Summary
Motility of sperm is crucial for their directed migration to the egg. The acquisition and modulation of motility are regulated to ensure that sperm move when and where needed, thereby promoting reproductive success. One specific example of this phenomenon occurs during differentiation of the amoeboid sperm of C. elegans as they activate from a round spermatid to a mature, crawling spermatozoon. Sperm activation is regulated by redundant pathways to occur at a specific time and place for each sex. Here, we report the identification of the solute carrier 6 (SLC6) transporter protein SNF-10 as a key regulator of C. elegans sperm activation in response to male protease activation signals. We find that SNF-10 is present in sperm and is required for activation by the male but not by the hermaphrodite. Loss of both snf-10 and a hermaphrodite activation factor render sperm completely insensitive to activation. Using in vitro assays, we find that snf-10 mutant sperm show a specific deficit in response to protease treatment but not to other activators. Prior to activation, SNF-10 is present in the plasma membrane, where it represents a strong candidate to receive signals that lead to subcellular morphogenesis. After activation, it shows polarized localization to the cell body region that is dependent on membrane fusions mediated by the dysferlin FER-1. Our discovery of snf-10 offers insight into the mechanisms differentially employed by the two sexes to accomplish the common goal of producing functional sperm, as well as how the physiology of nematode sperm may be regulated to control motility as it is in mammals.
Keywords: sperm, cell motility, signaling, reproduction, SLC6 transporter, C. elegans
Introduction
A key feature of sexual reproduction is the generation of motile sperm cells capable of migrating to and fertilizing an oocyte. Upon transfer to a female, sperm become motile and then must further modulate their motility in response to local cues during transit through the reproductive tract (Bloch Qazi et al., 2003; Suarez, 2008). Thus, as for many other cell types that migrate during development, sperm motility is tightly and dynamically regulated in response to the external environment. However, unlike other cell types, sperm must accomplish these tasks without new protein synthesis. Instead they rely on interactions between soluble factors and cell surface receptors that trigger alterations in cellular physiology, ultimately leading to remodeling of cell structure and/or motility. In mammalian sperm, ion channel activity promotes several crucial steps in remodeling the internal and external properties of the sperm cell to render it competent for directional migration and fertilization; these include capacitation, the acrosome reaction, chemotaxis and acquisition of hyperactivated motility (Darszon et al., 2011; Lishko et al., 2011; Santi et al., 2013; Shukla et al., 2012). Sperm from other species must undergo analogous processes of maturation and are similarly dependent on signals from the extracellular milieu (Darszon et al., 2008; Santella et al., 2012).
In the nematode C. elegans, sperm lack flagella and instead move by crawling using a pseudopod (L'Hernault, 2006; Ward and Carrel, 1979). Both motility and fertilization competence are acquired during a process termed activation, during which the pseudopod is generated and proteins important for fertilization are displayed on the cell surface. Both sexes make sperm, and activation is regulated in sex-specific ways to ensure that sperm become motile only when and where it is appropriate. For hermaphrodites, it is advantageous to initiate self sperm motility rapidly after its production, thus ensuring functional sperm are available to fertilize oocytes. For males, sperm motility must be prevented during storage within the gonad; otherwise, when mating occurs, sperm transfer fails (Stanfield and Villeneuve, 2006). However, after transfer to a hermaphrodite, male sperm also need to become motile rapidly to migrate into the reproductive tract and avoid being swept out by the outward passage of oocytes (Argon and Ward, 1980). Males accomplish this in part by including an activator(s) in seminal fluid, which is mixed with sperm during transfer (Shakes and Ward, 1989; Ward et al., 1983). The two sexes have distinct genetic requirements for sperm activation that facilitate these differential spatial and temporal requirements. In hermaphrodites, a set of sperm-specific proteins, the SPE-8 group, is required for self sperm activation, possibly in response to a zinc signal (L'Hernault et al., 1988; Liu et al., 2013). In males, the seminal fluid serine protease TRY-5 is a strong candidate to initiate activation (Smith and Stanfield, 2011), but sperm factors involved in this male-specific pathway have not been identified. Although male sperm do not normally require spe-8 group function to activate via the male-specific pathway, they express these genes, and they can activate via the hermaphrodite pathway if the male activator is not present (LaMunyon and Ward, 1994; Smith and Stanfield, 2011). Likewise, if hermaphrodite sperm lack spe-8 group function, they can “transactivate” in response to the male activator, which is transferred in seminal fluid during mating (Shakes and Ward, 1989). For both sexes, it is unclear how signals are transduced into changes in cellular morphology required for sperm function.
In this study, we report the identification of snf-10 (snf, sodium:neurotransmitter transporter family) as a key regulator of sperm motility in C. elegans. SNF-10 is a member of the SLC6 (solute carrier 6) family of transporters, which couple the import of cargo such as amino acids, osmolytes, or monoamine neurotransmitters to the influx of sodium (Broer and Gether, 2012; Kristensen et al., 2011). SLC6 transporter activity is crucial for cellular function and homeostasis in diverse tissues, from neurons to the mammalian kidney, and mutations in human transporters are implicated in hereditary forms of intellectual disability and aminoaciduria, among other diseases (Pramod et al., 2013). In Drosophila, the SLC6 transporter Ntl (Neurotransmitter transporter-like) is required for spermiogenesis (Chatterjee et al., 2011). Here we show that C. elegans SNF-10 is required in sperm for activation by protease signals via the male-specific pathway. The localization of SNF-10 is regulated during activation and dependent in part on the dysferlin FER-1, providing a link between signaling and cellular morphogenesis.
Materials and Methods
Nematode growth and strains
Unless indicated otherwise, C. elegans nematodes were grown at 20°C on NGM (nematode growth medium) seeded with E. coli strain OP50 (Brenner, 1974). The him-5(e1490) (Hodgkin et al., 1979) allele was present in all strains for which males were analyzed, including wild-type controls. Loss of him-5 does not affect sperm activation, but leads to nondisjunction of the X chromosome, resulting in ~30% male offspring from self-fertilizing hermaphrodites and ensuring the presence of males in a strain. All strains were derived from Bristol N2 with the exception of the strain CB4856 used for mapping. Previously described alleles used in this work were fer-1(hc1ts), spe-8(hc40, hc53), sem-4(n1378), unc-13(e51), ttTi5605, unc-119(ed3, ed9), dpy-18(e364), spe-6(hc163), cxTi10816, spe-27(it110), dpy-20(e1282), spe-17(ok2593), dpy-4(e1166), dpy-11(e224), swm-1(me66, me86, me87), him-5(e1490), unc-76(e911), fog-2(q71), nT1[unc(n754) let qIs50], nT1[qIs51], mIs11.
For analysis of spe-27; snf-10 double mutant hermaphrodites, Dumpy non-GFP-positive offspring were selected from spe-27 dpy-20/nT1; snf-10 him-5/nT1[unc(n754) let qIs50] mothers or control spe-27 dpy-20/nT1; him-5/nT1[unc(n754) let qIs50] mothers. For analysis of spe-27; snf-10 double mutant males, Dumpy non-GFP-positive offspring were selected from crosses between spe-27 dpy-20/nT1; snf-10 him-5/nT1[qIs51] parents or control spe-27 dpy-20/nT1; him-5/nT1[qIs51] parents. For analysis of spe-17 sperm expressing SNF-10::mCherry, GFP-negative male offspring were selected from crosses between jnSi96[Psnf-10::SNF-10:mCh]; unc-119; spe-17/nT1; snf-10 him-5/nT1[qIs51] parents.
Cloning of snf-10
The allele jn3 was shown to be linked to the swm-1 him-5 region of chromosome V by the following data: swm-1(me86) him-5; jn3 males were crossed to dpy-11 swm-1(me86) him-5 hermaphrodites, non-Dumpy F1 hermaphrodites were selected and allowed to self-fertilize, and 40/40 F2 Dumpy male offspring were non-Activated. For fine mapping, dpy-11(e224) swm-1(me86) snf-10(jn3) him-5(e1490) unc-76(e911) hermaphrodites were crossed to CB4856 males, heterozygous F1 hermaphrodites were allowed to self-fertilize, and F2 Dpy non-Unc and Unc non-Dpy recombinant hermaphrodites were selected. Homozygous recombinants were isolated and scored for single-nucleotide polymorphisms using either restriction digest or sequencing. If the recombination breakpoint was potentially informative, the recombinant strain was crossed to swm-1(me86) him-5 and swm-1(me86) snf-10(jn3) him-5 strains to test for complementation with swm-1 and snf-10. We thus iteratively identified a ~49 kb region between the polymorphisms WBVar00001560 and WBVar00001645 that contained jn3 (Supplementary Fig. 1; Wormbase release WS240).
Molecular biology and generation of transgenic strains
Standard procedures for molecular biology were used. RNA was isolated from mixed-stage him-5(e1490) worms using TRIzol (Life Technologies) and RNeasy (Qiagen). RACE (rapid amplification of cDNA ends) was performed using the GeneRacer system (Life Technologies) and RT-PCR was used to generate a full-length cDNA based on sequence data from RACE products.
For rescue experiments involving extrachromosomal arrays, a ~3.9 kb PCR fragment comprising the snf-10 locus was amplified using the primers 5’-GGGTCCACGAGGTATAGAAGG-3’ and 5’-CGGGAATTTCAATCGAGAAG-3’. This fragment was mixed with Psur-5::gfp plasmid (Yochem et al., 1998) at 75-100 ng/μl each and injected into swm-1(me86) snf-10(jn4) him-5(e1490) hermaphrodites (Mello et al., 1991). Independent lines were founded from individual transgenic F1 hermaphrodites.
Single-copy transgenic strains were generated using MosSCI (Frokjaer-Jensen et al., 2012; Frokjaer-Jensen et al., 2008). snf-10 constructs were generated in the pCFJ150 vector using Gateway cloning (Life Technologies) and injected into ttTi5605; unc-119 hermaphrodites. Details of primers used to generate expression constructs are available upon request.
