Abstract
Mutations of the isocitrate dehydrogenase 1 (IDH1) gene are among the most prevalent in low-grade glioma and secondary glioblastoma, represent an early pathogenic event, and are associated with epigenetically-driven modulations of metabolism. Of particular interest is the recently uncovered relationship between the IDH1 mutation and decreased activity of the branched-chain amino acid transaminase 1 (BCAT1) enzyme. Non-invasive imaging methods that can assess BCAT1 activity could therefore improve detection of mutant IDH1 tumors and aid in developing and monitoring new targeted therapies. BCAT1 catalyzes the transamination of branched-chain amino acids while converting α-ketoglutarate (α-KG) to glutamate. Our goal was to use 13C magnetic resonance spectroscopy to probe the conversion of hyperpolarized [1-13C] α-KG to hyperpolarized [1-13C] glutamate as a readout of BCAT1 activity. We investigated two isogenic glioblastoma lines that differed only in their IDH1 status, and performed experiments in live cells and in vivo in rat orthotopic tumors. Following injection of hyperpolarized [1-13C] α-KG, hyperpolarized [1-13C] glutamate production was detected both in cells and in vivo, and the level of hyperpolarized [1-13C] glutamate was significantly lower in mutant IDH1 cells and tumors compared to their IDH1-wild-type counterparts. Importantly however, in our cells the observed drop in hyperpolarized [1-13C] glutamate was likely mediated not only by a drop in BCAT1 activity, but also by reductions in aspartate transaminase and glutamate dehydrogenase activities, suggesting additional metabolic reprogramming at least in our model. Hyperpolarized [1-13C] glutamate could thus inform on multiple mutant IDH1-associated metabolic events that mediate reduced glutamate production.
Keywords: glioma, metabolism, IDH1 mutation, MR spectroscopy, hyperpolarized 13C
INTRODUCTION
Heterozygous mutations in the gene coding for the isocitrate dehydrogenase 1 (IDH1) enzyme have been reported in several cancer types, most notably in over 70% of diffuse low-grade gliomas and 80% of secondary glioblastomas (GBM) (1, 2). The resulting mutant IDH1 enzyme acquires the neomorphic activity of catalyzing the reduction of α-ketoglutarate (α-KG) to the oncometabolite 2-hydroxyglutarate (2-HG), leading to significantly elevated 2-HG levels in tumor tissue (3). The specific mechanisms through which IDH1 mutations and elevated 2-HG drive tumor development are still being uncovered. Nonetheless, it is clear that 2-HG inhibits a range of α-KG-dependent dioxygenases, resulting in a broad range of alterations in the cellular epigenome (4, 5), which ultimately lead to oncogenesis (6–11).
Amongst other effects, presence of the IDH1 mutation is associated with a range of alterations in the cellular metabolome. These have been reported in mutant IDH1-expressing cell models, ex vivo tumor samples, and in vivo patient studies, and include significant changes in choline-containing metabolites, N-acetyl aspartate, and glutamate (12–15). At the enzymatic level, a recent study showed that expression of the pyruvate carboxylase (PC) enzyme was significantly increased in mutant IDH1 cells and patient samples, suggesting that PC flux could serve as a source of TCA anaplerosis in mutant IDH1 cells that channel glutamine to 2-HG production (16). A separate study recently reported that several glycolytic enzymes were underexpressed in mutant IDH1 glioma patient samples, likely due to hypermethylation of their promoter regions. Most notably, the expression of lactate dehydrogenase A was silenced in mutant IDH1 tumors (7). Another enzyme that was recently reported as modulated in mutant IDH1 cells is branched-chain amino acid transaminase 1 (BCAT1) (17). BCAT1 is a cytosolic enzyme that catalyzes the catabolism of branched-chain L-amino acids (BCAAs) to branched chain α-keto acids (BCKAs), while concomitantly converting α-KG to glutamate. The expression of BCAT1 was significantly reduced in mutant IDH1 glioma cells compared to their wild-type counterparts, and this effect was associated with epigenetic silencing likely driven by the IDH1 mutation (17). Additionally, studies show that BCAT1 could serve as a novel therapeutic target for glioma (18). Innovative methods for non-invasive assessment of BCAT1 activity could therefore help refine the diagnosis and monitoring of tumors harboring the IDH1 mutation, and aid in the development and monitoring of BCAT1-targeting therapies (17, 18).
1H Magnetic Resonance Spectroscopy (MRS) is a non-invasive method that can probe the steady-state levels of several endogenous cellular metabolites (19). It has been widely used in the clinical setting as a diagnostic and prognostic tool for brain tumor patients (19, 20). More recently, a complementary metabolic neuroimaging approach, hyperpolarized 13C MRS, has been successfully developed and implemented. Through the use of Dynamic Nuclear Polarization (DNP), 13C-labeled compounds can be hyperpolarized, resulting in a 10,000 to 50,000-fold increase in their MR-detectable signal-to-noise ratio (SNR) when compared to thermally polarized compounds (21). Accordingly, hyperpolarized 13C MRS provides a non-invasive method to dynamically image metabolic fluxes. Over the past decade, this method has proven extremely useful in the field of oncology to monitor tumor metabolism, in the absence of ionizing radiation and with convenient integration to standard MR imaging techniques (22). In particular, [1-13C] pyruvate, the most commonly utilized hyperpolarized probe, has been widely used to detect the presence of tumor and response to treatment in several preclinical models of cancer (23, 24), including glioma (25–28). Furthermore, the first clinical trial of this technique was recently completed on prostate cancer patients, demonstrating the translational value of the hyperpolarized imaging approach (29).
In the context of the IDH1 mutation, both 1H and hyperpolarized 13C MRS have proven useful. 1H MRS methods have been used to monitor the presence of 2-HG in glioma patients in vivo (15, 30, 31), in preclinical rodent models of GBM (32), and in patient biopsy samples (12, 33). Additionally, we recently developed [1-13C] α-KG as a new hyperpolarized probe, and were able to detect the conversion of hyperpolarized [1-13C] α-KG to hyperpolarized [1-13C] 2-HG in real-time in mutant IDH1 cells and orthotopic tumors using 13C MRS (34).
Considering that BCAT1 requires α-KG as a substrate to generate glutamate while transaminating BCAAs to BCKAs, we sought to expand on the use of hyperpolarized [1-13C] α-KG as an imaging probe, and investigated its conversion to hyperpolarized [1-13C] glutamate as a method for monitoring BCAT1 activity. We studied two isogenic cell lines that differ only in their IDH1 status, and show that following injection of hyperpolarized [1-13C] α-KG, the production of hyperpolarized [1-13C] glutamate can be detected, and is reduced in cells and tumors that express the IDH1 mutation. However, in our model, the presence of the IDH1 mutation led not only to a drop in BCAT1 activity, but also to a drop in the activities of two additional enzymes that can catalyze the conversion of α-KG to glutamate, namely aspartate transaminase (AST) and glutamate dehydrogenase (GDH). Our metabolic imaging therefore informs not only on BCAT1 status but also on a broader metabolic reprogramming associated with the IDH1 mutation. As such, hyperpolarized glutamate could serve as an indirect imaging biomarker of IDH1 status that is complementary to 2-HG.
