Abstract
Objectives
To evaluate the independent and joint effects of genetic factors and environmental variables on advanced forms of age-related macular degeneration (AMD), including geographic atrophy and choroidal neovascularization, and to develop a predictive model with genetic and environmental factors included.
Methods
Demographic information, including age at onset, smoking status, and body mass index, was collected for 1844 participants. Genotypes were evaluated for 8 variants in 5 genes related to AMD. Unconditional logistic regression analyses were performed to generate a risk predictive model.
Results
All genetic variants showed a strong association with AMD. Multivariate odds ratios were 3.52 (95% confidence interval, 2.08-5.94) for complement factor H, CFH rs1061170 CC, 4.21 (2.30-7.70) for CFH rs2274700 CC, 0.46 (0.27-0.80) for C2 rs9332739 CC/CG, 0.44 (0.30-0.66) for CFB rs641153 TT/CT, 10.99 (6.04-19.97) for HTRA1/LOC387715 rs10490924 TT, and 2.66 (1.43-4.96) for C3 rs2230199 GG. Smoking was independently associated with advanced AMD after controlling for age, sex, body mass index, and all genetic variants.
Conclusion
CFH confers more risk to the bilaterality of geographic atrophy, whereas HTRA1/LOC387715 contributes more to the bilaterality of choroidal neovascularization. C3 confers more risk for geographic atrophy than choroidal neovascularization. Risk models with combined genetic and environmental factors have notable discrimination power.
Clinical Relevance
Early detection and risk prediction of AMD could help to improve the prognosis of AMD and to reduce the outcome of blindness. Targeting high-risk individuals for surveillance and clinical interventions may help reduce disease burden.
Age-related macular degeneration (AMD) is a complex disease with genetic and environmental factors contributing to its pathogenesis. Because AMD is one of the most studied common eye diseases of the past 5 years, knowledge of its genetic basis has increased exponentially. Genetic variants at 2 gene regions, complement factor H (CFH [OMIM 134370])1-3 and high-temperature requirement factor A-1 (HTRA1 [OMIM 602194])/LOC387715,4-7 confer major disease risks, together likely accounting for approximately 40% to 60% of the genetic risks of AMD in whites.8-12 A number of other genetic variants, such as complement component 2 (C2 [OMIM 217000]), complement factor B (CFB [OMIM 138470]), and complement component 3 (C3 [OMIM 120700]),13-15 have also been identified to be strongly and consistently associated with AMD. Of the environmental risk factors, age and smoking have most consistently been identified as major risks.16-18 However, it remains unclear to what extent these risk factors as a group could explain the occurrence of AMD.19 Early detection and risk prediction could potentially improve disease prognosis and outcomes by allowing for gene-based treatment or spurring patients to modify lifestyle habits. Joint effects of genetic variants and environmental factors are implicated to have better prediction of susceptibility to advanced AMD.8,9,16,20 In this study, we used a combined data set consisting of cohorts from Utah and the Age-Related Eye Disease Study (AREDS) to refine the association of known genetic and environmental factors with advanced AMD. Effects of potential gene-gene (G × G) and gene-environment (G × E) interactions were also estimated. We aimed to develop an AMD risk model to distinguish individuals who would be infected with advanced AMD from those who would not.
Methods
Phenotypes
The study was approved by the institutional review boards of the University of California, San Diego, and University of Utah. All participants signed informed consent statements; all research adhered to the tenets of the Declaration of Helsinki. Access to AREDS data was granted by the AREDS Access Committee. This study's cohort consisted of 1844 unrelated white individuals. A total of 723 participants, including 591 patients with advanced AMD and 132 healthy control individuals, were from AREDS.21 Phenotypic data were obtained from the database of genotypes and phenotypes.22 Demographic and risk factor data were taken at the baseline visit. Diagnoses of advanced AMD were based on the presence of geographic atrophy (GA) or choroidal neovascularization (CNV) according to the same criteria (AREDS category 4 or 5). Determination of unilateral (AREDS category 4) or bilateral (AREDS category 5) AMD was made at the final visit. The remaining 1121 white study participants, including 744 patients with advanced AMD and 377 healthy controls, were enrolled at the Moran Eye Center at the University of Utah and the Shiley Eye Center at the University of California, San Diego. Participants underwent a standard examination, which included visual acuity measurements, dilated slitlamp biomicroscopy, and stereoscopic color fundus photography. Diagnosis of AMD was determined using the criteria defined by AREDS.21 Age at enrollment, smoking status, and body mass index (BMI) were recorded based on AREDS corresponding variance definitions. Smoking status was assessed by whether the individual was a current smoker, had ever smoked for at least 6 months, or had never smoked (as assessed by self-report), with participants classified as current smokers, ever smokers, or never smokers. The BMI was calculated as the weight in kilograms divided by height in meters squared. Control individuals were 60 years or older, without drusen or retinal pigment epithelial abnormalities (Table 1).
