cytosolic and microsomal glutathione S-transferase isoenzymes in normal human liver and intestinal epithelium. Gut. 1989;30:854–9. doi: 10.1136/gut.30.6.854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vaughn MP, Biswal Shinohara D, Castagna N, Hicks JL, Netto G, De Marzo AM, et al. Humanizing pi-class glutathione S-transferase regulation in a mouse model alters liver toxicity in response to acetaminophen overdose. PLoS One. 2011;6:e25707. doi: 10.1371/journal.pone.0025707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McLellan LI, Hayes JD. Sex-specific constitutive expression of the pre-neoplastic marker glutathione S-transferase, YfYf, in mouse liver. The Biochemical journal. 1987;245:399–406. doi: 10.1042/bj2450399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang M, Coles BF, Delongchamp R, Lang NP, Kadlubar FF. Effects of the ADH3, CYP2E1, and GSTP1 genetic polymorphisms on their expressions in Caucasian lung tissue. Lung cancer. 2002;38:15–21. doi: 10.1016/s0169-5002(02)00150-2. [DOI] [PubMed] [Google Scholar]
- 38.Anttila S, Hirvonen A, Vainio H, Husgafvel-Pursiainen K, Hayes JD, Ketterer B. Immunohistochemical localization of glutathione S-transferases in human lung. Cancer Res. 1993;53:5643–8. [PubMed] [Google Scholar]
- 39.Wu C, Orozco C, Boyer J, Leglise M, Goodale J, Batalov S, et al. BioGPS: an extensible and customizable portal for querying and organizing gene annotation resources. Genome Biol. 2009;10:R130. doi: 10.1186/gb-2009-10-11-r130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hayes JD, Chanas SA, Henderson CJ, McMahon M, Sun C, Moffat GJ, et al. The Nrf2 transcription factor contributes both to the basal expression of glutathione S-transferases in mouse liver and to their induction by the chemopreventive synthetic antioxidants, butylated hydroxyanisole and ethoxyquin. Biochem Soc Trans. 2000;28:33–41. doi: 10.1042/bst0280033. [DOI] [PubMed] [Google Scholar]
- 41.Schroer KT, Gibson AM, Sivaprasad U, Bass SA, Ericksen MB, Wills-Karp M, et al. Downregulation of glutathione S-transferase pi in asthma contributes to enhanced oxidative stress. J Allergy Clin Immunol. 2011;128:539–48. doi: 10.1016/j.jaci.2011.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Primiano T, Sutter TR, Kensler TW. Antioxidant-inducible genes. Advances in pharmacology. 1997;38:293–328. doi: 10.1016/s1054-3589(08)60989-8. [DOI] [PubMed] [Google Scholar]
- 43.Venugopal R, Jaiswal AK. Nrf1 and Nrf2 positively and c-Fos and Fra1 negatively regulate the human antioxidant response element-mediated expression of NAD(P)H:quinone oxidoreductase1 gene. Proceedings of the National Academy of Sciences of the United States of America. 1996;93:14960–5. doi: 10.1073/pnas.93.25.14960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang Y, Gonzalez V, Xu MJ. Expression and regulation of glutathione S-transferase P1-1 in cultured human epidermal cells. J Dermatol Sci. 2002;30:205–14. doi: 10.1016/s0923-1811(02)00107-x. [DOI] [PubMed] [Google Scholar]
- 45.Friling RS, Bensimon A, Tichauer Y, Daniel V. Xenobiotic-inducible expression of murine glutathione S-transferase Ya subunit gene is controlled by an electrophile-responsive element. Proceedings of the National Academy of Sciences of the United States of America. 1990;87:6258–62. doi: 10.1073/pnas.87.16.6258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rushmore TH, Pickett CB. Transcriptional regulation of the rat glutathione S-transferase Ya subunit gene. Characterization of a xenobiotic-responsive element controlling inducible expression by phenolic antioxidants. The Journal of biological chemistry. 1990;265:14648–53. [PubMed] [Google Scholar]
- 47.Okuda A, Imagawa M, Sakai M, Muramatsu M. Functional cooperativity between two TPA responsive elements in undifferentiated F9 embryonic stem cells. Embo J. 1990;9:1131–5. doi: 10.1002/j.1460-2075.1990.tb08219.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Moffat GJ, McLaren AW, Wolf CR. Functional characterization of the transcription silencer element located within the human Pi class glutathione S-transferase promoter. J Biol Chem. 1996;271:20740–7. doi: 10.1074/jbc.271.34.20740. [DOI] [PubMed] [Google Scholar]
- 49.Moffat GJ, McLaren AW, Wolf CR. Sp1-mediated transcriptional activation of the human Pi class glutathione S-transferase promoter. Journal of Biological Chemistry. 1996;271:1054–60. doi: 10.1074/jbc.271.2.1054. [DOI] [PubMed] [Google Scholar]
- 50.Lo HW, Ali-Osman F. Genomic cloning of hGSTP1*C, an allelic human Pi class glutathione S-transferase gene variant and functional characterization of its retinoic acid response elements. J Biol Chem. 1997;272:32743–9. doi: 10.1074/jbc.272.52.32743. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.