MATERIAL and METHODS
Cell models and culture
U87 GBM cells were virally transduced with the wild-type IDH1 gene or the mutant IDH1 gene to produce U87IDHwt and U87IDHmut cells as previously described (34). Unique DNA “fingerprint” identities (i.e., variable number tandem repeat PCR products) established for these cell lines, were used to regularly confirm their identities. Both strains were cultured under standard conditions in high glucose Dulbecco’s Modified Eagle Medium (DMEM H-21, UCSF Cell Culture Facility, CA) supplemented with 10% heat-inactivated fetal bovine serum (Thermo Scientific Hyclone, Logan, UT), 2mM L-Glutamine (Invitrogen, Carlsbad, CA), 100u.mL−1 penicillin, and 100mg.mL−1 streptomycin (UCSF Cell Culture Facility, CA). Both cell lines were maintained as exponentially growing monolayers at 37°C in a humidified atmosphere of 95% air and 5% CO2.
Spectrophotometric enzyme assays
All spectrophotometric measurements were performed on an Infinite m200 spectrophotometer (Tecan Systems, Inc, San Jose, CA) and experiments performed in quadruplicate for each cell line.
The BCAT1 activity assay was adapted from methods developed by Cooper et al. (35). Briefly, 1×106 U87IDHwt and U87IDHmut cells were lysed using lysis buffer (Cell Signaling Technology, Danvers, MA) in the presence of protease inhibitor (Calbiochem, Merck KGaA, Darmstadt, Germany). A reaction mix was prepared using 10mM L-leucine, 5mM α-KG, 5mM ammonium sulfate, 50μM NADH, 0.5mM GTP, and 1.9units of leucine dehydrogenase in 100mM phosphate buffer (in dH2O; pH=7.4; all reagents from Sigma Aldrich, St Louis, MO). 200μL of reaction mix and 20μL of cell lysate were placed in each well of a 96-well plate. The decrease in absorbance at λ=340nm was measured every 30 seconds over 10 minutes. BCAT1 reaction rate RBCAT1 was calculated for each cell line and expressed in fmol of NADH/cell/min.
Glutamate dehydrogenase (GDH) and aspartate transaminase (AST) activities were measured using assay kits (BioVision, Inc., CA). Note that, for both enzymes, the assays reflect the combined activities of cytoplasmic and mitochondrial isoforms. For both assays, 1×106 cells were lysed in 200μL of kit buffer. After a 1:50 dilution, 50μL of sample was added to each well and the absorbance at λ=450nm was measured every 10 seconds over 70 minutes. The reaction rate of GDH RGDH was expressed in fmol of NADH/cell/min and the reaction rate of AST RAST in fmol of glutamate/cell/min.
Alanine aminotransferase (ALT) activity was measured using an endpoint assay kit (Bio Scientific pty. ltd., NZ). Briefly, 1×106 cells were lysed in 400μL of kit buffer and lysates centrifuged at 14,000rpm for 30 minutes at 4°C. 10μL of lysate was placed in each well and kit reagents were added. Absorbance at λ=510nm was monitored every 10 seconds for 30 minutes. The reaction rate of ALT RALT was calculated by linear regression and reported in fmol of pyruvate/cell/min.
Western blotting analysis
For each cell line, denatured proteins were electrophoresed on 4–15% Biorad Ready gels (Life Science Research, Hercules, CA) using the SDS-PAGE method and electrotransferred onto Polyvinylidene fluoride membranes. Blots were blocked and incubated with the primary antibodies anti-BCAT1 (1:1000 dilution, Cell Signaling Technology, Danvers, MA), anti-AST1 (cytosolic, 1:100 dilution, Abcam®, Cambridge, MA), anti-AST2 (mitochondrial, 1:500 dilution, Abcam®), anti-GDH1 (cytosolic, 1:1000 dilution, Novus USA, Littleton, CO), anti-GDH2 (mitochondrial, 1:500 dilution, Abcam®), anti-ALT1 (cytosolic, 1:500 dilution, Abcam®) and anti-ALT2 (mitochondrial, 1:100 dilution, Abcam®) overnight in TBS-t with 5% BSA. Blots were then incubated with secondary antibodies goat anti-rabbit horseradish peroxidase–linked (Cell Signaling Technology, Danvers, MA) or goat anti-mouse horseradish peroxidase–linked (Santa Cruz Biotechnology Inc., Dallas, TX) for 1 hour in TBS-t with 5% milk. The immunocomplexes were visualized using ECL Western Blotting Substrate (ThermoFisher Scientific, Waltham, MA). Bands were quantified using the ImageJ software (NIH) and normalized to beta-actin.
qRT-PCR analysis
mRNA was isolated from 5×106 U87IDHwt and U87IDHmut cells using RNeasy kit (Qiagen Inc., Valencia, CA). Quantitative real-time polymerase chain reaction (qRT-PCR) was performed by the Genome Analysis Core Facility at UCSF (16). Briefly, cDNA was generated from RNA using the qScript cDNA Synthesis Kit (Quanta Biosciences, Gaithersburg, MD) on a PTC-225 Thermocycler (MJ Research, St Bruno, QC, Canada). qRT-PCR was conducted in a 384-well plate with 20μL 1X Taqman buffer, 5.5mM MgCl2, 200nM of the corresponding Taqman probe, 0.2μM of each deoxynucleotide triphosphate, 0.025unit/μL AmpliTaq Gold, 5ng cDNA, and 0.5mM of each of the following TaqMan® primers (all reagents from Life Technologies, Grand Island, NY) for the following genes (n=9 per gene): BCAT1 coding for the cytosolic BCAT1 isoform; GOT1 and GOT2 (Glutamic-Oxaloacetic Transaminase 1 and 2) coding for the cytosolic and mitochondrial AST1 and AST2 isoforms, respectively; GLUD1 and GLUD2 (glutamate dehydrogenase 1 and 2) coding for the cytosolic and mitochondrial GDH1 and GDH2 isoforms, respectively; GPT and GPT2 (Glutamic-Pyruvate Transaminase 1 and 2) coding for the cytosolic and mitochondrial ALT1 and ALT2 isoforms, respectively. The ABI 7900HT instrument (Life Technologies, Grand Island, NY) was used with 1 cycle of 95°C for 10 minutes and 40 cycles of 95°C for 15 seconds. Analysis was carried out using the SDS software to determine Ct. Expression levels were normalized to the glyceraldehyde-3-phosphate dehydrogenase (GAPDH) transcript.
Perfusion system
For live cells studies, U87IDHwt and U87IDHmut cells were seeded on Biosiln® Microcarrier beads (NUNC, Rochester, NY). After 48 hours, ~3.5×107 cells on beads were loaded into a 10-mm NMR tube connected to a perfusion system which maintained an atmosphere of 5% CO2/95% air and circulated growth medium through the cells (flow rate 1.5mL.min−1). (25, 28, 36). A port on the inflow line allowed for injection of hyperpolarized material, during which time the perfusion was briefly stopped. All experiments were performed at 37°C.