Table 1. Phenotype Comparison of the Utah and AREDS Cohorts.
| Variable | Utah | AREDS | ||
|---|---|---|---|---|
|
|
|
|||
| Cases | Controls | Cases | Controls | |
| Total, No. | 744 (208 GA and 536 CNV) | 377 | 591 (133 GA and 458 CNV) | 132 |
| Sex, % | ||||
| Female | 53.6 | 60.7 | 55.5 | 57.6 |
| Male | 46.4 | 39.3 | 44.5 | 42.4 |
| Age, mean (SD), y | 79.1 (9.4) | 74.4 (7.1) | 70.2 (5.1) | 67.0 (4.3) |
| Smoking, % | ||||
| Never | 62.6 | 67.0 | 37.6 | 51.5 |
| Ever | 31.3 | 31.1 | 49.7 | 38.6 |
| Current | 6.1 | 1.9 | 12.7 | 9.8 |
Abbreviations: AREDS, Age-Related Eye Disease Study; CNV, choroidal neovascularization; GA, geographic atrophy.
Genotypes
Genomic DNA was extracted from peripheral blood leukocytes according to established protocols. The AREDS DNA samples were obtained from the AREDS Genetic Repository.
Eight single-nucleotide polymorphisms (SNPs) in 5 genes associated with AMD were selected according to the literature. They were rs1061170,1-3 rs2274700,23 andrs141099624 in CFH; rs933273913 in C2; rs64115313 in CFB; rs10490924 and rs11200638 in the HTRA1/LOC387715 region4,5,7; and rs2230199 in C3.14,15
All SNPs were genotyped using the SNaPshot method according to the manufacturer's recommendations. Briefly, an SNP was amplified by polymerase chain reaction, and the polymerase chain reaction product was purified by Exo I and shrimp alkaline phosphatase (New England Biolabs Inc, Ipswich, Massachusetts). The purified polymerase chain reaction product and the SNaPshot primer were then used to perform a single base-pair extension with the SNaPshot multiplex mix (Applied Biosystems Inc, Foster City, California). After an additional purification step using shrimp alkaline phosphatase, the product was analyzed on an ABI 3130xl genetic analyzer (Applied Biosystems Inc), and the genotyping results were obtained directly.
Statistical Analysis
Individuals with advanced AMD, as well as subtypes of CNV and GA, were compared with controls with regard to genetic (CFH rs1061170, CFH rs2274700, CFH rs1410996, C2 rs9332739, CFB rs641153, HTRA1/LOC387715 rs10490924, HTRA1/LOC387715 rs11200638, and C3 rs2230199) and environmental (age, sex, smoking, and BMI) risk factors. Fisher exact tests and Cochran-Armitage tests for multiplicative models and additive models over genotypes or alleles were performed to assess evidence of an association by using PEPI, version 4.04 (Brixton Health, London, England). Adaptive permutation tests were performed by using PLINK, version 1.06 (http://pngu.mgh.harvard.edu/purcell/plink/). Haplotypes were deduced by using HaploView, version 4.1 (Broad Institute, Cambridge, Massachusetts). Deviations from Hardy-Weinberg equilibrium were tested with the standard χ2 test.
Multivariate unconditional logistic regression analysis was performed to evaluate the relationship between AMD and all the genotypes plus environmental risk factors, including age (<65, 65-69, 70-74, 75-79, 80-84, or ≥85 years), sex, smoking (never smoker, former smoker, or current smoker), and BMI (<25, 25-29, or ≥30). Tests for multiplicative interactions between each of the genotypes vs smoking and BMI were calculated by using cross-product terms according to genotype and the individual risk factors. Similar analyses were performed to assess interactions for each combination of different genotypes. Odds ratios (ORs) and 95% confidence intervals (CIs) were calculated for each genotype, and significant variant stepwise backward logistic regression (terms with P > .05 were removed) was used to survey for the best fit risk model. Sensitivities and specificities were calculated to maximize the sum of the 2 values using receiver operating characteristic (ROC) curves constructed using STATA 8.0 statistical software (Stata-Corp LP, College Station, Texas).
Results
A total of 1844 unrelated white individuals were involved in this study. There were 341 patients with GA, 994 patients with CNV, and 509 controls. There was a slight preponderance of women in each group. The mean (SD) age of patients and controls was 75.1 (9.0) and 72.5 (7.3) years, respectively.
Unadjusted association between genetic variables and AMD was evaluated for each SNP (Table 2). All SNPs exhibit a strongly significant association with AMD on allelic P values under the multiplicative model and trend P values under the additive model (P < .001 for all). The ORs of the risk alleles on CFH (rs1061170, rs2274700, and rs1410996) and HTRA1/LOC387715 (rs10490924 and rs11200638) were roughly 2.50 to 3.00, and the OR of the risk allele of C3 rs2230199 was 1.62. In addition, the protective alleles of C2 rs9332739 and CFB rs641153 had ORs of 0.55 and 0.54, respectively (Table 2).