Fertility and sperm competition
To measure hermaphrodite fertility, single L4 hermaphrodites were placed on plates and transferred daily until eggs were no longer produced. To measure male fertility, L4 males were placed with either spe-8(hc40); dpy-4(e1166) or fog-2(q71) L4 hermaphrodites in 1:1 crosses (i.e., one male was placed with one hermaphrodite) for 48 hr; males were then removed and hermaphrodites were transferred daily. spe-8 hermaphrodites allow for detection of mating in all cases where seminal fluid is transferred, even if sperm are not functional (Shakes and Ward, 1989), and fog-2(q71) hermaphrodites lack sperm and thus are essentially female (Schedl and Kimble, 1988). To assay sperm competition, L4 males were placed with L4 dpy-4(e1166) hermaphrodites in 1:1 crosses for 24 hr, males were removed, and hermaphrodites were transferred to new plates daily until egg laying ceased. For all fertility assays, non-Dumpy cross progeny and/or Dumpy self progeny were counted after reaching at least the L4 stage. Only broods for which hermaphrodites survived for at least four days were included in analyses. For these and other assays involving measurements of progeny, sperm migration, or mating, assays for control strains were done in parallel, assay data were compared among matched samples to control for variation in factors such as temperature and media quality, and each experiment was repeated 2-4 times.
Transactivation
To assay transactivation by males, fer-1 or fer-1; snf-10 L4 males raised at 25°C were placed with L4 spe-8(hc53); dpy-4(e1166) hermaphrodites in 1:1 crosses for 48 hr at 25°C. After the mating period, adults were removed, progeny were allowed to develop until at least the L4 stage, and Dumpy self progeny were counted. Only matings lacking non-Dumpy (cross) progeny were included in analysis. To assay the ability of hermaphrodite sperm to transactivate, swm-1(me87) him-5(e1490); mIs11 males were placed with spe-27 dpy-20; him-5 or spe-27 dpy-20; snf-10 him-5 hermaphrodites in 1:1 crosses for 48 hr; progeny counts and analyses were performed as for the male assays.
Sperm migration and mating frequency
Sperm transfer and migration rates were assayed in 1:1 crosses between MitoTracker-labeled males and spe-8(hc40); dpy-4(e1166) hermaphrodites as described in Stanfield (2006). For infertile male strains, mating frequency was assessed by placing MitoTracker (Life Technologies) labeled males with sem-4 unc-13 or fog-2 hermaphrodites for 5 hr and scoring for sperm transfer as described in Smith (2011).
Sperm activation assays
The activation state of individual sperm cells was scored based on the absence or presence of a pseudopod visible by DIC microscopy. To measure the frequency of sperm activation within the male gonad, L4 males were isolated from hermaphrodites and scored 48 hr later for the presence of activated or non-activated sperm within the seminal vesicle. Males were considered non-activated if 100% of sperm lacked a pseudopod, fully activated if fewer than 1% of cells lacked a pseudopod, and partially-activated at intermediate frequencies.
Assays of in vitro activation were performed essentially as described in Shakes (1989). Virgin 24-48 hr post-L4 males were dissected in Sperm Medium (SM) containing 5 mM HEPES pH7.0 or 7.4, 50 mM NaCl, 25 mM KCl, 5 mM CaCl2, 1 mM MgSO4, and 10 mM dextrose. Additional medium with (SM+) or without (SM-) activator was added to the cells and images were collected every 3-5 min for 24-42 min. For each activator, wild-type controls and controls lacking activator were performed in parallel for each repeat, at least two slides were examined for each condition per experiment, and each experiment was repeated at least three times. Essentially no activation was ever observed on SM- slides. Between 42-181 cells were examined on each slide. Concentrations of activators used were: Pronase, 200 μg/μL; triethanolamine (TEA), 60 mM; monensin, 5×10-7 M; 4,4’-diisothiocyano-2,2’-stilbenedisulfonic acid (DIDS), 0.4 mM; ZnCl2, 1mM.
Immunocytochemistry
Sperm from 1-2 day post-L4 virgin males was dissected into SM onto Colorfrost Plus slides (Fisher). Fixation was performed by a freeze-crack procedure followed by immersion in ice-cold methanol for 15 min (Gleason et al., 2012). Washes were performed in PBS pH 7.2 with 0.1% Tween 20 (PBSTw). Blocking (30 min) and antibody incubations (2 hr each) were performed in PBSTw with 1% BSA. Antibodies used were: 1CB4 anti-MO monoclonal at 1:500 (Arduengo et al., 1998; Okamoto and Thomson, 1985), rabbit anti-RFP at 1:1000 (Rockland #600-401-379), Alexa Fluor 568 goat anti-rabbit IgG at 1:500 and Alexa Fluor 488 goat anti-mouse IgG at 1:200 to 1:500 (Life Technologies).
Results
The SLC6 family transporter SNF-10 is required for sperm activation in swm-1 mutant males
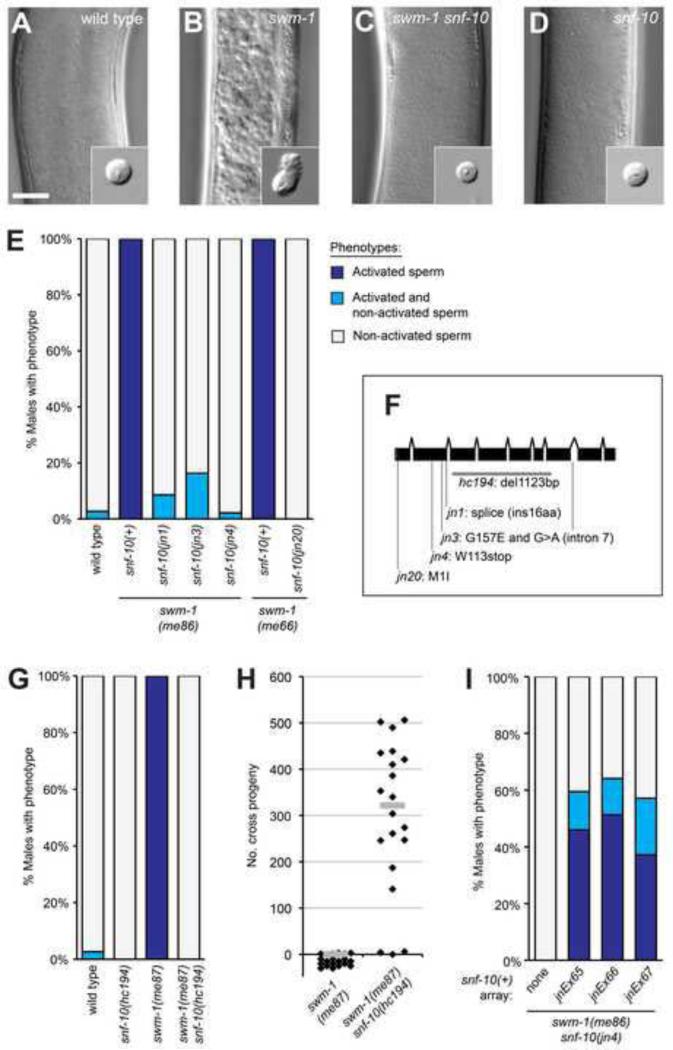
Males with a mutation in the protease inhibitor gene swm-1 show premature sperm activation in the absence of an opportunity to mate. We took advantage of this ectopic sperm activation phenotype of swm-1 mutant males to perform suppressor screens searching for genes that promote sperm activation caused by protease signaling (Smith and Stanfield, 2011; Stanfield and Villeneuve, 2006 and data not shown). In wild-type males, sperm are stored in the seminal vesicle in the form of non-activated, round spermatids (Fig. 1A). In swm-1 mutants, male sperm become prematurely activated prior to sperm transfer (Fig. 1B), leading to infertility. We mutagenized strains carrying a mutation in swm-1 as well as in him-5 to ensure the presence of males (Hodgkin et al., 1979), screened visually for males containing non-activated sperm, and isolated strains harboring suppressor mutations. One complementation group comprised four strong suppressors (jn1, jn3, jn4, jn20), for which nearly all swm-1; suppressor double mutant males showed a non-activated, wild-type phenotype (Fig. 1E). All these mutations were recessive and mutant animals exhibited no obvious phenotypic defects apart from suppression of swm-1 (data not shown).
Fig. 1. Suppressor mutations prevent early sperm activation and loss of fertility caused by loss of swm-1 in males.
(A-D) Images of seminal vesicles and dissected sperm (insets) from 48 hr post-L4 males. Genotypes shown: (A) wild type (B) swm-1(me87) (C) swm-1(me87) snf-10(hc194), and (D) snf-10(hc194). All strains also contained him-5(e1490). Scale bar, 20 microns. (E) Quantitation of suppression of swm-1 sperm activation by mutations in snf-10. For this and other whole-worm activation data, column shading indicates the percent of males containing only activated sperm (dark blue), a mixture of spermatids and activated sperm (light blue), or only non-activated sperm (grey). n, 34-74. (F) Gene model diagram for snf-10 showing the locations of mutant alleles isolated in the screen and the deletion hc194. The jn1 allele alters the splice donor of the second intron and is expected to result in an insertion of 16 amino acids (YLSEPNFSFFQNPQFR) between Arg169 and Thr170. The jn3 strain harbors two mutations, a missense change and a G to A change within intron 7 that is predicted to be silent. (G) Quantitation of suppression of the sperm activation phenotype of the null swm-1(me87) allele by the snf-10 deletion hc194. n, 31-74. (H) Male fertility is restored in a swm-1 snf-10 double mutant. Each point represents the total cross-progeny (non-Dumpy) brood of a single mated spe-8; dpy-4 hermaphrodite; bars represent medians. To permit depiction of all data points, some “0” data points are shown below the X axis. (I) Rescue of the sperm activation phenotype of snf-10 in three strains carrying independently-derived extrachromosomal arrays. Categories for stacked column as in Fig. 1E; n, 32-39.