Hyperpolarized 13C MR studies of live cells
A volume of 30μL of [1-13C] α-KG solution (5.9M, 3:1 water:glycerol, 17.3mM OX63 radical, 0.4mM Dotarem) was polarized using a hypersense polarizer (Oxford Instruments) for approximately 1 hour (34). Hyperpolarized [1-13C] α-KG was then rapidly dissolved in isotonic buffer (40mM Tris, 30mM NaOH, 3.0μmol/L Na2EDTA) and injected into the perfusion medium of U87IDHwt cells (n=5) and U87IDHmut cells (n=5) to a final concentration of 15mM. Dynamic sets of hyperpolarized 13C spectra were acquired on a 11.7 Tesla INOVA spectrometer (Agilent Technologies, Inc., Palo Alto, CA) starting at the beginning of the injection using a pulse-acquire sequence (13 degrees flip angle (FA), 3 second repetition time (TR), acquisition time 300 seconds)(25).
MR data analysis of perfused cells experiments
All spectral assignments were based on literature reports (e.g. www.hmdb.ca). The integrals of hyperpolarized [1-13C] α-KG and hyperpolarized [1-13C] glutamate were quantified by peak integration of the dynamic 13C spectra using ACD/Spec Manager 9. The integral of hyperpolarized [1-13C] glutamate was normalized to noise and to cell number.
Normalized glutamate kinetics were further analyzed using a gamma-variate analysis (37) to derive the following parameters: R-value, area under the curve (AUC, expressed in a.u.), time of maximum peak height (Peak Time, expressed in seconds), maximum peak height (Peak Height, expressed in a.u.), and full width at half maximum (FMHW, expressed in seconds).
Tumor-bearing animals
All animal research was approved by the Institutional Animal Care and Use Committee of the University of California, San Francisco. Athymic rats (n=2 U87IDHmut, n=2 U87IDHwt, average weight 200g, male, rnu/rnu homozygous, 5–6-week-old; Harlan Laboratories, Indianapolis, IN) were used in this study. An hour before the intracranial injection, U87IDHwt or U87IDHmut cells were washed once with PBS solution, collected by trypsinization, counted and resuspended in serum-free McCoy’s medium to a concentration of 3×105 cells per 10μl. Rats were then anaesthetized by an intraperitoneal injection of a mixture of ketamine/xylazine (100/20mg.kg−1, respectively), and 10μl of cell suspension was slowly injected into the right putamen of the animal brain using the freehand technique (34). Buprenorphine (0.03mg kg−1, V=600μl) and bupivacaine (5mg.kg−1, V=300μl) were injected subcutaneously before injection for optimal pain management.
In vivo hyperpolarized 13C MR studies
In vivo experiments were performed on a 3 Tesla clinical MR system (GE Healthcare Little Chalfont, United Kingdom) equipped with a dual-tuned 1H–13C transmit/receive volume coil (ØI=40 mm). Rats were anesthetized using isoflurane (1–2% in O2, 1.5L.min−1) and a 23 gauge catheter was secured in the tail vein. For all in vivo studies, B0 shimming was performed using the automatic GE shimming routine. Anatomical imaging was performed using a 2D fast spin echo sequence (axial/coronal, echo time (TE) =20ms, TR=1200ms, field of view=40×40mm2, matrix 256×256, 20 slices, thickness=1 mm, acquisition time 5 minutes 7 seconds, number of transients (NT)=1). After 1 hour, hyperpolarized [1-13C] α-KG was rapidly dissolved in isotonic buffer (40mM Tris, 200mM NaOH, 0.1 mg.L−1 Na2EDTA) to obtain a 100mM solution. Within 15 seconds post dissolution, a volume of 2ml of hyperpolarized solution was injected over 12 seconds.
13C 2D dynamic CSI was acquired starting 5 seconds after the beginning of hyperpolarized [1-13C] α-KG injection. The sequence was optimized for glutamate detection using a specialized variable flip-angle (VFA) multi-band spectral-spatial RF excitation pulse designed to provide an initial 4-degree FA for [1-13C] α-KG (to preserve substrate magnetization) and 25-degree for [1-13C] glutamate. These FAs were progressively increased over time to efficiently use the hyperpolarized magnetization in the presence of metabolic conversion, with both VFA schemes ramping up to 90-degree FA (38). An echo-planar spectroscopic imaging read-out gradient was used for increased imaging speed. Additional parameters were: adiabatic double spin-echo acquisition, bandwidth 543Hz, resolution 10.4Hz, 52points, TE/TR=140/215ms, matrix 8×18, 5×5mm2 resolution, slice thickness 2cm, 1.7 seconds per image, images every 5 seconds) (34, 39, 40).
MR data analysis of in vivo experiments
MR datasets were analyzed using the in-house SIVIC software (http://sourceforge.net/apps/trac/sivic/). T2-weighted anatomical images were superimposed to the 2D CSI grid to derive the location of the tumor and normal brain voxels (34). The integral values of the hyperpolarized [1-13C] α-KG and hyperpolarized [1-13C] glutamate peaks were calculated for each voxel type and for each tumor type. The ratios of hyperpolarized [1-13C] glutamate to hyperpolarized [1-13C] α-KG were calculated for normal brain voxels and tumor voxels as the ratio of the integral values. Color heatmaps of hyperpolarized [1-13C] α-KG and hyperpolarized [1-13C] glutamate were generated for the 25 seconds time point using a sinc-based interpolation of the 13C 2D CSI data to the resolution of the anatomical images using SIVIC.
Statistical analysis
All results are expressed as mean ± standard deviation. Two-tailed Student t-test was used to determine the statistical significance of the results, with a p-value below 0.05 considered significant.