Table 2. Association of 8 SNPs Between Patients With Advanced Age-Related Macular Degeneration and Control Individualsa.
| Gene | SNP | Allele | Cases (n = 1335) | Controls (n=509) | OR (95% CI) |
|---|---|---|---|---|---|
| CFH | rs1061170 | C | 0.59 | 0.37 | 2.45 (2.11-2.84) |
| CFH | rs2274700 | C | 0.78 | 0.56 | 2.76 (2.36-3.22) |
| CFH | rs1410996 | C | 0.78 | 0.59 | 2.52 (2.16-2.94) |
| C2 | rs9332739 | C | 0.03 | 0.05 | 0.55 (0.38-0.81) |
| CFB | rs641153 | T | 0.05 | 0.09 | 0.54 (0.41-0.71) |
| HTRA1/LOC387715 | rs10490924 | T | 0.41 | 0.19 | 2.94 (2.47-3.50) |
| HTRA1/LOC387715 | rs11200638 | A | 0.42 | 0.22 | 2.61 (0.21-3.09) |
| C3 | rs2230199 | G | 0.28 | 0.19 | 1.62 (1.36-1.93) |
Abbreviations: CI, confidence interval; OR, odds ratio; SNP, single-nucleotide polymorphism.
P< .001 for all.
To determine whether association results differed between 2 subtypes of late AMD, comparisons were also made between GA and CNV (wet) AMD. Only C3 rs2230199 was demonstrated to have a statistically significant difference between the 2 subtypes (P < .001) adjusted for age and sex. The risk allele G showed a 6.0% higher allele frequency in GA (32.4%) than in CNV (26.4%). None of the other SNPs were found to have a significantly different frequency between GA and CNV.
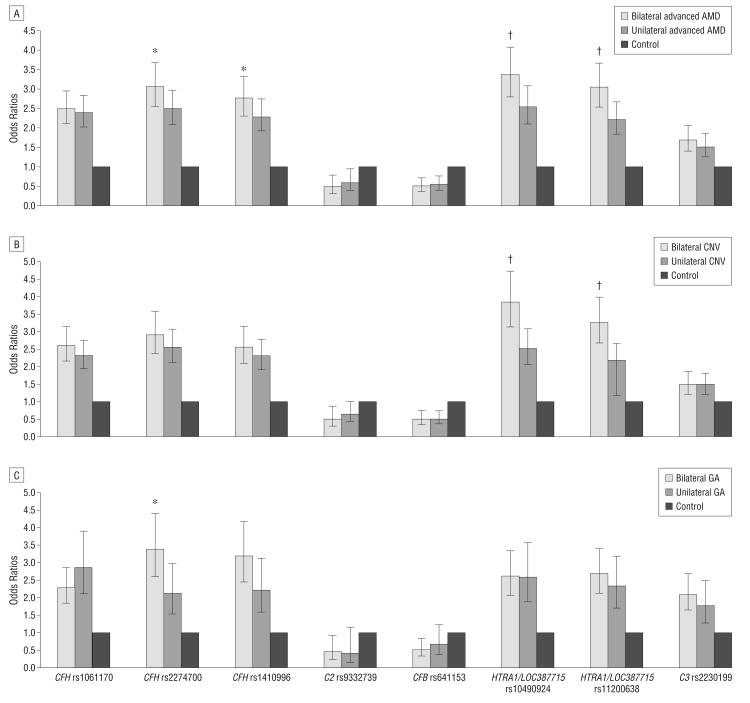
Furthermore, CFH rs2274700, CFH rs1410996, HTRA1/LOC387715 rs10492094, and HTRA1/LOC387715 rs11200638 showed significant P values (P=.01, P=.03, P=< .001, and P=< .001, respectively) when their association with AMD was compared between bilaterally affected patients and unilaterally affected patients (Figure 1). They all remained statistically significant (P = .03, P = .04, P<.001, and P<.001, respectively) after adaptive permutation tests (PLINK, version 1.06). However, in further analysis with the 2 subtypes of CNV and GA, we found that SNPs on HTRA1/LOC387715 (rs10490924 and rs11200638) showed a significant difference (P< .001) only between bilateral CNV and unilateral CNV, respectively, whereas SNPs on CFH (rs2274700 and rs1410996) showed only a slightly significant difference (P=.01 and P=.06) between bilateral GA and unilateral GA, respectively (Figure 1) (P=.02 and P=.07 after adaptive permutation tests).
Figure 1.
Association of age-related macular degeneration (AMD), choroidal neovascularization (CNV), and geographic atrophy (GA) in bilaterally affected and unilaterally affected patients. *Indicates P<.05. †Indicates P<.001. Odds ratios of bilateral and unilateral AMD (A), bilateral and unilateral CNV (B), and bilateral and unilateral GA (C) compared with control individuals. Error bars indicate 95% confidence intervals of the odds ratios.
Haplotype analyses25 demonstrated that rs1410996 and rs2274700 were located in the same linkage disequilibrium, with a D′ of 0.95 and an R2 of 0.87. Also, rs10490924 and rs11200638 have a D′ of 0.93 and an R2 of 0.82 as well. Therefore, only rs2274700 and rs10490924 were included in the regression model screening because they were surrogates for rs1410996 and rs11200638, respectively.