To identify the gene affected in these suppressor strains, we performed meiotic mapping of the jn3 suppressor mutation against the polymorphic Hawaiian strain CB4856 and localized jn3 to a 49 kb region of chromosome V (Material and Methods, Supplementary Fig. 1). Within this region, one gene, snf-10, had been shown previously to have sperm-enriched expression (Reinke et al., 2004; Reinke et al., 2000). We determined the sequence of snf-10 in the four non-complementing mutant strains and identified specific lesions in each of them (Fig. 1F). Additionally, we obtained a snf-10 deletion allele, hc194 (gift of Harold Smith), and constructed a snf-10(hc194) swm-1(me87) double mutant strain to test for suppression of the null Swm-1 sperm activation phenotype. We found that hc194 males appeared wild-type and showed full suppression of this allele of swm-1 in assays of both sperm activation and male fertility (Fig. 1C,D,G,H). To confirm that snf-10 was the affected gene, we introduced snf-10 transgenes into swm-1 snf-10 animals and scored sperm activation in males. Males harboring transgenes containing a ~3.9 kb region encompassing the snf-10 coding sequence showed strong rescue of sperm activation (Material and Methods; Fig. 1I). Thus, function of the snf-10 gene is required for sperm activation resulting from misregulation of protease activity in C. elegans males.
snf-10 encodes a member of the SLC6 family of sodium- and sometimes chloride-dependent transporters (Beuming et al., 2008; Broer and Gether, 2012; Pramod et al., 2013). In mammals, SLC6 transporters are functionally important in a variety of tissues, including the nervous system and kidney, and their loss is implicated in a number of inherited human diseases including Hartnup disease, hyperekplexia, and X-linked intellectual disability. The best-characterized family members reside on the plasma membrane and function to import specific cargo into cells. Different family members transport cargo such as amino acids, neurotransmitters, or osmolytes, and this activity is coupled to symport of sodium ions, as well as chloride in some cases. A high-resolution structure has been determined for a prokaryotic SLC6 leucine transporter, LeuT (Yamashita et al., 2005), which has facilitated biochemical analyses of transport and conductance activities of eukaryotic family members (reviewed in Kristensen et al., 2011). Eukaryotic SLC6 proteins contain 12 transmembrane (TM) spanning regions, of which TM1-5 and TM6-10 subdomains show two-fold symmetry across the membrane and together compose the channel. Regions surrounding TM1, TM3, TM6 and TM8 are the most conserved among family members and are involved in substrate and sodium binding, as well as a number of highly conserved gating interactions.
Although clearly a member of the SLC6 family, SNF-10 is divergent by several criteria. SNF-10 is well-conserved in related Caenorhabditis nematodes, but we could not identify a clear ortholog from more distant species (data not shown). When compared against all family members, SNF-10 clusters with other SLC6 proteins from C. elegans but does not show particularly strong similarity to any specific SLC6 protein from mammals (Boudko, 2012 and data not shown). While the overall sequence of SNF-10 shows closest similarity to glycine transporters (e.g., it shares 58% similarity with human SLC6A5/GlyT2), its substrate-binding residues are not well-conserved with any SLC6 transporter with a known cargo (Supplementary Fig. 2). In addition, residues directly implicated in sodium passage are divergent in SNF-10. Thus, SNF-10 falls within the orphan class of this large transporter family.
snf-10 is not required for fertility or sperm competition
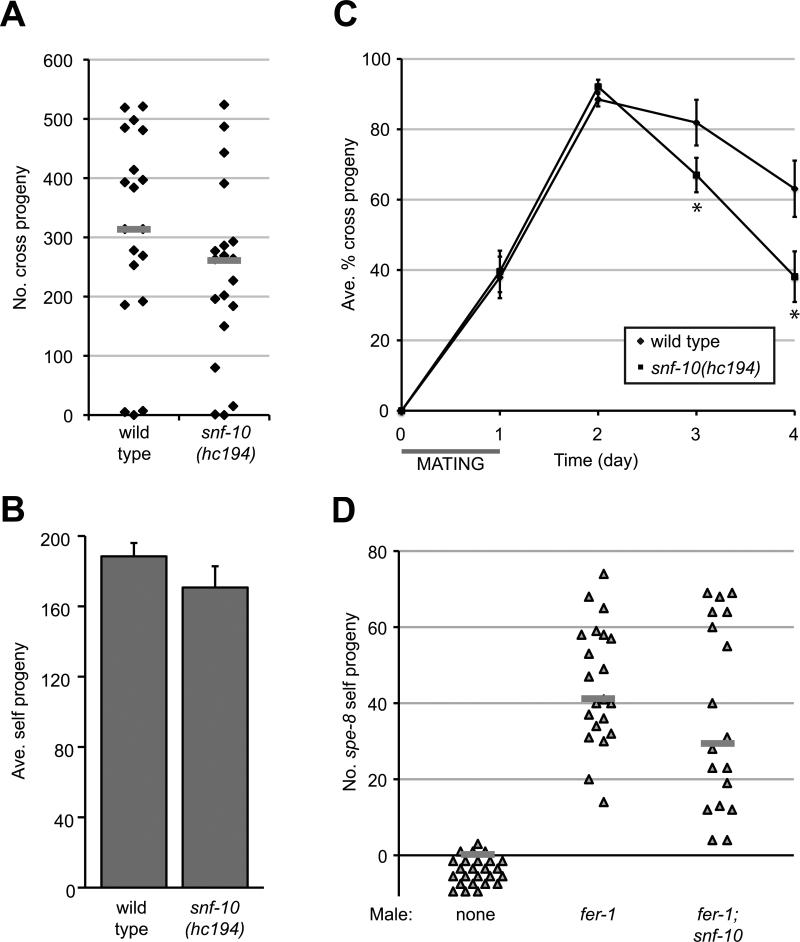
Since snf-10 is required for the ectopic sperm activation that occurs in swm-1 mutant males, we reasoned that snf-10 might be required generally for males and/or hermaphrodites to activate their sperm. Sperm motility is required for fertility, as non-activated sperm cannot maintain their position within the female reproductive tract and are incompetent for fertilization, so we assayed fertility as a proxy to measure activation. To assess male fertility, we measured cross progeny produced by wild-type and snf-10(hc194) mutant males in crosses to spe-8; dpy-4 hermaphrodites. snf-10 mutant males were highly fertile and showed minor, if any, defects in progeny production (Fig. 2A). Thus, snf-10 male sperm are transferred, become activated, and fertilize oocytes at essentially normal levels. To assess hermaphrodite fertility, we counted progeny generated by self-fertilizing hermaphrodites. In C. elegans, virtually 100% of the self sperm generated during development go on to fertilize an egg, so measurement of self progeny is equivalent to quantitation of functional sperm. We found that snf-10(hc194) hermaphrodite brood sizes were only slightly reduced relative to those of wild-type controls, essentially within the variability observed for typical wild-type strains (Fig. 2B and data not shown). Thus, snf-10 is dispensable for fertility in both males and hermaphrodites, consistent with involvement in the male-specific pathway for sperm activation (Smith and Stanfield, 2011).
Fig. 2. snf-10 is not required for sperm function or for transfer of male activator.
(A) snf-10 mutant males showed no significant decrease in fertility as compared to the wild type (P>0.05, Kolmogorov-Smirnov test). Each point represents the total cross-progeny brood of a single spe-8; dpy-4 hermaphrodite; bars represent medians. (B) snf-10 hermaphrodite brood sizes were indistinguishable from those of the wild type (P>0.05, Student's t test; error bars, standard error of the mean). (C) snf-10 males show a normal competitive advantage in crosses to self-fertile dpy-4 hermaphrodites at early time points, but long-term success is decreased as compared to wild-type males. Error bars, 95% confidence intervals; *, P<0.05, Kolmogorov-Smirnov test. (D) Transactivation by snf-10 mutant males. Each point represents self progeny generated by an individual spe-8; dpy-4 hermaphrodite after crossing to the indicated male strain; bars represent medians. No significant difference between fer-1 and fer-1; snf-10 was detected (P>0.05, Kolmogorov-Smirnov test).
While motility is generally required for sperm function, males transfer large numbers of sperm and can be highly fertile even if a subset of cells are defective. However, assays of competition can reveal more subtle defects. We thus took advantage of the male-hermaphrodite mode of reproduction in C. elegans to further evaluate snf-10 sperm function and determine their success in a competitive situation. In wild-type C. elegans, male sperm show strong preferential usage as compared to hermaphrodite sperm. After male sperm are transferred, the fraction of offspring sired by male sperm rises rapidly and often reaches close to 100% (LaMunyon and Ward, 1995). If loss of snf-10 reduced activation, this male precedence effect might be reduced or delayed. To test this possibility, we performed crosses of snf-10(hc194) and wild-type males to dpy-4 recipient hermaphrodites and measured production of both cross and self progeny over time. We found that as compared to the wild type, snf-10 mutant males showed an equivalent level of early success, as measured by preferential generation of cross progeny (Fig. 2C), indicating that snf-10 male sperm activate rapidly and then show normal precedence over hermaphrodite self sperm. However, at later time points, cross-progeny production was slightly decreased. Visual inspection of sperm transfer and migration using MitoTracker-labeled males failed to reveal any obvious differences between snf-10 and wild-type males (data not shown), suggesting that snf-10 males transfer a normal number of sperm, which activate rapidly and crawl to the spermathecae. It is possible that snf-10 sperm have reduced long-term viability as compared to the wild type. Regardless of these minor defects, the fertility and competition data indicate that snf-10 sperm are highly competent to activate, migrate, and fertilize oocytes.
snf-10 is not required for the production or transfer of male activator
A sperm activator is transferred from males to hermaphrodites during mating (Ward and Carrel, 1979). Previous analyses of suppressors of swm-1 identified TRY-5, a seminal fluid protease that likely functions as the male activator (Smith and Stanfield, 2011). One possibility for snf-10 function was that it might be required for production and/or transfer of the male activator. As a specific assay for the male activator, males that fail to transfer functional sperm, but do transfer seminal fluid (e.g., a fer-1 mutant), can be crossed to spe-8 mutant hermaphrodites, whose sperm are defective for response to hermaphrodite activator, but not the male one. Self progeny, which arise from “transactivation” of hermaphrodite sperm by male seminal fluid, are then counted (Shakes and Ward, 1989). To assay transactivation by snf-10 males, we crossed fer-1; snf-10 or fer-1 control males to spe-8; dpy-4 hermaphrodites and measured induction of Dumpy self progeny after mating. There was no statistical difference between transactivation levels caused by males mutant for snf-10 as compared to wild-type (Fig. 2D). In addition, we looked directly at the pattern of TRY-5 expression and localization in animals mutant for snf-10. In wild-type animals, TRY-5::GFP is expressed within the male somatic gonad in cells of the seminal vesicle, valve and vas deferens, where it localizes to large vesicle-like structures; this pattern is unchanged in snf-10 mutant males (Smith and Stanfield, 2011 and data not shown). These data indicate that snf-10 is not required for production or transfer of male activator but rather functions in a downstream step of activation.