RESULTS
BCAT1 activity and expression are significantly decreased in U87 mutant IDH1 cells
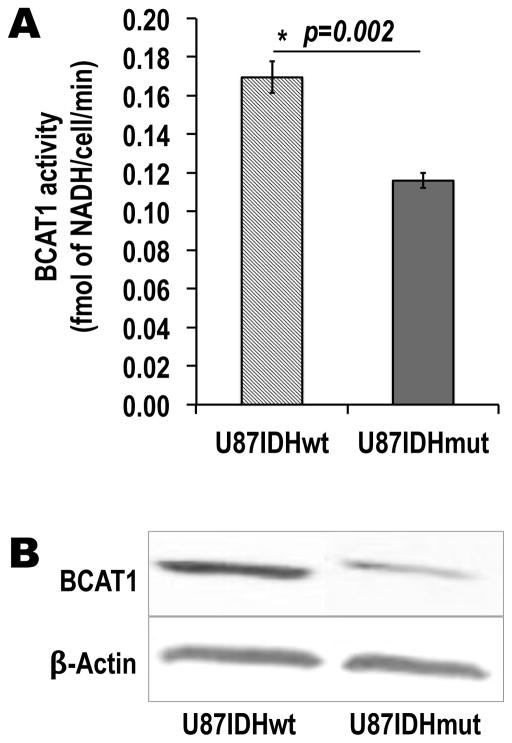
The goal of this study was to develop a method to non-invasively assess BCAT1 activity as an indirect readout of IDH1 mutational status. First, we therefore had to verify that, in our model, the presence of mutant IDH1 was associated with a drop in BCAT1 activity and expression, as described in other systems (17). As presented in Figure 1A and Table 1, our results show that BCAT1 activity was significantly decreased by 32±7% in U87IDHmut cells as compared to U87IDHwt: RBCAT1=0.17±0.01 fmol of NADH/cell/min for U87IDHwt cells versus RBCAT1=0.12±0.004 fmol of NADH/cell/min for U87IDHmut cells (p=0.002, n=4 per cell line). BCAT1 western blots are shown in Figure 1B. Consistent with the activity assay, BCAT1 protein levels were significantly lower in U87IDHmut cells, down to 38±12% of U87IDHwt (p=0.01; n=3 per cell line; Table 1). qRT-PCR analysis confirmed this finding: mRNA levels were also significantly lower in U87IDHmut cells, down to 48±19% of U87IDHwt (p=0.01; n=9 per cell line; Table 1) and within experimental error of the observed drop in protein levels.
Figure 1. IDH1 mutation is associated with a drop in BCAT1 activity and expression in our cell model.
(A) BCAT1 enzymatic activity, as measured by a spectrophotometric assay, is significantly decreased in mutant IDH1 cells as compared to wild-type cells (*p<0.002; n=4 per cell line). (B) Western blots of the BCAT1 enzyme, showing the drop in BCAT1 protein levels in U87IDHmut cells when compared to U87IDHwt.
Table 1. Enzymatic activities, protein levels and mRNA levels for the four enzymes catalyzing the α-KG to glutamate conversion.
Enzymatic activities, protein levels and mRNA levels were measured by spectrophotometric assays, western blot and qRT-PCR analysis, respectively (first row: enzyme; second row: corresponding gene).
| Enzyme | BCAT1 | AST1 | AST2 | GDH1 | GDH2 | ALT1 | ALT2 |
|---|---|---|---|---|---|---|---|
| Gene | BCAT1 | GOT1 | GOT2 | GLUD1 | GLUD2 | GPT | GPT2 |
| Activity levels (% of U87IDHwt) | 68 ± 7** | 83 ± 10* | 69 ± 15* | b.d. | |||
|
| |||||||
| Protein levels (% of U87IDHwt) | 38 ± 12** | 55 ± 11* | 112 ± 30 | b.d. | 131 ± 33 | 103 ± 18 | 131 ± 32 |
|
| |||||||
| mRNA levels (% of U87IDHwt) | 48 ± 19** | 73 ± 9** | 79 ± 5** | 91 ± 18 | b.d. | b.d. | 132 ± 30 |
All values are for U87IDHmut cells and expressed in percent of U87IDHwt levels (b.d.= below detection level; *p<0.05;**p<0.01; n=3 per protein per cell line; n=9 per gene per cell line).
Formation of hyperpolarized [1-13C] glutamate from hyperpolarized [1-13C] α-KG can be observed in live U87IDHwt perfused cells, but not in U87IDHmut cells
Before performing preclinical animal studies, we assessed the feasibility of monitoring the conversion of hyperpolarized [1-13C] α-KG to hyperpolarized [1-13C] glutamate in live perfused cells using 13C MRS.
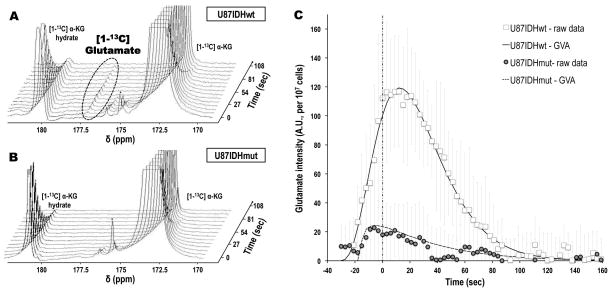
As shown in Figure 2A, injection of hyperpolarized [1-13C] α-KG (chemical shift (δ) δα-KG=172.6ppm) into the perfusion medium of U87IDHwt cells resulted in the detectable build-up of a new hyperpolarized metabolite at δ=177.5ppm – the chemical shift of [1-13C] glutamate. Furthermore, the maximum peak height of this new compound was observed at 12.2±0.1 seconds after the maximum peak height of hyperpolarized [1-13C] α-KG, in line with an expected delay when metabolism occurs (Peak Time, Table 2). Based on chemical shift value and the observed delay in metabolite build-up, we attributed this newly detected resonance to hyperpolarized [1-13C] glutamate produced from hyperpolarized [1-13C] α-KG.
Figure 2. Hyperpolarized [1-13C] glutamate formation from hyperpolarized [1-13C] α-KG can be detected in live cells, and is decreased in U87IDHmut cells as compared to U87IDHwt.
Stack plots of dynamic 13C MR spectra acquired at 11.7 Tesla following injection of hyperpolarized [1-13C] α-KG in live U87IDHwt (A) and U87IDHmut (B) perfused cells (temporal resolution 9 seconds), showing the formation of hyperpolarized [1-13C] glutamate in U87IDHwt cells. Note the absence of detectable hyperpolarized [1-13C] glutamate in U87IDHmut cells. (C) Intensities of hyperpolarized [1-13C] glutamate in U87IDHwt (□) and U87IDHmut (
 ) perfused cells, showing the significantly higher level of glutamate in U87IDHwt cells as compared to U87IDHmut. Also of importance is the delayed formation of hyperpolarized [1-13C] glutamate versus the time of maximum hyperpolarized [1-13C] α-KG (vertical dashed line), as expected when metabolism occurs. The fit derived from the gamma-variate analysis (GVA) is displayed as a continuous line for U87IDHwt and as a dashed line for U87IDHmut perfused cells.
) perfused cells, showing the significantly higher level of glutamate in U87IDHwt cells as compared to U87IDHmut. Also of importance is the delayed formation of hyperpolarized [1-13C] glutamate versus the time of maximum hyperpolarized [1-13C] α-KG (vertical dashed line), as expected when metabolism occurs. The fit derived from the gamma-variate analysis (GVA) is displayed as a continuous line for U87IDHwt and as a dashed line for U87IDHmut perfused cells.
Table 2.
Gamma-variate analysis of the hyperpolarized [1-13C] glutamate kinetics observed in live cells.
|
|
|||||
|---|---|---|---|---|---|
| R-value | Area | Peak Time (sec) | Peak Height | FWHM (sec) | |
| U87IDHwt (n=5) | 0.98±0.11 | 2444±211 | 12.2±0.1 | 119±4 | 57±3 |
| U87IDHmut (n=5) | 0.82±0.21 | 494±8** | 0.2±1.1** | 24±0.2** | 51±3* |
All values are expressed as mean ± sd (FWHM=full width at half-maximum; *p<0.05;**p<0.01; n=5 per cell line).