The multivariate adjusted ORs for advanced AMD were obtained by using logistic regression, starting from a full model with 4 environmental and 6 genotyping variables (Table 3). CFH variants (rs1061170: ORhetero, 1.45 [95% CI, 1.02-2.08]; ORhomo, 3.52 [2.08-5.94]; rs2274700: OR hetero, 1.98 [1.15-3.41]; ORhomo, 4.21 [2.30-7.70]), HTRA1/LOC387715 variant (rs10490924: ORhetero, 2.59 [1.94-3.45]; ORhomo, 10.99 [6.04-19.97]), and C3 variant (rs2230199: ORhetero, 1.80 [1.34-2.42]; ORhomo, 2.66 [1.43-4.96]) were shown to be associated with an increased risk of AMD. The protective effects of the C2 variant (rs9332739: OR, 0.46; 95% CI, 0.27-0.80) and the CFB variant (rs641153: 0.44; 0.30-0.66) were shown to be significantly associated with AMD. Smoking was independently associated with advanced AMD (OR, 1.80 [95% CI, 1.32-2.45] for ever smokers and 3.71 [2.02-6.80] for current smokers), and BMI was shown to have a marginal association (P=.09 for 25-29 and P = .08 for ≥30) with combined advanced AMD, although this was barely significant in the CNV subgroup.
Table 3. Multivariate Adjusted ORs for Advanced AMD, CNV, and GA in Logistic Regression Modela.
| Variable | Combined Patients With Advanced AMD (n = 1057) | Patients With CNV vs Control Individuals (n=780) | Patients With GA vs Controls (n=277) | Regression Coefficient | |||
|---|---|---|---|---|---|---|---|
|
|
|
|
|||||
| OR (95% CI) | P Value | OR (95% CI) | P Value | OR (95% CI) | P Value | ||
| Age, y | |||||||
| <65 | 1 [Reference] | 1 [Reference] | 1 [Reference] | ||||
| Every ≥5 | 1.42 (1.29-1.57) | ≤.001 | 1.43 (1.28-1.59) | ≤.001 | 1.44 (1.27-1.64) | ≤.001 | 0.34 |
| Sex | |||||||
| Male | 1 [Reference] | 1 [Reference] | 1 [Reference] | ||||
| Female | 0.79 (0.58-1.06) | .11 | 0.82 (0.60-1.12) | .21 | 0.82 (0.55-1.23) | .34 | |
| BMI | |||||||
| <25 | 1 [Reference] | 1 [Reference] | 1 [Reference] | ||||
| 25-29 | 1.32 (0.95-1.84) | .09 | 1.45 (1.02-2.05) | .04 | 1.13 (0.73-1.76) | .58 | |
| ≥30 | 1.37 (0.96-1.96) | .08 | 1.52 (1.04-2.22) | .03 | 1.12 (0.70-1.81) | .64 | |
| Smoking | |||||||
| Never | 1 [Reference] | 1 [Reference] | 1 [Reference] | ||||
| Ever | 1.80 (1.32-2.45) | ≤.001 | 1.95 (1.41-2.70) | ≤.001 | 1.38 (0.90-2.11) | .14 | 0.69 |
| Current | 3.71 (2.02-6.80) | ≤.001 | 3.82 (2.01-7.25) | ≤.001 | 3.59 (1.61-8.02) | .002 | 1.36 |
| rs1061170 | |||||||
| TT | 1 [Reference] | 1 [Reference] | 1 [Reference] | ||||
| CT | 1.45 (1.02-2.08) | .04 | 1.58 (1.07-2.33) | .02 | 1.28 (0.78-2.08) | .33 | 0.36 |
| CC | 3.52 (2.08-5.94) | ≤.001 | 3.75 (2.14-6.56) | ≤.001 | 3.12 (1.61-6.05) | .001 | 1.25 |
| rs2274700 | |||||||
| TT | 1 [Reference] | 1 [Reference] | 1 [Reference] | ||||
| CT | 1.98 (1.15-3.41) | .01 | 1.75 (0.96-3.19) | .07 | 2.49 (1.09-5.72) | .03 | 0.70 |
| CC | 4.21 (2.30-7.70) | ≤.001 | 3.82 (1.97-7.40) | ≤.001 | 5.24 (2.17-12.67) | ≤.001 | 1.46 |
| rs9332739 | |||||||
| GG | 1 [Reference] | 1 [Reference] | 1 [Reference] | ||||
| CC/CG | 0.46 (0.27-0.80) | .005 | 0.53 (0.30-0.93) | .03 | 0.32 (0.13-0.77) | .01 | −0.79 |
| rs641153 | |||||||
| CC | 1 [Reference] | 1 [Reference] | 1 [Reference] | ||||
| TT/CT | 0.44 (0.30-0.66) | ≤.001 | 0.39 (0.25-0.60) | ≤.001 | 0.48 (0.27-0.85) | .01 | −0.78 |
| rs10490924 | |||||||
| GG | 1 [Reference] | 1 [Reference] | 1 [Reference] | ||||
| TG | 2.59 (1.94-3.45) | ≤.001 | 2.81 (2.07-3.82) | ≤.001 | 2.29 (1.55-3.38) | ≤.001 | 0.94 |
| TT | 10.99 (6.04-19.97) | ≤.001 | 13.49 (7.29-24.98) | ≤.001 | 6.57 (3.13-13.78) | ≤.001 | 2.37 |
| rs2230199 | |||||||
| CC | 1 [Reference] | 1 [Reference] | 1 [Reference] | ||||
| CG | 1.80 (1.34-2.42) | ≤.001 | 1.56 (1.14-2.13) | .006 | 2.38 (1.61-3.54) | ≤.001 | 0.57 |
| GG | 2.66 (1.43-4.96) | .002 | 2.39 (1.23-4.63) | .01 | 3.86 (1.77-8.40) | .001 | 0.94 |
Abbreviations: AMD, age-related macular degeneration; BMI, body mass index; CI, confidence interval; CNV, choroidal neovascularization; GA, geographic atrophy; OR, odds ratio.