snf-10 functions cell-autonomously in sperm to promote sperm activation
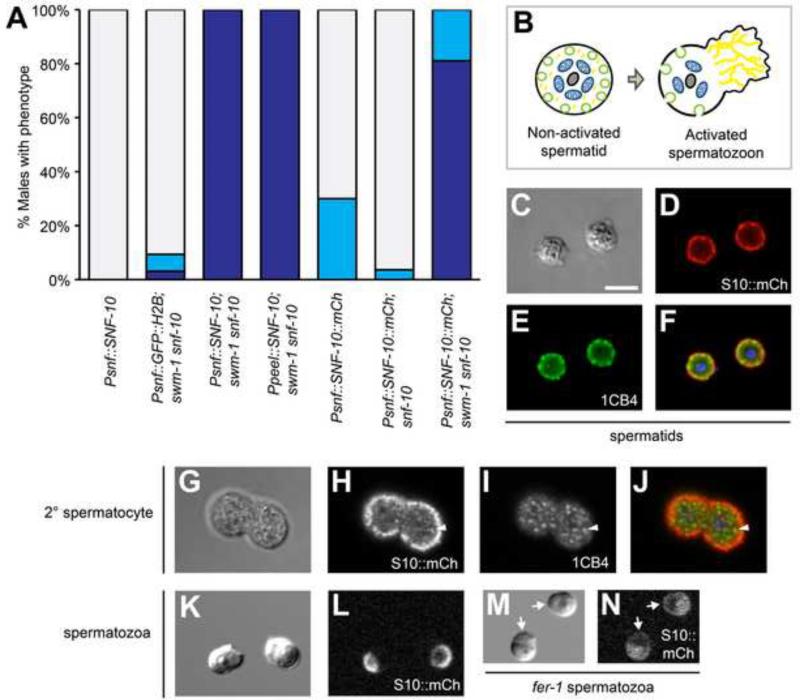
To address how SNF-10 regulates sperm activation, we sought to determine in which cells SNF-10 functions. snf-10 was previously reported to show sperm-enriched expression in genome-wide expression analyses (Reinke et al., 2004; Reinke et al., 2000). To determine whether SNF-10 indeed functions in sperm, we performed tissue-specific rescue and inactivation experiments. First, we generated animals expressing the SNF-10 protein under the control of the sperm-specific peel-1 promoter (Seidel et al., 2011) and assayed for rescue of sperm activation in swm-1(me87) snf-10(hc194) males. Expression of SNF-10 in sperm fully restored the Swm-1 ectopic sperm activation phenotype, equivalent to expression using the snf-10 promoter (Fig. 3A). Importantly, swm-1(+) animals carrying snf-10 transgenes contained non-activated sperm, indicating that the activation observed in the swm-1 mutant background represented rescue of snf-10 rather than a gain-of-function effect. To confirm that snf-10 function is required in sperm, we performed germ line-specific RNAi of snf-10, using a mutation in rrf-1 to reduce RNAi efficacy in somatic tissues (Kumsta and Hansen, 2012; Sijen et al., 2001). When fed snf-10 RNAi bacteria, both swm-1 and rrf-1; swm-1 males showed equivalent levels of suppression of the Swm-1 sperm activation phenotype, consistent with strong reduction of snf-10 function in both genotypes (Supplementary Fig. 3). Thus, activity of snf-10 in the germ line is both sufficient and necessary to promote sperm activation in males.
Fig. 3. snf-10 is expressed in and functions in sperm.
(A) Rescue of the sperm activation phenotype by snf-10 transgenes. Either a snf-10 or sperm-specific peel-1 promoter can confer rescue, as can expression of SNF-10::mCherry reporter protein. As in Fig. 1E, column shading indicates the percent of males containing only activated sperm (dark blue), a mixture of spermatids and activated sperm (light blue), or only non-activated sperm (grey); n, 30-55. (B) Schematic of spermatid and spermatozoon structure. Grey, chromatin; blue, mitochondria; green, MOs; yellow, MSP cytoskeleton. (C-J) In jnSi96[Psnf-10::SNF-10::mCh] spermatids, SNF-10::mCh and 1CB4 are both localized primarily to the cell periphery; some concentrations of mCh and 1CB4 are clearly distinct from one another. In 2° spermatocytes, most mCh is on the cell cortex and fails to colocalize with 1CB4, but some cytoplasmic puncta are visible. Arrowhead indicates a region of overlap between mCh and 1CB4. Images are fixed cells visualized by (C,G) DIC, (D,H) anti-RFP staining, (E,I) 1CB4 staining, and (F,J) a merge of anti-RFP, 1CB4, and DAPI. (K-N) SNF-10::mCh is polarized to the cell body plasma membrane in activated spermatozoa, but distributed throughout the plasma membrane in a fer-1 mutant. Live sperm were visualized by (K,M) DIC and (L,N) mCherry fluorescence. Arrows indicate SNF-10::mCh in fer-1 pseudopodia. (A-N) All strains were mutant for unc-119 and him-5, as well as snf-10(hc194) except as indicated in (A). (M,N) Worms were raised at 25°C, the non-permissive temperature for fer-1(hc1). Scale bar for all images, 5 microns.
SNF-10 localizes to the sperm plasma membrane and is polarized to the cell body of mature spermatozoa
To examine snf-10 expression and protein localization within the germ line, we generated transgenic animals expressing snf-10 reporters under the control of the genomic region sufficient for rescue activity. To identify cells expressing snf-10, we used transcriptional Psnf-10::htas-1::mCh (mCherry) and Psnf-10::GFP::H2B (histone H2B) reporters with fluorescent proteins targeted to sperm chromatin (Chu et al., 2006; Merritt et al., 2008). Expression was visible in the male and hermaphrodite germ line in cells undergoing differentiation into spermatocytes as well as at later stages of spermatogenesis (Supplementary Fig. 4 and data not shown). No fluorescence was visible in somatic tissues, in the mitotic or meiotic-prophase regions of the germ line, or in germline cells developing into oocytes.
To determine where in sperm the SNF-10 protein localizes, we examined Psnf-10::SNF-10::mCh worms expressing full-length SNF-10 fused to mCherry. Use of either the snf-10 genomic coding region or a snf-10 cDNA was sufficient for rescue of the sperm activation phenotype of swm-1 snf-10 mutant males (Fig. 3A and data not shown), indicating that the fusion protein was functional. C. elegans sperm show a stereotypical arrangement of cell structures (L'Hernault, 2006) (shown in Fig. 3B). Non-activated spermatids are spherical and contain a central chromatin mass surrounded by mitochondria; the sperm-specific membranous organelles (MOs) reside adjacent to the plasma membrane. In activated, polarized spermatozoa, the cell body retains the nucleus, mitochondria, and MO fusion sites while the pseudopod contains the network of MSP filaments that power cell movement. We found that in spermatids, SNF-10::mCh was primarily localized to the cell cortex, with brighter concentrations in some regions, as well as in occasional puncta deeper within the cell (Fig. 3C,D; Supplementary Fig. 5A,B and data not shown). Due to the close apposition of MOs and the plasma membrane in spermatids, it is difficult to distinguish between them by conventional fluorescence microscopy, so this cortical localization could indicate the presence of SNF-10 on the plasma membrane, the MOs, or both. Thus we performed additional experiments to distinguish between these cellular compartments. First, we performed coincident imaging of SNF-10::mCh with two different MO markers, the monoclonal antibody 1CB4 (Okamoto and Thomson, 1985) and PEEL-1::GFP (Seidel et al., 2011). We found that while the SNF-10::mCh and 1CB4 signals were very close and potentially overlapping, the areas of high cortical SNF-10 concentration were often distinct from MO puncta (Fig. 3C-F). Similarly, in live cells, SNF-10::mCherry was peripheral to PEEL-1::GFP (Supplementary Fig. 5A-D). Second, we examined SNF-10 localization in situations where MOs are located farther from the plasma membrane. In cells at earlier stages of spermatogenesis, MOs are distributed throughout the cytoplasm, and they move to abut the plasma membrane after the meiotic divisions (L'Hernault, 2006). In secondary spermatocytes and immature spermatids, the majority of SNF-10 was localized to the cell cortex as at later stages, and this pattern was largely distinct from that of 1CB4; occasional cytoplasmic puncta appeared to show overlap (Fig. 3G-J and data not shown). We also examined spe-17 mutant spermatids, which contain MOs and other organelles asymmetrically mislocalized within the cytoplasm (L'Hernault et al., 1993). In spe-17 spermatids, SNF-10::mCh was present not only on the cell cortex but also in asymmetric, bright cytoplasmic foci, suggesting that some SNF-10 is indeed present on MOs, though it is also possible that SNF-10 is mislocalized in this mutant (Supplementary Fig. 5E,F). Taken together, these data suggest that in spermatids, the majority of SNF-10 is present in the plasma membrane, but we cannot exclude the possibility that at least a portion may localize to MOs as well.