In contrast to U87IDHwt cells, hyperpolarized [1-13C] glutamate was detected within 0.2±1.1 seconds after the maximum of hyperpolarized [1-13C] α-KG, and was generally barely above the noise level in U87IDHmut cells, as shown in Figure 2B. The time courses of hyperpolarized [1-13C] glutamate formation in U87IDHwt and U87IDHmut cells (normalized to noise and cell number) are plotted in Figure 2C, and show the substantial and delayed build-up of glutamate in U87IDHwt cells only. These kinetics were further analyzed using a gamma-variate analysis, as summarized in Table 2. Importantly, all estimated parameters were significantly different between U87IDHwt and U87IDHmut cells. In particular, the area under the curve and the peak height were significantly decreased by 84.1±5.1% and 80.0±5.0% in U87IDHmut cells as compared to U87IDHwt, respectively (p<0.01, n=5 per cell line), demonstrating that significantly less glutamate is produced from α-KG in mutant IDH1 cells.
Hyperpolarized [1-13C] glutamate can be detected in vivo in U87IDHwt tumors, but not in U87IDHmut tumors
Next, we translated the method developed in the perfused cell experiments, and performed an in vivo proof-of-principle study to confirm the feasibility and potential of using hyperpolarized [1-13C] α-KG to monitor hyperpolarized [1-13C] glutamate production in orthotopic brain tumors at clinical field strength (3 Tesla).
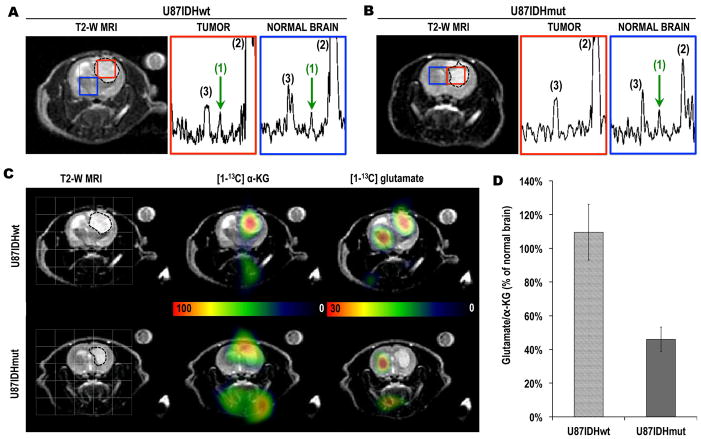
Figures 3A&B illustrate the MR images and spectra obtained from U87IDHwt and U87IDHmut tumor-bearing rats with comparable tumor sizes. The tumors appear as hypersignals on the T2-weighted images (borders as dotted lines). Following injection of hyperpolarized [1-13C] α-KG, hyperpolarized [1-13C] glutamate could be detected at the 25-second time point. Spectra acquired at this time point are illustrated in Figures 3A&B, and demonstrate that, in U87IDHwt animals, hyperpolarized [1-13C] glutamate was detected both in normal brain and in the tumor. In contrast, in U87IDHmut animals, hyperpolarized [1-13C] glutamate was detected in normal brain, but was barely detectable in the tumor. Heatmaps from the 25-second time point (Figure 3C) further illustrate high levels of hyperpolarized [1-13C] α-KG in the tumors regardless of IDH1 status, as well as the presence of hyperpolarized [1-13C] glutamate in the normal brain of all animals (n=4), and the presence of hyperpolarized [1-13C] glutamate in U87IDHwt tumors but not in U87IDHmut tumors. Quantification (Figure 3D) shows that the ratio of hyperpolarized [1-13C] glutamate to hyperpolarized [1-13C] α-KG in U87IDHwt tumors was comparable to the ratio observed in normal contralateral brain (110±17%; n=2), but was lower than normal contralateral brain in U87IDHmut tumors (46±7%; n=2).
Figure 3. Hyperpolarized [1-13C] glutamate from hyperpolarized [1-13C] α-KG can be detected in vivo in normal brain and in U87IDHwt tumors only.
T2-weighted MR images of (A) a U87IDHwt and (B) a U87IDHmut tumor-bearing animals overlaid with tumor (red) and normal brain (blue) voxels from the 2D CSI acquisition. The tumors appear as hypersignals and are circled with dotted lines. The corresponding hyperpolarized 13C MR spectra from tumor and normal brain voxels are shown for both animals. The observed resonances are: (1) hyperpolarized [1-13C] glutamate (green arrow, δGLU=177.5ppm); (2) hyperpolarized [1-13C] α-KG (δα-KG=172.6ppm); (3) hyperpolarized [1-13C] α-KG hydrate (δα-KG-Hyd=180.9ppm). Note the presence of hyperpolarized [1-13C] glutamate in normal brain for both animals, but only in the tumor tissue of the U87IDHwt animal. (C) T2-weighted MR images (first column) of the head of a U87IDHwt (top row) and a U87IDHmut (bottom row) tumor-bearing animal overlaid with the grid used for 2D 13C CSI acquisition. Corresponding heatmaps of hyperpolarized [1-13C] α-KG and hyperpolarized [1-13C] glutamate acquired 25 seconds post injection of hyperpolarized [1-13C] α-KG. These heatmaps illustrate the presence of hyperpolarized [1-13C] glutamate in normal brain and tumor in U87IDHwt animals, and in normal brain only in U87IDHmut animals. (D) Ratio of hyperpolarized [1-13C] glutamate to hyperpolarized [1-13C] α-KG (integral values) expressed as a percent of the same ratio in normal brain. The normalized glutamate/α-KG ratio is lower in U87IDHmut tumors as compared to U87IDHwt (n=2 per tumor type).
The activities and expression of other enzymes catalyzing the α-KG-to-glutamate conversion are significantly reduced in mutant IDH1 cells
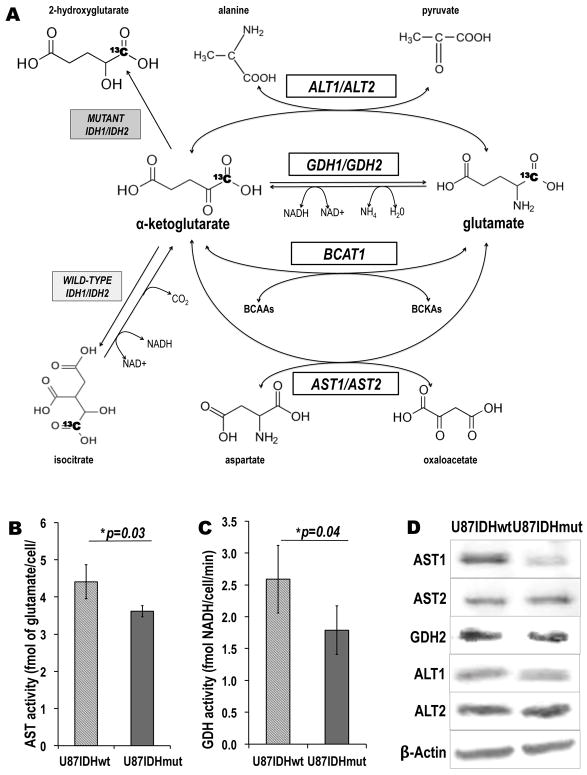
In order to determine the specificity of our imaging approach, further biochemical analyses were performed to measure the activity and expression of three additional enzymes known to catalyze the α-KG-to-glutamate conversion, namely aspartate transaminase (AST), glutamate dehydrogenase (GDH) and alanine aminotransferase (ALT) (Figure 4A).