Of 1844 study participants, 1432 individuals have full information regarding environmental data contributed-risk modeling.
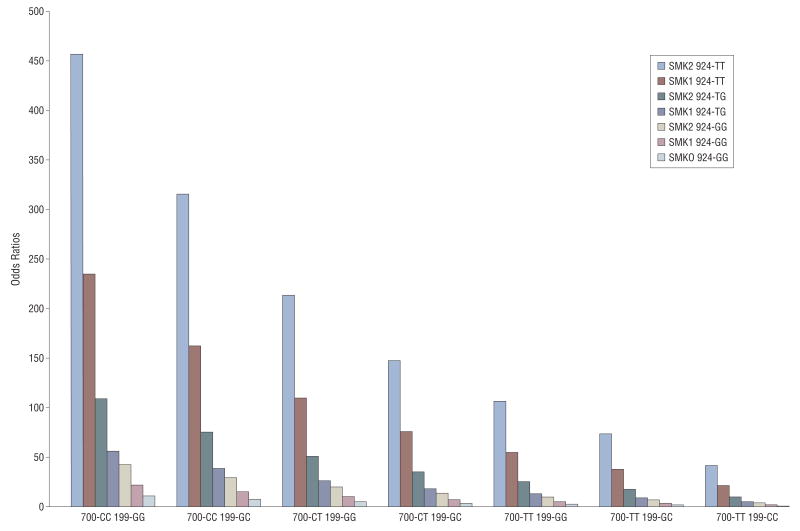
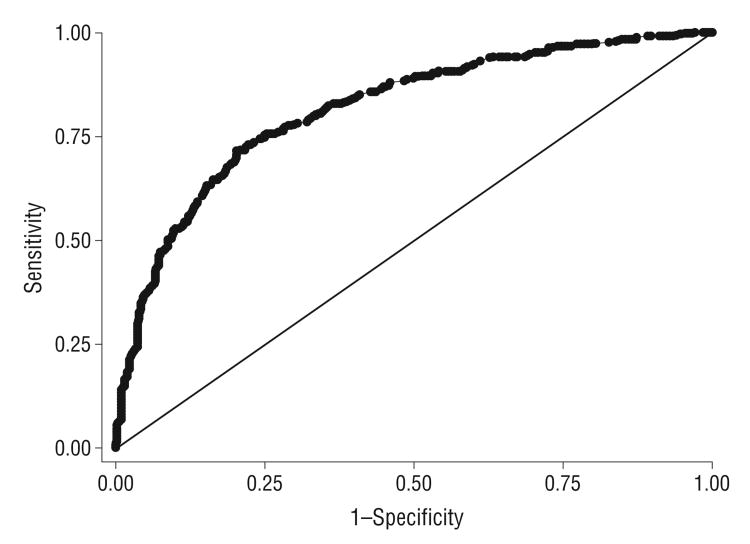
Interactions between each genotype and smoking and each genotype and BMI in the case-control comparison were also evaluated by the cross-products of each other. No significant interactions were found between any of the genotypes and smoking or BMI. In terms of interactions among the genotypes, only CFH rs1061170CT × HTRA1/LOC387715 rs10490924TT and CFH rs2274700CT × HTRA1/LOC387715 rs10490924TG showed a weak effect of interaction, with P values of .03 for both. However, the risk model was not improved by introduction of these interaction factors. No interactions among the other genotypes were found. The best fit model was achieved with age, smoking, and 6 genetic markers (CFH rs1061170, CFH rs2274700, C2 rs9332739, CFB rs641153, HTRA1/LOC387715 rs10490924, and C3 rs2230199) as a multiplicative model (Table 3). The joint effects of CFH rs2274700, HTRA1/LOC387715 rs10490924, C3 rs2230199, and smoking status were shown by ORs calculated by the risk model, adjusted for age, sex, BMI, and other genetic variants (Figure 2). Given that P is the probability of AMD, the logistic regression model is , where α = −2.209, i = 1,…,13, β is the coefficient for each variant being listed in Table 3, and the OR=exp(β)/[1+exp(β)]. Because of insufficient statistical power caused by limited study participants in a particular genotyping and environmental combination, some genotyping categories, such as rs10490924 TT and smk0, are not shown in Figure 2. A ROC curve, which is a graphic plot of the sensitivity, vs (1 − specificity), was constructed (Figure 3). The area under the ROC curve presented in Figure 3 was equal to 0.82. Various cutoff values were tested to acquire best sensitivity and specificity in diagnostic decision making. The cutoff of 0.73 yielded 75.5% sensitivity and 74.7% specificity. The highest discrimination accuracy of the model is 78.8%.
Figure 2.