In activated spermatozoa, SNF-10::mCh became restricted to the cell body region of the plasma membrane and was absent from the pseudopod (Fig. 3K,L). In many cells, brighter cortical puncta were visible. These concentrations likely corresponded to the sites of fused MOs, as activated sperm retain stable invaginations at these sites that show elevated staining with plasma membrane markers, including dyes such as FM1-43, due to the increased concentration of membrane there (Washington and Ward, 2006 and data not shown). We sought to determine whether SNF-10::mCh localization would be altered in the mutant fer-1, in which MOs associate with the plasma membrane but fail to fuse with it during activation (Ward and Miwa, 1978; Washington and Ward, 2006). We found that in activated fer-1 sperm, SNF-10::mCh was visible not only in cortical and intracellular puncta of the cell body but also on the pseudopod plasma membrane, from which it is normally excluded (Fig. 3M,N). Thus, SNF-10 shows a restricted localization in spermatozoa, which is dependent on either FER-1 or some other protein that is absent from the cell surface when MOs fail to fuse.
snf-10 is required for sperm to respond to the male protease activator
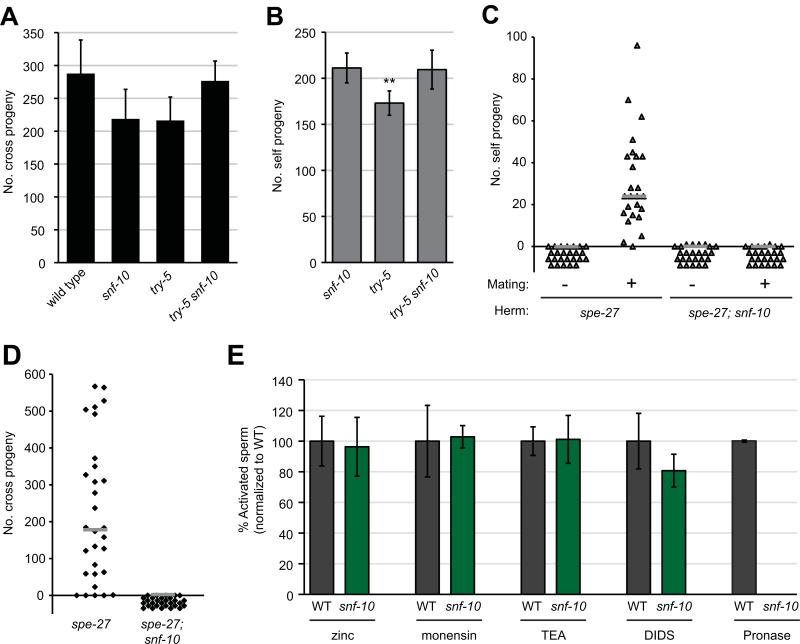
To understand how SNF-10 promotes sperm activation, we next sought to distinguish between a role in the “male” pathway downstream of protease signaling or a role in the “hermaphrodite” pathway, which requires spe-8 group activity and can be triggered by extracellular zinc (Liu et al., 2013). Loss of the hermaphrodite pathway is sufficient to result in hermaphrodite self sterility, though males are fertile (L'Hernault et al., 1988); the activity of both pathways must be lost to result in male sterility (Smith and Stanfield, 2011). Our swm-1 suppression screen could identify factors in either pathway, as loss of either leads to reduced activation of male sperm (Stanfield and Villeneuve, 2006 and G.M.S., unpublished results). However, spe-8 group; swm-1 double mutants show a characteristic “spiky” morphology associated with partial activation (Shakes and Ward, 1989; Stanfield and Villeneuve, 2006), so our finding that swm-1 snf-10 double mutants show no sign of activation suggested snf-10 was in the male pathway. To test this explicitly, we assayed fertility in double mutants lacking the activity of both snf-10 and either try-5 or spe-8 group function. First, we generated try-5(tm3813) snf-10(hc194) double mutant males. Their fertility was at least as high as that of either single mutant and indistinguishable from that of wild-type worms (Fig. 4A). try-5 snf-10 hermaphrodites were also fully fertile (Fig. 4B). We then generated animals doubly mutant for both snf-10 and the spe-8 group gene spe-27. spe-27(it110); snf-10(hc194) hermaphrodites were self-sterile, as expected due to the spe-27 mutation. To assay response to male activator, we performed transactivation assays by crossing these hermaphrodites to swm-1 mutant males, which are defective for sperm transfer but competent for seminal fluid transfer (Stanfield and Villeneuve, 2006 and data not shown). We found that spe-27; snf-10 double mutant hermaphrodite sperm were incapable of transactivation (Fig. 4C). In addition, spe-27; snf-10 males were completely sterile in crosses to fog-2 “females” (Fig. 4D). We confirmed that spe-27; snf-10 males were capable of mating and transferring sperm by staining them with MitoTracker dye and assaying transfer of labeled sperm after a 5 hr mating period. While the frequency of sperm transfer for spe-27; snf-10 males was at least as high as that for the control (37/59 or 63% transferred sperm, as compared to 30/56 or 54% for spe-27), double mutant sperm were never observed to migrate towards the spermathecae (data not shown), consistent with failure to activate. Thus, loss of both snf-10 and spe-8 group activities results in profound sterility in both sexes, suggesting that all pathways to activation are blocked.
Fig. 4. snf-10 functions in the male sperm activation pathway downstream of the protease activator.
(A) try-5 snf-10 double mutant males show levels of fertility indistinguishable from those of the wild type or single mutants (P>0.05, Kolmogorov-Smirnov test). The indicated males were crossed to spe-8; dpy-4 hermaphrodites and total offspring were counted. Error bars, 95% confidence intervals. (B) try-5 snf-10 double mutant hermaphrodites show no reduction in fertility relative to single mutants. Note that the try-5 strain showed slightly reduced fertility in this experiment (P<0.01, Kolmogorov-Smirnov test). (C) spe-27; snf-10 double mutant hermaphrodite sperm are unresponsive to transactivation, in contrast to spe-27 hermaphrodites whose self fertility is rescued after transfer of male seminal fluid. The symbol “-“ designates controls in which no males were present (hermaphrodites were allowed to self-fertilize); “+” indicates that swm-1 males were present (transactivation conditions). (D) spe-27; snf-10 double mutant males are completely infertile. The indicated males were crossed to fog-2 hermaphrodites for 48 hr and total offspring were counted. (C,D) Bars represent medians. (E) snf-10 mutant sperm are resistant to in vitro activation by Pronase, but respond normally to other activators. Wild-type (WT) or snf-10(hc194) spermatids were dissected from males and incubated in activators as indicated. Graph shows percent of activated cells normalized to that observed in wild-type for each experiment. Error bars, 95% confidence intervals.
To further place snf-10 in the male pathway for sperm activation, we sought to distinguish between a role in the response of sperm to extracellular signals or a role in more downstream events. While sperm normally activate in response to extracellular cues, mutations in genes involved in spermatogenesis can lead to signal-independent activation (Liau et al., 2013; Muhlrad and Ward, 2002). In particular, certain alleles of the casein kinase 1 gene spe-6 or other genes in this class suppress the sterility of spe-8 group mutants and lead to ectopic sperm activation within males. To determine whether snf-10 is required for this activation, we examined males mutant for both snf-10 and the activating mutation spe-6(hc163). spe-6; snf-10 double mutants showed a fully-activated phenotype indistinguishable from that of spe-6 males (Table 1). Thus, snf-10 likely functions in the response of sperm to activator.
Table 1.
snf-10 is not required for activation in spe-6 animals.
| Genotype1 | % Act2 | n |
|---|---|---|
| wild type | 2.8 | 36 |
| spe-6(hc163) | 100 | 38 |
| snf-10(hc194) | 2.1 | 47 |
| spe-6(hc163); snf-10(hc194) | 100 | 38 |
All strains also contained the mutations dpy-18(e364) and him-5(e1490).
Percent of 48hr post-L4 males containing activated sperm.
snf-10 is required for in vitro activation by proteases
The presence of SNF-10 in sperm suggests a role in reception or transduction of the activation signal. Little is known about how extracellular signals result in the morphological changes that occur during activation. However, sperm can be activated in vitro by a variety of different compounds that presumably alter cell physiology in ways that mimic or otherwise feed into the normal in vivo pathways (Liu et al., 2013; Machaca et al., 1996; Nelson and Ward, 1980; Shakes and Ward, 1989; Ward et al., 1983). To probe the mechanism by which SNF-10 promotes sperm activation, we sought to determine whether its activity was required for response to these activators. Most activators were fully effective on snf-10(hc194) mutant sperm (Fig. 4D). Treatment of snf-10 sperm with zinc, which is thought to activate sperm via the hermaphrodite pathway (Liu et al., 2013), resulted in normal levels of activation as compared to the wild type. We observed similar results with the sodium-selective ionophore monensin and the weak base TEA, both of which lead to alkalinization of sperm cytoplasm (Ward et al., 1983). snf-10 mutant sperm also activated normally with DIDS (4,4’-diisothiocyano-2,2’-stilbenedisulfonic acid), a chloride channel blocker that has been shown to activate sperm with similar dose dependence as seen for inhibition of a spermatid Clir channel activity (Machaca et al., 1996). Thus, all of these activators apparently bypass SNF-10's function in sperm activation. However, snf-10(hc194) mutant sperm failed to activate in Pronase, a mixture of protease activities that are thought to mimic the male activator (Smith and Stanfield, 2011). Wild-type sperm activate rapidly to high levels (often >95%) when incubated in Pronase, but even when snf-10 cells were subjected to extended treatment (42 min, over twice as long as normally required), almost no activation was observed. Furthermore, Pronase-treated snf-10 spermatids remained spherical and lacked any signs of spiking or other cytoskeletal extensions. To assess whether MO fusions might still occur in the absence of other morphological rearrangements, we incubated spermatids in the vital membrane dye FM1-43 during Pronase treatment, but observed no fusion events in snf-10 mutant spermatids (0/268 snf-10 cells showed one or more fusions after 15 min, as compared to 169/198 wild-type cells). Therefore, the results of both our genetic analysis and in vitro assays indicate that SNF-10 functions specifically in the male pathway and is required downstream of TRY-5 for sperm to respond to protease sperm activation signals.
Discussion
Like flagellate sperm, the amoeboid sperm of C. elegans and other nematodes require motility to migrate to and fertilize an oocyte. Successful sperm motility requires both execution of a complex differentiation program during cellular development as well as appropriate responses to environmental cues encountered during the migration process. In C. elegans, the regulation of sperm motility is further influenced by its male-hermaphrodite mode of reproduction. As a result, C. elegans harbors a surprisingly complex network of factors that control the activation of sperm to a motile state. Males and hermaphrodites utilize sex-biased but redundant pathways to ensure that their sperm become motile in the correct time and place to promote reproductive success. In this study, we describe a role for an SLC6 family transporter, SNF-10, in C. elegans sperm activation. SNF-10 is required for sperm to respond to protease signaling for activation via the male-specific pathway. However, it is completely dispensable in hermaphrodites as well as in most known physiological activation assays in vitro. SNF-10 functions in sperm and localizes to the plasma membrane, placing it in a prime position to transduce signals from extracellular protease activity to effect subcellular morphogenesis.