Figure 4. In addition to BCAT1, the presence of IDH1 mutation is associated with decreased activities and protein levels of additional enzymes catalyzing α-KG-to-glutamate conversion.
(A) Schematic of [1-13C] α-KG metabolism illustrating four enzymatic pathways through which α-KG can be metabolized to glutamate: BCAT1 (branched chain aminotransferase 1; BCAAs: branched-chain aminoacids; BCKAs: branched-chain ketoacids), AST1 and AST2 (aspartate aminotransferase 1 and 2), GDH1 and GDH2 (glutamate dehydrogenase 1 and 2) and ALT1 and ALT2 (Alanine Aminotransferase 1 and 2). The reactions catalyzed by wild-type and mutant IDH1 and IDH2 are also shown (For all enzymes: 1=cytoplasmic isoform; 2=mitochondrial isoform). The 13C label at the C1 position of α-KG is highlighted in bold. (B) AST and (C) GDH enzymatic activities as measured by spectrophotometric assays. The activities of both enzymes are significantly decreased in mutant IDH1 cells as compared to wild-type cells (*p<0.05; **p<0.01; n=4 per cell line and per enzyme). (D) Western blots for the AST1, AST2, GDH2, ALT1 and ALT2 enzymes. BCAT1 and AST1 protein levels were significantly decreased in U87IDHmut cells, whereas the rest of the enzymes levels were unchanged. β-Actin was used as a loading control. (Note: GDH1 was below detection level).
Results show that total cellular AST and GDH activities were significantly decreased by 17±10% and 31±15%, respectively, in U87IDHmut cells as compared to U87IDHwt (Figure 4B&C; Table 1): RAST=4.41±0.45 fmol of glutamate/cell/min for U87IDHwt cells versus RAST=3.62±0.15 fmol of glutamate/cell/min for U87IDHmut cells (p=0.03, n=4 per cell line); RGDH=1.79±0.38 fmol of NADH/cell/min for U87IDHwt cells versus RGDH=2.59±0.53 fmol of NADH/cell/min for U87IDHmut cells (p=0.04, n=4 per cell line). ALT activity was below detection level in both cell lines.
When considering protein levels, the western blots for the three enzymes of interest (cytoplasmic and mitochondrial isoforms) are presented in Figure 4D. The quantified protein and mRNA levels are summarized in Table 1. In mutant IDH1 cells, the protein and mRNA levels of the cytoplasmic isoform AST1 were reduced significantly to 55±11% (p=0.04; n=3 per cell line) and 73±9% (p=0.009; n=9 per cell line) of U87IDHwt, respectively. In contrast, for the mitochondrial isoform AST2, only the mRNA levels were significantly reduced to 79±5% of U87IDHwt (p=0.008; n=9 per cell line) in U87IDHmut, whereas any changes in protein levels were below detection. In the case of GDH and ALT, the protein and mRNA levels were either below detection, or did not change significantly (Table 1).
DISCUSSION
The IDH1 mutation, which is the most common somatic mutation in low grade gliomas (3), is considered an early oncogenic event (41) and is associated with global modulations in the methylome, transcriptome and metabolome (6–11, 14). Consequently, the IDH1 mutation is being used for patient stratification and prognosis, and mutant IDH1 inhibitors are being developed as therapies (42, 43). Non-invasive methods are therefore needed to monitor IDH1 status, and to help develop and monitor new targeted treatments. In this context, and in light of the recently uncovered link between BCAT1 expression and the IDH1 mutation (17), we investigated the potential of hyperpolarized [1-13C] α-KG as an imaging probe to monitor the BCAT1-driven α-KG-to-glutamate conversion and its modulation in the presence of the IDH1 mutation in cells and in vivo.
Decreased BCAT1 activity, protein levels and mRNA expression have previously been reported in four cell models harboring the IDH1 mutation (17). Importantly, our results are in line with this previous report. We show that BCAT1 activity, protein levels and mNRA expression were significantly decreased in our U87IDHmut cells as compared to their wild-type counterpart, in addition to our previously reported elevation in 2-HG levels in the same model (34).
In an initial cell study, we investigated the potential of hyperpolarized [1-13C] α-KG to provide a non-invasive readout of the confirmed drop in BCAT1 activity in U87IDHmut cells. Injection of hyperpolarized [1-13C] α-KG into the perfusion medium of U87IDHwt cells resulted in a significant build-up of hyperpolarized [1-13C] glutamate at δGLU=177.5ppm, with a maximum of glutamate production at 12.2±0.1 seconds after the maximum of hyperpolarized [1-13C] α-KG. This result is, to our knowledge, the first time the detection of hyperpolarized [1-13C] glutamate from hyperpolarized [1-13C] α-KG is reported in live cells. This result also confirms that enough hyperpolarized [1-13C] α-KG permeates the cells to enable the detection of its conversion to hyperpolarized [1-13C] glutamate within a time frame compatible with a hyperpolarized 13C MRS experiment, as previously observed in our 2-HG study (34). Finally and most importantly, whereas hyperpolarized [1-13C] glutamate was detectable in U87IDHwt cells for up to 80 seconds post injection of hyperpolarized [1-13C] α-KG into their perfusion medium, hyperpolarized [1-13C] glutamate formation was almost undetectable in U87IDHmut cells, reflecting mutant IDH1-driven metabolic alterations in our model.
When injected intravenously in vivo in orthotopic tumor-bearing rats, hyperpolarized [1-13C] α-KG conversion to hyperpolarized [1-13C] glutamate could also be observed in situ. Hyperpolarized [1-13C] glutamate could be detected in the tumor of U87IDHwt animals 25 seconds post injection of the hyperpolarized substrate, or 10 seconds after the maximum of hyperpolarized [1-13C] α-KG, in line with the live cells results. In contrast, hyperpolarized [1-13C] glutamate was barely detected in U87IDHmut tumors, highlighting the translational potential of our approach.
Hyperpolarized [1-13C] glutamate was also detected in normal brain voxels in animals bearing both tumor types. We cannot rule out that a small amount of hyperpolarized [1-13C] glutamate is produced elsewhere in the body and flows to the normal brain through the blood stream. However, because we did not observe significant levels of glutamate in U87IDHmut tumors, glutamate produced elsewhere is likely below detection in our animals. In contrast, the presence of hyperpolarized [1-13C] glutamate in the normal brain is consistent with our previous work showing that, although hyperpolarized [1-13C] α-KG levels detected in normal brain are lower than in tumor, significant levels are observed in the normal brain (34). Thus, it is likely that hyperpolarized [1-13C] glutamate observed in the normal brains of our rats was produced in situ and reflects normal brain metabolism.