Joint effects of CFH rs2274700, HTRA1/LOC387715 rs10490924, C3 rs2230199, and smoking status were shown with odds ratios calculated by logistic regression model, adjusted for age, sex, body mass index, and other genetic variants. SMK0, SMK1, and SMK2 indicate never smoker, ever smoker, and current smoker, respectively; 700, 924, and 199 indicate CFH rs2274700, HTRA1/LOC387715 rs10490924, and C3 rs2230199, respectively.
Figure 3.
Sensitivities and specificities for a variety of risk score cutoffs and receiver operating characteristic (ROC) curve for risk of age-related macular degeneration. Area under the ROC curve=0.82.
Comment
We demonstrate a significant association between AMD and known genetic polymorphisms of CFH, HTRA1/LOC387715, C2, CFB, and C3. The results of allele frequencies and the ORs for each marker confirmed the findings of previously published reports.2,4,7,15,26,27
Dewan et al4 reported HTRA1/LOC387715 as a risk gene for CNV in AMD, implicating that polymorphisms in the HTRA1/LOC387715 region are responsible for this specific phenotype of late AMD. Yet most other groups have not found a difference in the association data between GA and CNV.7,28 We were not able to find a difference in allele frequency of the 2 major susceptibility genes CFH and HTRA1/LOC387715 with respect to GA and CNV. However, the risk allele of C3 rs2230199 was significantly higher in GA (32.4%) than in CNV (26.4%) (P < .001) when adjusted for age and sex. This result, for the first time to our knowledge, shows that C3 rs2230199 predisposes individuals to GA more than CNV. A similar trend was also observed in an earlier study.27 The ways in which C3 contributes differently to the pathogenesis of GA vs CNV require further investigation.
Vision-related quality of life is strongly associated with visual acuity and the presence of bilateral AMD. Bilateral AMD corresponds to a more severe stage of the disease and is a sign of progression. It is not surprising to find that all the risk alleles are more common in the bilaterally affected group than in the unilaterally affected group but only significantly for CFH rs2274700, CFH rs1410996, HTRA1/LOC387715 rs10490924, and HTRA1/LOC387715 rs11200638. Interestingly, if the study participants were confined to a specific subtype of AMD, CFH rs2274700 and CFH rs1410996 only remain significant in comparison with bilateral GA and unilateral GA, and HTRA1/LOC387715 rs10490924 and rs11200638 only remain significant in comparison with bilateral CNV and unilateral CNV, respectively. Although not statistically significant, SNPs in CFH still showed a tendency to have a higher risk allele frequency in GA, whereas SNPs in HTRA1/LOC387715 have higher allele frequencies in CNV. Overall, neither CFH nor HTRA1/LOC387715 has been shown to be responsible for directing AMD toward a specific late phenotype (GA or CNV). However, both genes may play a role in increasing its severity once a late phenotype develops. Our results show that CFH increases the severity of GA, whereas HTRA1/LOC387715 heightens CNV. This is in agreement with the findings from other authors20,29 that the HTRA1/LOC387715 gene is more strongly related to the progression of CNV than to GA.
Surprisingly, CFH rs1061170 did not show a statistically significant difference between bilateral GA and unilateral GA. Although a recent report30 suggested that the CC genotype of rs1061170 was associated with an increased likelihood of bilateral vs unilateral findings of soft drusen and pigmentary abnormalities, they did not establish significant associations with bilaterality of GA or CNV. In further analysis of haplotypes consisting of rs1061170 and rs2274700, we found there were 3 haplotypes in our combined data set with greater than 5% frequency. -T-T-is the protective haplotype and -C-C- is the risk haplotype associated with AMD. -T-C- was a neutral haplotype when AMD patients and controls were compared. However, -T-C- became a risk haplotype when bilateral GA (25.4%) and unilateral GA (12.8%) were compared (P<.001), which explained why CFH rs1061170 was not significantly associated with the bilaterality of GA in our dataset.
Our results showed that of the environmental risk factors, smoking and age were identified as major risk factors, which was consistent with the combined analysis of population-based eye studies from 3 continents.18 Smoking was confirmed as an independent risk factor for AMD in this study. Patients have a 1.8-fold higher chance of developing AMD if they ever smoked compared with those who never smoked. The risk was elevated to 3.7-fold for current smokers. As another risk factor,29 BMI showed a weak contribution to the occurrence of AMD. Neither smoking nor BMI was found to have a significant interaction with genotypes. Although a single study9 found an interaction between smoking and HTRA1/LOC387715 rs10490924, interaction between smoking and genotypes was eliminated when stepwise logistic regression was performed, which was consistent with data from multiple reports.16,20,29 However, an interaction is still a possibility because logistic regression has only modest power for distinguishing interactions.31
In terms of interactions among genotypes, we found weak interactions of CFH rs1061170CT × HTRA1/LOC387715 rs10490924TT and CFH rs2274700CT × HTRA1/LOC387715 rs10490924TG. Because our model was not improved by inclusion of these interactions, for the sake of simplicity, no interaction term was included in our risk model. This result is similar to that of a study8 in Finland in which a tentative interaction between CFH and LOC387715 with a marginal P value (.06) was observed. However, most studies7,9,20,29,32,33 have not found an interaction between these 2 genes. Our final model supported the notion that CFH and HTRA1/LOC387715 act independently, and the log-linear additive model fits well for the joint effects of these 2 genes.