The SLC6 transporters (also known as NTT for neurotransmitter transporters, NSS for neurotransmitter sodium symporters, or NAT for nutrient amino acid transporters) are members of the larger superfamily of APC (amino acid-polyamine-organocation) transporters (Vangelatos et al., 2009), which comprises several protein families with similar structure but unrelated primary sequence. Characterized SLC6 family transporters couple the import of molecules such as amino acids, monoamine neurotransmitters, or osmolytes to the import of sodium and often chloride ions. However, channel activity uncoupled from cargo movement also has been observed (DeFelice and Goswami, 2007). Much focus has been on the monoamine transporters, taking advantage of pharmacological approaches to analyze transport activity and its regulation by trafficking, post-translational modifications, and interactions with other proteins (Broer and Gether, 2012).
Is SNF-10 an active transporter, and if so then what is its cargo? In keeping with its function in reproduction, SNF-10 is a relatively divergent member of the SLC6 family. Its substrate-binding region is not well conserved, making it difficult to predict specificity. In addition, none of the typical molecules transported by SLC6 proteins are present in the sperm medium used for in vitro activation studies, suggesting that such a cargo is not required for activation or is generated in situ at the cell surface and works in a paracrine fashion. Given the variety of transport activities exhibited by the family, it is also possible that SNF-10 promotes sperm activation via channel activity rather than transport of larger cargo. The influx of ions is well known to regulate motility of flagellate sperm in a variety of ways (Darszon et al., 2011; Lishko et al., 2011; Santi et al., 2013; Shukla et al., 2012). Less is known about the relationship between cellular ion physiology and motility of nematode sperm, although activation to motility requires specific ions and can be induced by compounds that challenge cellular ion homeostasis (Liu et al., 2013; Machaca et al., 1996; Nelson and Ward, 1980; Shakes and Ward, 1989). Furthermore, pH gradients within the cell may act to regulate dynamics of the pseudopodial MSP cytoskeleton, as shown for the larger sperm of Ascaris suum (Italiano et al., 1999; King et al., 1994). Thus, it is likely that amoeboid nematode sperm use ion flux as a way to sense and respond to their environment, just as flagellate sperm do. Alternatively, SNF-10 might lack transport activity and instead function in signal transduction, perhaps in a receptor or scaffolding role. Functions for SLC6 family members that may be based on protein-protein interactions rather than any form of transport or channel activity have been identified previously. For example, the Drosophila SLC6 protein Bedraggled has been shown to play a role in tissue polarity in the eye (Rawls et al., 2007). Recently, C. elegans SNF-12 was shown to function in innate immunity by binding to STA-2, a STAT transcription factor, promoting transcriptional responses to fungal exposure (Dierking et al., 2011). In both of these cases, it is unclear whether any canonical transport activity is involved.
In activated spermatozoa, SNF-10 becomes restricted to the cell body region of the plasma membrane. This localization pattern is so far unique, but a number of other spermatozoan cell surface proteins (SPE-9, SPE-38) are restricted to the pseudopod, while others (FER-1, TRP-3/SPE-41) are found on both the pseudopod and cell body (Chatterjee et al., 2005; Washington and Ward, 2006; Xu and Sternberg, 2003; Zannoni et al., 2003). Little is known about how nematode sperm polarization is regulated, including how specific protein domains are established, but several membrane proteins are sequestered in MOs prior to activation and could be involved in the polarization process. For example, the function of SPE-38 is required for the proper distribution of TRP-3/SPE-41 (Singaravelu et al., 2012). Since SNF-10 does not appear to play a major role in sperm after activation, its polarized localization might arise from earlier interaction with other proteins that remain restricted to the cell body region. Related to this idea, we find that in fer-1 spermatozoa, SNF-10 is no longer confined to the cell body but rather is distributed throughout the plasma membrane. FER-1 and other dysferlins are involved in membrane fusion in a variety of contexts including muscle repair as well as neurotransmission (Lek et al., 2011). While activated fer-1 sperm are grossly polarized, specific proteins involved in fertilization are trapped in unfused MOs and thus absent from the sperm surface (Chatterjee et al., 2005; Xu and Sternberg, 2003). Our data suggest that restriction of SNF-10 to the cell body is dependent on interaction with either FER-1 or possibly another protein derived from MOs, since FER-1 is not itself restricted to the same domain as SNF-10 (Lek et al., 2011).
We have found that SNF-10 is primarily present on the plasma membrane in spermatids, but our data also suggest that some SNF-10 could be present on MOs. There is precedent for such a site of action: while most SLC6 proteins are present on the plasma membrane, some other family members are localized to intracellular vesicles (e.g., Parra et al., 2008; Renick et al., 1999). Their function in this location has not been analyzed, though when expressed on the plasma membrane at least one shows a typical sodium-dependent cargo import activity. We speculate that SNF-10 could function to alter cytoplasmic ion or osmolyte homeostasis via the release of pools present in MOs.
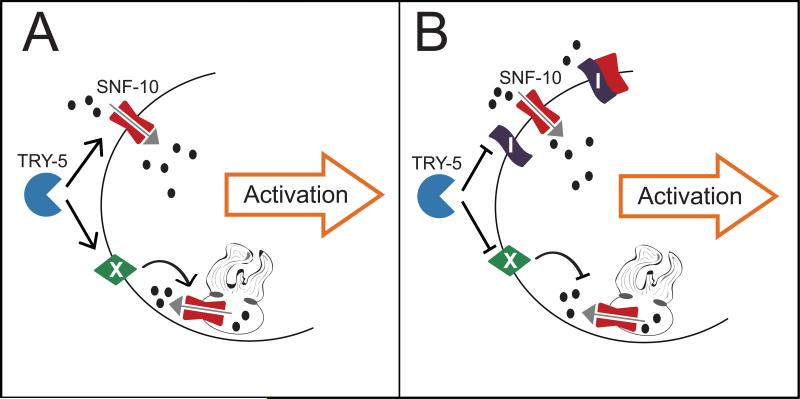
How does SNF-10 function downstream of protease activity to promote the morphological changes associated with sperm activation? One simplistic but intriguing model is that SNF-10 is cleaved directly by the serine protease TRY-5 (Fig. 5A). Importantly, such cleavage should induce or alter, but not eliminate, SNF-10's activity. If so, this would define a novel regulatory mechanism for SLC6 family proteins. Several of the monoamine transporters have been shown to be cleaved by calpain-family proteases, but this resulted either in reduced activity or in no functional consequence (e.g., Baliova and Jursky, 2010; Baliova et al., 2009). Overall, little data exists to address if positive or negative regulation of the family by proteolysis could be more widespread. Another possibility is that in spermatids, SNF-10 associates with an inhibitor that is itself cleaved by TRY-5, leading to SNF-10's activity (Fig. 5B). Finally, if SNF-10 functions on the MOs, activation or de-inhibition by TRY-5 would necessarily be indirect (Fig. 5A,B).
Fig. 5. Models for the role of SNF-10 in protease-initiated sperm activation.
(A) Model 1: TRY-5 positively regulates SNF-10 activity in spermatids. TRY-5 might act directly on SNF-10 localized to the plasma membrane; it also might cleave an intermediate (X) that promotes SNF-10 activity on the MO membrane. Ultimately, SNF-10 transport and/or channel activity (grey arrow) induces maturation of the spermatid into a motile, active spermatozoon. Black arrows represent positive regulation; black ovals indicate substrates and/or ions to which SNF-10 is permeable. (B) Model 2: TRY-5 cleaves an inhibitor (I) of SNF-10 activity, which prevents SNF-10 activity either directly or through an intermediate (X). With the inhibitor disabled, SNF-10 functions to promote sperm activation. Black bars represent inhibition.
Another SLC6 transporter, Ntl, has been shown recently to be required for spermiogenesis in Drosophila (Chatterjee et al., 2011). Some mammalian SLC6 transporters show testis expression (Farmer et al., 2000; Hoglund et al., 2005), including the protein SLC6A16, which also shows divergent sequence features as compared to family members expressed in other tissues. However, the mammalian testis-expressed genes have not been analyzed genetically so the nature of their role in spermatogenesis is unknown. Our identification of SNF-10 as a regulator of sperm activation in C. elegans suggests a shared role for SLC6 proteins in sperm physiology and function from invertebrates to mammals.
Supplementary Material
Highlights.
Identification of an SLC6 transporter that functions in sperm to promote motility
SNF-10 is required for sperm to respond to the male protease activator
snf-10 mutants are fertile and have competitive sperm
SNF-10 localization is regulated during sperm activation
Acknowledgements
Some strains were provided by the Caenorhabditis Genetics Center, which is funded by the NIH Office of Research Infrastructure Programs (P40 OD010440). We thank Wormbase; the University of Utah Imaging and Sequencing Core Facilities; M. Ailion and C. Frokjaer-Jensen for reagents; E. Jorgensen and M. Metzstein for helpful discussions; and S. Mansour and C. Murtaugh for comments on the manuscript. This work was supported by NIH R01-GM087705.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ahringer J, editor. Reverse genetics, WormBook. The [i]C. elegans[/i] Research Community, WormBook. 2006 doi/10.1895/wormbook.1.47.1, http://www.wormbook.org.