We cannot rule out that the level of hyperpolarized [1-13C] glutamate reflects not only its production, but also its conversion into multiple downstream metabolites such as glutamine, γ-Glu-Cys etc. However, within the relatively short lifetime of the hyperpolarized species, any downstream metabolites were below detection level in our study. Thus the level of hyperpolarized [1-13C] glutamate observed in our cells likely reflects primarily glutamate production. Hyperpolarized [1-13C] glutamate can be produced from hyperpolarized [1-13C] α-KG through the activity of BCAT1, but also via three other enzymes, namely AST, GDH and ALT. In addition to the expected drop in BCAT1 activity, we show that, in our mutant IDH1 cells, GDH and AST activities were significantly reduced, suggesting additional metabolic reprogramming. In the case of AST, as in the case of BCAT1, this effect is likely mediated by a drop in expression of the cytoplasmic isoform of AST, AST1. In the case of GDH, its unchanged mRNA expression indicates that our observed drop in cellular activity in mutant IDH1 cells is likely mediated by a post-translational modification of the enzyme, such as modulation of its acetylation (44, 45). Further investigations are therefore needed to fully explore the underlying mechanisms of our findings. Nonetheless, and independent of the mechanisms, our results are in line with the observed modulations of the enzymes catalyzing the α-KG to glutamate conversion, as well as previous studies reporting that most of the glutamine-derived α-KG pool is channeled toward 2-HG production rather than glutamate production in mutant IDH1 cells (3, 34, 46). Finally, our results are also in line with reports of decreased steady state levels of glutamate in mutant IDH1 tumors when compared to wild-type, including results in our U87-based model (12–14).
From a metabolic imaging perspective, this study is the first report of the detection of hyperpolarized [1-13C] glutamate produced from hyperpolarized [1-13C] α-KG both in cells and in vivo. However, whereas the α-KG-to-glutamate conversion had not been previously reported, other metabolic imaging studies have investigated the reverse metabolic reaction, namely glutamate-to-α-KG conversion. In a recent report, hyperpolarized [1-13C] glutamate was used as the injected hyperpolarized substrate, and detection of hyperpolarized [1-13C] α-KG was reported in hepatoma cells and tumors (47). Interestingly, hyperpolarized [1-13C] α-KG could be detected only when hyperpolarized [1-13C] glutamate was co-injected with pyruvate, the co-factor needed for transamination by the ALT enzyme (Figure 1). Another study indirectly investigated the glutamate-to-α-KG reaction by monitoring the transamination of hyperpolarized α-keto-[1-13C] isocaproate (KIC) to leucine by BCAT1, a reaction which concomitantly converts glutamate to α-KG (48). Interestingly, when comparing these two studies, hyperpolarized [1-13C] glutamate conversion to hyperpolarized [1-13C] α-KG appears to be driven mainly by the ALT enzyme, whereas hyperpolarized [1-13C] KIC only reflects BCAT1 activity. In our study, we show that hyperpolarized [1-13C] α-KG to hyperpolarized [1-13C] glutamate conversion reflects the activity of several enzymes, namely BCAT1, GDH, and AST. Accordingly, the technique presented in this study is reaction-specific (α-KG-to-glutamate conversion) rather that enzyme-specific. Future studies combining our approach with other specific metabolic probes, such as hyperpolarized [1-13C] KIC or hyperpolarized [1-13C] glutamate, could prove useful to evaluate the contributions of the different enzymes to the detected hyperpolarized [1-13C] glutamate production.
On a technical level, this study benefits from innovative dedicated pulse sequences that allow preservation of substrate magnetization while enhancing the signal of the detected metabolic product, namely hyperpolarized [1-13C] glutamate (38, 40). This glutamate-specific sequence was adapted from a previously reported 2-HG-specific sequence that we recently used to detect hyperpolarized [1-13C] 2-HG production from hyperpolarized [1-13C] α-KG (34). The sequences used in each of those two studies were metabolite-specific. Therefore, in each study, the detection of only one α-KG-derived metabolite (either 2-HG or glutamate) was fully optimized and only one metabolite could reliably and consistently be observed. In the context of tumor characterization, because 2-HG is produced by mutant IDH1, the 2-HG-specific sequence provides a direct and positive readout of mutant IDH1 enzyme activity, and thus specifically detects cells harboring the IDH1 mutation. In contrast, hyperpolarized [1-13C] glutamate informs on mutant IDH1-associated metabolic modulations and, as such, provides an indirect confirmation of mutant IDH1 status. Further development of advanced pulse sequences is expected to lead to simultaneous detection of both hyperpolarized [1-13C] 2-HG and hyperpolarized [1-13C] glutamate following hyperpolarized [1-13C] α-KG injection, providing complementary direct and indirect metabolic information with regard to IDH1 mutational status. The ability to detect both metabolites simultaneously, in a single sequence, would likely enhance the accuracy of mutant IDH1 imaging.
In summary, this study shows that 13C MRS of hyperpolarized [1-13C] α-KG can be used to monitor hyperpolarized [1-13C] glutamate production. Most importantly, the observed level of hyperpolarized [1-13C] glutamate was significantly lower in mutant IDH1 cells and tumors, in line with decreased BCAT1, AST, and GDH activities. Hyperpolarized [1-13C] glutamate could therefore serve as a secondary metabolic biomarker of IDH1 mutational status in gliomas, together with 2-HG.
Acknowledgments
Financial support (for each author): This work was supported by NIH R21CA161545 (SMR), NIH R01CA172845 (SMR), NIH R01CA154915 (SMR), NIH center grant P41EB013598 (DBV) and a fellowship from the American Brain Tumor Association (ABTA) (MMC).
The authors acknowledge Dr. Robert Bok for help with in vivo studies.
Footnotes
Conflict of interest: Funding from GE Healthcare (to PEZL).