We developed a risk model that predicts the individual's risk for AMD. Targeting high-risk individuals could lead to more frequent surveillance and clinical interventions. Patients would benefit from more targeted education regarding a healthy lifestyle. However, the risk predictions resulting from this model are directly applicable only to the population from which it was developed; we still need to be careful when extending the results to other populations. Sensitivities and specificities for a variety of risk factors were evaluated to assess the optimal use of the model for individual risk prediction. The sensitivity, specificity, and area under the ROC curve established in this study were analogous to those reported by previous studies.16,29 To improve the AMD prediction model, more genetic- or environmental-influencing factors need to be clarified.
Acknowledgments
Funding/Support: Funding support for the Age-Related Eye Disease Study (AREDS) was provided by grant N01-EY-0-2127 from the National Eye Institute. Dr Zhang is supported by grants from the National Institutes of Health, the Veterans Affairs Merit Award, the Foundation Fighting Blindness, the Macula Vision Research Foundation, the Ruth and Milton Steinbach Fund Inc, Research to Prevent Blindness, a Burroughs Wellcome Fund Clinical Scientist Award in Translational Research, and grants from Chinese National 985 Project to Sichuan University and West China Hospital.
Footnotes
Financial Disclosure: None reported.
References
- 1.Edwards AO, Ritter R, III, Abel KJ, Manning A, Panhuysen C, Farrer LA. Complement factor H polymorphism and age-related macular degeneration. Science. 2005;308(5720):421–424. doi: 10.1126/science.1110189. [DOI] [PubMed] [Google Scholar]
- 2.Haines JL, Hauser MA, Schmidt S, et al. Complement factor H variant increases the risk of age-related macular degeneration. Science. 2005;308(5720):419–421. doi: 10.1126/science.1110359. [DOI] [PubMed] [Google Scholar]
- 3.Klein RJ, Zeiss C, Chew EY, et al. Complement factor H polymorphism in age-related macular degeneration. Science. 2005;308(5720):385–389. doi: 10.1126/science.1109557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dewan A, Liu M, Hartman S, et al. HTRA1 promoter polymorphism in wet age-related macular degeneration. Science. 2006;314(5801):989–992. doi: 10.1126/science.1133807. [DOI] [PubMed] [Google Scholar]
- 5.Yang Z, Camp NJ, Sun H, et al. A variant of the HTRA1 gene increases susceptibility to age-related macular degeneration. Science. 2006;314(5801):992–993. doi: 10.1126/science.1133811. [DOI] [PubMed] [Google Scholar]
- 6.Jakobsdottir J, Conley YP, Weeks DE, Mah TS, Ferrell RE, Gorin MB. Susceptibility genes for age-related maculopathy on chromosome 10q26. Am J Hum Genet. 2005;77(3):389–407. doi: 10.1086/444437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rivera A, Fisher SA, Fritsche LG, et al. Hypothetical LOC387715 is a second major susceptibility gene for age-related macular degeneration, contributing independently of complement factor H to disease risk. Hum Mol Genet. 2005;14(21):3227–3236. doi: 10.1093/hmg/ddi353. [DOI] [PubMed] [Google Scholar]
- 8.Seitsonen SP, Onkamo P, Peng G, et al. Multifactor effects and evidence of potential interaction between complement factor H Y402H and LOC387715 A69S in age-related macular degeneration. PLoS One. 2008;3(12):e3833. doi: 10.1371/journal.pone.0003833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schmidt S, Hauser MA, Scott WK, et al. Cigarette smoking strongly modifies the association of LOC387715 and age-related macular degeneration. Am J Hum Genet. 2006;78(5):852–864. doi: 10.1086/503822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schaumberg DA, Hankinson SE, Guo Q, Rimm E, Hunter DJ. A prospective study of 2 major age-related macular degeneration susceptibility alleles and interactions with modifiable risk factors. Arch Ophthalmol. 2007;125(1):55–62. doi: 10.1001/archopht.125.1.55. [DOI] [PubMed] [Google Scholar]
- 11.Swaroop A, Branham KE, Chen W, Abecasis G. Genetic susceptibility to age-related macular degeneration: a paradigm for dissecting complex disease traits. Hum Mol Genet. 2007;16(spec No. 2):R174–R182. doi: 10.1093/hmg/ddm212. [DOI] [PubMed] [Google Scholar]
- 12.Swaroop A, Chew EY, Rickman CB, Abecasis GR. Unraveling a multifactorial late-onset disease: from genetic susceptibility to disease mechanisms for age-related macular degeneration. Annu Rev Genomics Hum Genet. 2009;10:19–43. doi: 10.1146/annurev.genom.9.081307.164350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gold B, Merriam JE, Zernant J, et al. AMD Genetics Clinical Study Group. Variation in factor B (BF) and complement component 2 (C2) genes is associated with age-related macular degeneration. Nat Genet. 2006;38(4):458–462. doi: 10.1038/ng1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maller JB, Fagerness JA, Reynolds RC, Neale BM, Daly MJ, Seddon JM. Variation in complement factor 3 is associated with risk of age-related macular degeneration. Nat Genet. 2007;39(10):1200–1201. doi: 10.1038/ng2131. [DOI] [PubMed] [Google Scholar]