- Arduengo PM, Appleberry OK, Chuang P, L'Hernault SW. The presenilin protein family member SPE-4 localizes to an ER/Golgi derived organelle and is required for proper cytoplasmic partitioning during [i]Caenorhabditis elegans[/i] spermatogenesis. J Cell Sci. 1998;111(Pt 24):3645–54. doi: 10.1242/jcs.111.24.3645. [DOI] [PubMed] [Google Scholar]
- Argon Y, Ward S. [i]Caenorhabditis elegans[/i] fertilization-defective mutants with abnormal sperm. Genetics. 1980;96:413–33. doi: 10.1093/genetics/96.2.413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baliova M, Jursky F. Calcium dependent modification of distal C-terminal sequences of glycine transporter GlyT1. Neurochem Int. 2010;57:254–61. doi: 10.1016/j.neuint.2010.06.003. [DOI] [PubMed] [Google Scholar]
- Baliova M, Knab A, Franekova V, Jursky F. Modification of the cytosolic regions of GABA transporter GAT1 by calpain. Neurochem Int. 2009;55:288–94. doi: 10.1016/j.neuint.2009.03.012. [DOI] [PubMed] [Google Scholar]
- Beuming T, Kniazeff J, Bergmann ML, Shi L, Gracia L, Raniszewska K, Newman AH, Javitch JA, Weinstein H, Gether U, Loland CJ. The binding sites for cocaine and dopamine in the dopamine transporter overlap. Nat Neurosci. 2008;11:780–9. doi: 10.1038/nn.2146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloch Qazi MC, Heifetz Y, Wolfner MF. The developments between gametogenesis and fertilization: ovulation and female sperm storage in [i]Drosophila melanogaster[/i]. Dev Biol. 2003;256:195–211. doi: 10.1016/s0012-1606(02)00125-2. [DOI] [PubMed] [Google Scholar]
- Boudko DY. Molecular basis of essential amino acid transport from studies of insect nutrient amino acid transporters of the SLC6 family (NAT-SLC6). J Insect Physiol. 2012;58:433–49. doi: 10.1016/j.jinsphys.2011.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S. The genetics of [i]Caenorhabditis elegans[/i]. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broer S, Gether U. The solute carrier 6 family of transporters. Br J Pharmacol. 2012;167:256–78. doi: 10.1111/j.1476-5381.2012.01975.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y, Li M, Mager S, Lester HA. Amino acid residues that control pH modulation of transport-associated current in mammalian serotonin transporters. J Neurosci. 1998;18:7739–49. doi: 10.1523/JNEUROSCI.18-19-07739.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee I, Richmond A, Putiri E, Shakes DC, Singson A. The [i]Caenorhabditis elegans spe-38[/i] gene encodes a novel four-pass integral membrane protein required for sperm function at fertilization. Development. 2005;132:2795–808. doi: 10.1242/dev.01868. [DOI] [PubMed] [Google Scholar]
- Chatterjee N, Rollins J, Mahowald AP, Bazinet C. Neurotransmitter Transporter-Like: a male germline-specific SLC6 transporter required for [i]Drosophila[/i] spermiogenesis. PLoS One. 2011;6:e16275. doi: 10.1371/journal.pone.0016275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu DS, Liu H, Nix P, Wu TF, Ralston EJ, Yates JR, 3rd, Meyer BJ. Sperm chromatin proteomics identifies evolutionarily conserved fertility factors. Nature. 2006;443:101–5. doi: 10.1038/nature05050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ClustalO. http://www.ebi.ac.uk/Tools/msa/clustalo/, date May 12, 2014.
- Darszon A, Guerrero A, Galindo BE, Nishigaki T, Wood CD. Sperm-activating peptides in the regulation of ion fluxes, signal transduction and motility. Int J Dev Biol. 2008;52:595–606. doi: 10.1387/ijdb.072550ad. [DOI] [PubMed] [Google Scholar]
- Darszon A, Nishigaki T, Beltran C, Trevino CL. Calcium channels in the development, maturation, and function of spermatozoa. Physiol Rev. 2011;91:1305–55. doi: 10.1152/physrev.00028.2010. [DOI] [PubMed] [Google Scholar]
- DeFelice LJ, Goswami T. Transporters as channels. Annu Rev Physiol. 2007;69:87–112. doi: 10.1146/annurev.physiol.69.031905.164816. [DOI] [PubMed] [Google Scholar]
- Dierking K, Polanowska J, Omi S, Engelmann I, Gut M, Lembo F, Ewbank JJ, Pujol N. Unusual regulation of a STAT protein by an SLC6 family transporter in [i]C. elegans[/i] epidermal innate immunity. Cell Host Microbe. 2011;9:425–35. doi: 10.1016/j.chom.2011.04.011. [DOI] [PubMed] [Google Scholar]
- Farmer MK, Robbins MJ, Medhurst AD, Campbell DA, Ellington K, Duckworth M, Brown AM, Middlemiss DN, Price GW, Pangalos MN. Cloning and characterization of human NTT5 and v7-3: two orphan transporters of the Na+/Cl− -dependent neurotransmitter transporter gene family. Genomics. 2000;70:241–52. doi: 10.1006/geno.2000.6387. [DOI] [PubMed] [Google Scholar]
- Frokjaer-Jensen C, Davis MW, Ailion M, Jorgensen EM. Improved Mos1-mediated transgenesis in [i]C. elegans[/i]. Nat Methods. 2012;9:117–8. doi: 10.1038/nmeth.1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frokjaer-Jensen C, Davis MW, Hopkins CE, Newman BJ, Thummel JM, Olesen SP, Grunnet M, Jorgensen EM. Single-copy insertion of transgenes in [i]Caenorhabditis elegans[/i]. Nat Genet. 2008;40:1375–83. doi: 10.1038/ng.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gleason EJ, Hartley PD, Henderson M, Hill-Harfe KL, Price PW, Weimer RM, Kroft TL, Zhu GD, Cordovado S, L'Hernault SW. Developmental genetics of secretory vesicle acidification during [i]Caenorhabditis elegans[/i] spermatogenesis. Genetics. 2012;191:477–91. doi: 10.1534/genetics.112.139618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goujon M, McWilliam H, Li W, Valentin F, Squizzato S, Paern J, Lopez R. A new bioinformatics analysis tools framework at EMBL-EBI. Nucleic Acids Res. 2010;38:W695–9. doi: 10.1093/nar/gkq313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobert O. PCR fusion-based approach to create reporter gene constructs for expression analysis in transgenic [i]C. elegans[/i]. Biotechniques. 2002;32:728–30. doi: 10.2144/02324bm01. [DOI] [PubMed] [Google Scholar]
- Hodgkin J, Horvitz HR, Brenner S. Nondisjunction Mutants of the Nematode [i]Caenorhabditis elegans[/i]. Genetics. 1979;91:67–94. doi: 10.1093/genetics/91.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann K, Stoffel W. TMbase - A database of membrane spanning proteins segments. Biol. Chem. Hoppe-Seyler. 1993:374. [Google Scholar]
- Hoglund PJ, Adzic D, Scicluna SJ, Lindblom J, Fredriksson R. The repertoire of solute carriers of family 6: identification of new human and rodent genes. Biochem Biophys Res Commun. 2005;336:175–89. doi: 10.1016/j.bbrc.2005.08.048. [DOI] [PubMed] [Google Scholar]
- Italiano JE, Jr., Stewart M, Roberts TM. Localized depolymerization of the major sperm protein cytoskeleton correlates with the forward movement of the cell body in the amoeboid movement of nematode sperm. J Cell Biol. 1999;146:1087–96. doi: 10.1083/jcb.146.5.1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamath RS, Fraser AG, Dong Y, Poulin G, Durbin R, Gotta M, Kanapin A, Le Bot N, Moreno S, Sohrmann M, Welchman DP, Zipperlen P, Ahringer J. Systematic functional analysis of the [i]Caenorhabditis elegans[/i] genome using RNAi. Nature. 2003;421:231–7. doi: 10.1038/nature01278. [DOI] [PubMed] [Google Scholar]
- King KL, Stewart M, Roberts TM. Supramolecular assemblies of the [i]Ascaris suum[/i] major sperm protein (MSP) associated with amoeboid cell motility. J Cell Sci. 1994;107(Pt 10):2941–9. doi: 10.1242/jcs.107.10.2941. [DOI] [PubMed] [Google Scholar]
- Kristensen AS, Andersen J, Jorgensen TN, Sorensen L, Eriksen J, Loland CJ, Stromgaard K, Gether U. SLC6 neurotransmitter transporters: structure, function, and regulation. Pharmacol Rev. 2011;63:585–640. doi: 10.1124/pr.108.000869. [DOI] [PubMed] [Google Scholar]
- Kumsta C, Hansen M. [i]C. elegans rrf-1[/i] mutations maintain RNAi efficiency in the soma in addition to the germline. PLoS One. 2012;7:e35428. doi: 10.1371/journal.pone.0035428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- L'Hernault SW. Spermatogenesis, WormBook. The [i]C. elegans[/i] Research Community, WormBook. 2006 doi: 10.1895/wormbook.1.85.1. doi/10.1895/wormbook.1.85.1, http://www.wormbook.org. [DOI] [PMC free article] [PubMed]
- L'Hernault SW, Benian GM, Emmons RB. Genetic and molecular characterization of the [i]Caenorhabditis elegans[/i] spermatogenesis-defective gene [i]spe-17[/i]. Genetics. 1993;134:769–80. doi: 10.1093/genetics/134.3.769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- L'Hernault SW, Shakes DC, Ward S. Developmental genetics of chromosome I spermatogenesis-defective mutants in the nematode [i]Caenorhabditis elegans[/i]. Genetics. 1988;120:435–52. doi: 10.1093/genetics/120.2.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaMunyon CW, Ward S. Assessing the viability of mutant and manipulated sperm by artificial insemination of [i]Caenorhabditis elegans[/i]. Genetics. 1994;138:689–92. doi: 10.1093/genetics/138.3.689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaMunyon CW, Ward S. Sperm precedence in a hermaphroditic nematode ([i]Caenorhabditis elegans[/i]) is due to competitive superiority of male sperm. Experientia. 1995;51:817–23. doi: 10.1007/BF01922436. [DOI] [PubMed] [Google Scholar]
- Lek A, Evesson FJ, Sutton RB, North KN, Cooper ST. Ferlins: regulators of vesicle fusion for auditory neurotransmission, receptor trafficking and membrane repair. Traffic. 2011;13:185–94. doi: 10.1111/j.1600-0854.2011.01267.x. [DOI] [PubMed] [Google Scholar]
- Liau WS, Nasri U, Elmatari D, Rothman J, LaMunyon CW. Premature sperm activation and defective spermatogenesis caused by loss of [i]spe-46[/i] function in [i]Caenorhabditis elegans[/i]. PLoS One. 2013;8:e57266. doi: 10.1371/journal.pone.0057266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lishko PV, Kirichok Y, Ren D, Navarro B, Chung JJ, Clapham DE. The control of male fertility by spermatozoan ion channels. Annu Rev Physiol. 2011;74:453–75. doi: 10.1146/annurev-physiol-020911-153258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Chen L, Shang Y, Huang P, Miao L. The micronutrient element zinc modulates sperm activation through the SPE-8 pathway in [i]Caenorhabditis elegans[/i]. Development. 2013;140:2103–7. doi: 10.1242/dev.091025. [DOI] [PubMed] [Google Scholar]
- Loland CJ, Granas C, Javitch JA, Gether U. Identification of intracellular residues in the dopamine transporter critical for regulation of transporter conformation and cocaine binding. J Biol Chem. 2004;279:3228–38. doi: 10.1074/jbc.M304755200. [DOI] [PubMed] [Google Scholar]
- Loland CJ, Norregaard L, Litman T, Gether U. Generation of an activating Zn(2+) switch in the dopamine transporter: mutation of an intracellular tyrosine constitutively alters the conformational equilibrium of the transport cycle. Proc Natl Acad Sci U S A. 2002;99:1683–8. doi: 10.1073/pnas.032386299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machaca K, DeFelice LJ, L'Hernault SW. A novel chloride channel localizes to [i]Caenorhabditis elegans[/i] spermatids and chloride channel blockers induce spermatid differentiation. Dev Biol. 1996;176:1–16. doi: 10.1006/dbio.1996.9999. [DOI] [PubMed] [Google Scholar]
- Mello CC, Kramer JM, Stinchcomb D, Ambros V. Efficient gene transfer in [i]C. elegans[/i]: extrachromosomal maintenance and integration of transforming sequences. EMBO J. 1991;10:3959–70. doi: 10.1002/j.1460-2075.1991.tb04966.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merritt C, Rasoloson D, Ko D, Seydoux G. 3′ UTRs are the primary regulators of gene expression in the [i]C. elegans[/i] germline. Curr Biol. 2008;18:1476–82. doi: 10.1016/j.cub.2008.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhlrad PJ, Ward S. Spermiogenesis initiation in [i]Caenorhabditis elegans[/i] involves a casein kinase 1 encoded by the [i]spe-6[/i] gene. Genetics. 2002;161:143–55. doi: 10.1093/genetics/161.1.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson GA, Ward S. Vesicle fusion, pseudopod extension and amoeboid motility are induced in nematode spermatids by the ionophore monensin. Cell. 1980;19:457–64. doi: 10.1016/0092-8674(80)90520-6. [DOI] [PubMed] [Google Scholar]
- Okamoto H, Thomson JN. Monoclonal antibodies which distinguish certain classes of neuronal and supporting cells in the nervous tissue of the nematode [i]Caenorhabditis elegans[/i]. J Neurosci. 1985;5:643–53. doi: 10.1523/JNEUROSCI.05-03-00643.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pantanowitz S, Bendahan A, Kanner BI. Only one of the charged amino acids located in the transmembrane alpha-helices of the gamma-aminobutyric acid transporter (subtype A) is essential for its activity. J Biol Chem. 1993;268:3222–5. [PubMed] [Google Scholar]
- Parra LA, Baust T, El Mestikawy S, Quiroz M, Hoffman B, Haflett JM, Yao JK, Torres GE. The orphan transporter Rxt1/NTT4 (SLC6A17) functions as a synaptic vesicle amino acid transporter selective for proline, glycine, leucine, and alanine. Mol Pharmacol. 2008;74:1521–32. doi: 10.1124/mol.108.050005. [DOI] [PubMed] [Google Scholar]
- Pramod AB, Foster J, Carvelli L, Henry LK. SLC6 transporters: structure, function, regulation, disease association and therapeutics. Mol Aspects Med. 2013;34:197–219. doi: 10.1016/j.mam.2012.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawls AS, Schultz SA, Mitra RD, Wolff T. Bedraggled, a putative transporter, influences the tissue polarity complex during the R3/R4 fate decision in the [i]Drosophila[/i] eye. Genetics. 2007;177:313–28. doi: 10.1534/genetics.107.075945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinke V, Gil IS, Ward S, Kazmer K. Genome-wide germline-enriched and sex-biased expression profiles in [i]Caenorhabditis elegans[/i]. Development. 2004;131:311–23. doi: 10.1242/dev.00914. [DOI] [PubMed] [Google Scholar]
- Reinke V, Smith HE, Nance J, Wang J, Van Doren C, Begley R, Jones SJ, Davis EB, Scherer S, Ward S, Kim SK. A global profile of germline gene expression in [i]C. elegans[/i]. Mol Cell. 2000;6:605–16. doi: 10.1016/s1097-2765(00)00059-9. [DOI] [PubMed] [Google Scholar]
- Renick SE, Kleven DT, Chan J, Stenius K, Milner TA, Pickel VM, Fremeau RT., Jr. The mammalian brain high-affinity L-proline transporter is enriched preferentially in synaptic vesicles in a subpopulation of excitatory nerve terminals in rat forebrain. J Neurosci. 1999;19:21–33. doi: 10.1523/JNEUROSCI.19-01-00021.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santella L, Vasilev F, Chun JT. Fertilization in echinoderms. Biochem Biophys Res Commun. 2012;425:588–94. doi: 10.1016/j.bbrc.2012.07.159. [DOI] [PubMed] [Google Scholar]
- Santi CM, Orta G, Salkoff L, Visconti PE, Darszon A, Trevino CL. K+ and Cl− channels and transporters in sperm function. Curr Top Dev Biol. 2013;102:385–421. doi: 10.1016/B978-0-12-416024-8.00014-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schedl T, Kimble J. [i]fog-2[/i], a germ-line-specific sex determination gene required for hermaphrodite spermatogenesis in [i]Caenorhabditis elegans[/i]. Genetics. 1988;119:43–61. doi: 10.1093/genetics/119.1.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seidel HS, Ailion M, Li J, van Oudenaarden A, Rockman MV, Kruglyak L. A novel sperm-delivered toxin causes late-stage embryo lethality and transmission ratio distortion in [i]C. elegans[/i]. PLoS Biol. 2011;9:e1001115. doi: 10.1371/journal.pbio.1001115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shakes DC, Ward S. Initiation of spermiogenesis in [i]C. elegans[/i]: a pharmacological and genetic analysis. Dev Biol. 1989;134:189–200. doi: 10.1016/0012-1606(89)90088-2. [DOI] [PubMed] [Google Scholar]
- Shukla KK, Mahdi AA, Rajender S. Ion channels in sperm physiology and male fertility and infertility. J Androl. 2012;33:777–88. doi: 10.2164/jandrol.111.015552. [DOI] [PubMed] [Google Scholar]
- Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Soding J, Thompson JD, Higgins DG. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol. 2011;7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sijen T, Fleenor J, Simmer F, Thijssen KL, Parrish S, Timmons L, Plasterk RH, Fire A. On the role of RNA amplification in dsRNA-triggered gene silencing. Cell. 2001;107:465–76. doi: 10.1016/s0092-8674(01)00576-1. [DOI] [PubMed] [Google Scholar]
- Singaravelu G, Chatterjee I, Rahimi S, Druzhinina MK, Kang L, Xu XZ, Singson A. The sperm surface localization of the TRP-3/SPE-41 Ca2+ - permeable channel depends on SPE-38 function in [i]Caenorhabditis elegans[/i]. Dev Biol. 2012;365:376–83. doi: 10.1016/j.ydbio.2012.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JR, Stanfield GM. TRY-5 is a sperm-activating protease in [i]Caenorhabditis elegans[/i] seminal fluid. PLoS Genet. 2011;7:e1002375. doi: 10.1371/journal.pgen.1002375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanfield GM, Villeneuve AM. Regulation of sperm activation by SWM-1 is required for reproductive success of [i]C. elegans[/i] males. Curr Biol. 2006;16:252–63. doi: 10.1016/j.cub.2005.12.041. [DOI] [PubMed] [Google Scholar]
- Suarez SS. Regulation of sperm storage and movement in the mammalian oviduct. Int J Dev Biol. 2008;52:455–62. doi: 10.1387/ijdb.072527ss. [DOI] [PubMed] [Google Scholar]
- TMPred. http://www.ch.embnet.org/software/TMPRED_form.html, date May 12, 2014.
- Vangelatos I, Vlachakis D, Sophianopoulou V, Diallinas G. Modelling and mutational evidence identify the substrate binding site and functional elements in APC amino acid transporters. Mol Membr Biol. 2009;26:356–70. doi: 10.1080/09687680903170546. [DOI] [PubMed] [Google Scholar]
- Ward S, Carrel JS. Fertilization and sperm competition in the nematode [i]Caenorhabditis elegans[/i]. Dev Biol. 1979;73:304–21. doi: 10.1016/0012-1606(79)90069-1. [DOI] [PubMed] [Google Scholar]
- Ward S, Hogan E, Nelson GA. The initiation of spermiogenesis in the nematode [i]Caenorhabditis elegans[/i]. Dev Biol. 1983;98:70–9. doi: 10.1016/0012-1606(83)90336-6. [DOI] [PubMed] [Google Scholar]
- Ward S, Miwa J. Characterization of temperature-sensitive, fertilization-defective mutants of the nematode [i]Caenorhabditis elegans[/i]. Genetics. 1978;88:285–303. doi: 10.1093/genetics/88.2.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Washington NL, Ward S. FER-1 regulates Ca2+ -mediated membrane fusion during [i]C. elegans[/i] spermatogenesis. J Cell Sci. 2006;119:2552–62. doi: 10.1242/jcs.02980. [DOI] [PubMed] [Google Scholar]
- WormBase. http://www.wormbase.org, release WS240, date January 20, 2014.
- Xu XZ, Sternberg PW. A [i]C. elegans[/i] sperm TRP protein required for sperm-egg interactions during fertilization. Cell. 2003;114:285–97. doi: 10.1016/s0092-8674(03)00565-8. [DOI] [PubMed] [Google Scholar]
- Yamashita A, Singh SK, Kawate T, Jin Y, Gouaux E. Crystal structure of a bacterial homologue of Na+/Cl−-dependent neurotransmitter transporters. Nature. 2005;437:215–23. doi: 10.1038/nature03978. [DOI] [PubMed] [Google Scholar]
- Yochem J, Gu T, Han M. A new marker for mosaic analysis in [i]Caenorhabditis elegans[/i] indicates a fusion between hyp6 and hyp7, two major components of the hypodermis. Genetics. 1998;149:1323–34. doi: 10.1093/genetics/149.3.1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zannoni S, L'Hernault SW, Singson AW. Dynamic localization of SPE-9 in sperm: a protein required for sperm-oocyte interactions in [i]Caenorhabditis elegans[/i]. BMC Dev Biol. 2003;3:10. doi: 10.1186/1471-213X-3-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.