- 15.Yates JR, Sepp T, Matharu BK, et al. Genetic Factors in AMD Study Group. Complement C3 variant and the risk of age-related macular degeneration. N Engl J Med. 2007;357(6):553–561. doi: 10.1056/NEJMoa072618. [DOI] [PubMed] [Google Scholar]
- 16.Seddon JM, Reynolds R, Maller J, Fagerness JA, Daly MJ, Rosner B. Prediction model for prevalence and incidence of advanced age-related macular degeneration based on genetic, demographic, and environmental variables. Invest Ophthalmol Vis Sci. 2009;50(5):2044–2053. doi: 10.1167/iovs.08-3064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Evans JR. Risk factors for age-related macular degeneration. Prog Retin Eye Res. 2001;20(2):227–253. doi: 10.1016/s1350-9462(00)00023-9. [DOI] [PubMed] [Google Scholar]
- 18.Smith W, Assink J, Klein R, et al. Risk factors for age-related macular degeneration: pooled findings from three continents. Ophthalmology. 2001;108(4):697–704. doi: 10.1016/s0161-6420(00)00580-7. [DOI] [PubMed] [Google Scholar]
- 19.Jakobsdottir J, Gorin MB, Conley YP, Ferrell RE, Weeks DE. Interpretation of geetic association studies: markers with replicated highly significant odds ratios may be poor classifiers. PLoS Genet. 2009;5(2):e1000337. doi: 10.1371/journal.pgen.1000337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hughes AE, Orr N, Patterson C, et al. Neovascular age-related macular degeneration risk based on CFH, LOC387715/HTRA1, and smoking. PLoS Med. 2007;4(12):e355. doi: 10.1371/journal.pmed.0040355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Age-Related Eye Disease Study Research Group. The Age-Related Eye Disease Study (AREDS): design implications. AREDS report No. 1. Control Clin Trials. 1999;20(6):573–600. doi: 10.1016/s0197-2456(99)00031-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mailman MD, Feolo M, Jin Y, et al. The NCBI dbGaP database of genotypes and phenotypes. Nat Genet. 2007;39(10):1181–1186. doi: 10.1038/ng1007-1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li M, Atmaca-Sonmez P, Othman M, et al. CFH haplotypes without the Y402H coding variant show strong association with susceptibility to age-related macular degeneration. Nat Genet. 2006;38(9):1049–1054. doi: 10.1038/ng1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maller J, George S, Purcell S, et al. Common variation in three genes, including a noncoding variant in CFH, strongly influences risk of age-related macular degeneration. Nat Genet. 2006;38(9):1055–1059. doi: 10.1038/ng1873. [DOI] [PubMed] [Google Scholar]
- 25.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21(2):263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 26.Thakkinstian A, Han P, McEvoy M, et al. Systematic review and meta-analysis of the association between complement factor H Y402H polymorphisms and age-related macular degeneration. Hum Mol Genet. 2006;15(18):2784–2790. doi: 10.1093/hmg/ddl220. [DOI] [PubMed] [Google Scholar]
- 27.Despriet DD, van Duijn CM, Oostra BA, et al. Complement component C3 and risk of age-related macular degeneration. Ophthalmology. 2009;116(3):474–480. doi: 10.1016/j.ophtha.2008.09.055. [DOI] [PubMed] [Google Scholar]
- 28.Scholl HPN, Fleckenstein M, Fritsche LG, et al. CFH, C3 and ARMS2 are significant risk loci for susceptibility but not for disease progression of geographic atrophy due to AMD. PLoS One. 2009;4(10):e7418. doi: 10.1371/journal.pone.0007418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Seddon JM, Francis PJ, George S, Schultz DW, Rosner B, Klein ML. Association of CFH Y402H and LOC387715 A69S with progression of age-related macular degeneration. JAMA. 2007;297(16):1793–1800. doi: 10.1001/jama.297.16.1793. [DOI] [PubMed] [Google Scholar]
- 30.Pai ASI, Mitchell P, Rochtchina E, Iyengar S, Wang JJ. Blue Mountains Eye Study. Complement factor H and the bilaterality of age-related macular degeneration. Arch Ophthalmol. 2009;127(10):1339–1344. doi: 10.1001/archophthalmol.2009.239. [DOI] [PubMed] [Google Scholar]
- 31.Zhao J, Jin L, Xiong M. Test for interaction between two unlinked loci. Am J Hum Genet. 2006;79(5):831–845. doi: 10.1086/508571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Francis PJ, Hamon SC, Ott J, Weleber RG, Klein ML. Polymorphisms in C2, CFB and C3 are associated with progression to advanced age-related macular degeneration associated with visual loss. J Med Genet. 2009;46(5):300–307. doi: 10.1136/jmg.2008.062737. [DOI] [PubMed] [Google Scholar]
- 33.Conley YP, Jakobsdottir J, Mah T, et al. CFH, ELOVL4, PLEKHA1 and LOC387715 genes and susceptibility to age-related maculopathy: AREDS and CHS cohorts and meta-analyses. Hum Mol Genet. 2006;15(21):3206–3218. doi: 10.1093/hmg/ddl396. [DOI] [PubMed] [Google Scholar]