References
- 1.Parsons DW, Jones S, Zhang X, Lin JC, Leary RJ, Angenendt P, et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321:1807–12. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yan H, Parsons DW, Jin G, McLendon R, Rasheed BA, Yuan W, et al. IDH1 and IDH2 mutations in gliomas. The New England journal of medicine. 2009;360:765–73. doi: 10.1056/NEJMoa0808710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dang L, White DW, Gross S, Bennett BD, Bittinger MA, Driggers EM, et al. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature. 2009;462:739–44. doi: 10.1038/nature08617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chowdhury R, Yeoh KK, Tian YM, Hillringhaus L, Bagg EA, Rose NR, et al. The oncometabolite 2-hydroxyglutarate inhibits histone lysine demethylases. EMBO Rep. 2011;12:463–9. doi: 10.1038/embor.2011.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xu W, Yang H, Liu Y, Yang Y, Wang P, Kim SH, et al. Oncometabolite 2-hydroxyglutarate is a competitive inhibitor of alpha-ketoglutarate-dependent dioxygenases. Cancer Cell. 2011;19:17–30. doi: 10.1016/j.ccr.2010.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blough MD, Al-Najjar M, Chesnelong C, Binding CE, Rogers AD, Luchman HA, et al. DNA hypermethylation and 1p Loss silence NHE-1 in oligodendroglioma. Ann Neurol. 2012;71:845–9. doi: 10.1002/ana.23610. [DOI] [PubMed] [Google Scholar]
- 7.Chesnelong C, Chaumeil MM, Blough MD, Al-Najjar M, Stechishin OD, Chan JA, et al. Lactate dehydrogenase A silencing in IDH mutant gliomas. Neuro Oncol. 2013 doi: 10.1093/neuonc/not243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huse JT, Phillips HS, Brennan CW. Molecular subclassification of diffuse gliomas: seeing order in the chaos. Glia. 2011;59:1190–9. doi: 10.1002/glia.21165. [DOI] [PubMed] [Google Scholar]
- 9.Lu C, Ward PS, Kapoor GS, Rohle D, Turcan S, Abdel-Wahab O, et al. IDH mutation impairs histone demethylation and results in a block to cell differentiation. Nature. 2012;483:474–8. doi: 10.1038/nature10860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Turcan S, Rohle D, Goenka A, Walsh LA, Fang F, Yilmaz E, et al. IDH1 mutation is sufficient to establish the glioma hypermethylator phenotype. Nature. 2012;483:479–83. doi: 10.1038/nature10866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yen K, Wang F, Schalm S, Hansen E, Straley K, Kernytsky A, et al. Mutation Selective IDH Inhibitors Mediate Histone and DNA Methylation Changes. Blood. 2012:120. [Google Scholar]
- 12.Elkhaled A, Jalbert LE, Phillips JJ, Yoshihara HA, Parvataneni R, Srinivasan R, et al. Magnetic resonance of 2-hydroxyglutarate in IDH1-mutated low-grade gliomas. Sci Transl Med. 2012;4:116ra5. doi: 10.1126/scitranslmed.3002796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Izquierdo Garcia JL, Eriksson P, Chaumeil MM, Pieper RO, Phillips JJ, Ronen SM. Metabolic reprograming in IDH mutant glioma cells. ISMRM; 2014; Milan, Italy. [Google Scholar]
- 14.Reitman ZJ, Jin G, Karoly ED, Spasojevic I, Yang J, Kinzler KW, et al. Profiling the effects of isocitrate dehydrogenase 1 and 2 mutations on the cellular metabolome. Proc Natl Acad Sci U S A. 2011;108:3270–5. doi: 10.1073/pnas.1019393108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pope WB, Prins RM, Albert Thomas M, Nagarajan R, Yen KE, Bittinger MA, et al. Non-invasive detection of 2-hydroxyglutarate and other metabolites in IDH1 mutant glioma patients using magnetic resonance spectroscopy. J Neurooncol. 2012;107:197–205. doi: 10.1007/s11060-011-0737-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cai L, Izquierdo Garcia JL, Chaumeil MM, Eriksson P, Robinson AR, Pieper RO, et al. Glioma cells with the IDH1 mutation increase metabolic fractional flux through pyruvate carboxylase. ISMRM; 2014; Milan, Italy. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tonjes M, Barbus S, Park YJ, Wang W, Schlotter M, Lindroth AM, et al. BCAT1 promotes cell proliferation through amino acid catabolism in gliomas carrying wild-type IDH1. Nat Med. 2013;19:901–8. doi: 10.1038/nm.3217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goto M, Miyahara I, Hirotsu K, Conway M, Yennawar N, Islam MM, et al. Structural determinants for branched-chain aminotransferase isozyme-specific inhibition by the anticonvulsant drug gabapentin. J Biol Chem. 2005;280:37246–56. doi: 10.1074/jbc.M506486200. [DOI] [PubMed] [Google Scholar]
- 19.Nelson SJ. Assessment of therapeutic response and treatment planning for brain tumors using metabolic and physiological MRI. NMR in biomedicine. 2011;24:734–49. doi: 10.1002/nbm.1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Julia-Sape M, Coronel I, Majos C, Candiota AP, Serrallonga M, Cos M, et al. Prospective diagnostic performance evaluation of single-voxel 1H MRS for typing and grading of brain tumours. NMR in biomedicine. 2012;25:661–73. doi: 10.1002/nbm.1782. [DOI] [PubMed] [Google Scholar]
- 21.Ardenkjaer-Larsen JH, Fridlund B, Gram A, Hansson G, Hansson L, Lerche MH, et al. Increase in signal-to-noise ratio of > 10,000 times in liquid-state NMR. Proc Natl Acad Sci U S A. 2003;100:10158–63. doi: 10.1073/pnas.1733835100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kurhanewicz J, Vigneron DB, Brindle K, Chekmenev EY, Comment A, Cunningham CH, et al. Analysis of cancer metabolism by imaging hyperpolarized nuclei: prospects for translation to clinical research. Neoplasia. 2011;13:81–97. doi: 10.1593/neo.101102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dafni H, Larson PE, Hu S, Yoshihara HA, Ward CS, Venkatesh HS, et al. Hyperpolarized 13C spectroscopic imaging informs on hypoxia-inducible factor-1 and myc activity downstream of platelet-derived growth factor receptor. Cancer research. 2010;70:7400–10. doi: 10.1158/0008-5472.CAN-10-0883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Albers MJ, Bok R, Chen AP, Cunningham CH, Zierhut ML, Zhang VY, et al. Hyperpolarized 13C lactate, pyruvate, and alanine: noninvasive biomarkers for prostate cancer detection and grading. Cancer Res. 2008;68:8607–15. doi: 10.1158/0008-5472.CAN-08-0749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ward CS, Venkatesh HS, Chaumeil MM, Brandes AH, Vancriekinge M, Dafni H, et al. Noninvasive detection of target modulation following phosphatidylinositol 3-kinase inhibition using hyperpolarized 13C magnetic resonance spectroscopy. Cancer research. 2010;70:1296–305. doi: 10.1158/0008-5472.CAN-09-2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Park I, Larson PE, Zierhut ML, Hu S, Bok R, Ozawa T, et al. Hyperpolarized 13C magnetic resonance metabolic imaging: application to brain tumors. Neuro Oncol. 2010;12:133–44. doi: 10.1093/neuonc/nop043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chaumeil MM, Ozawa T, Park I, Scott K, James CD, Nelson SJ, et al. Hyperpolarized 13C MR spectroscopic imaging can be used to monitor Everolimus treatment in vivo in an orthotopic rodent model of glioblastoma. Neuroimage. 2012;59:193–201. doi: 10.1016/j.neuroimage.2011.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Venkatesh HS, Chaumeil MM, Ward CS, Haas-Kogan DA, James CD, Ronen SM. Reduced phosphocholine and hyperpolarized lactate provide magnetic resonance biomarkers of PI3K/Akt/mTOR inhibition in glioblastoma. Neuro Oncol. 2012;14:315–25. doi: 10.1093/neuonc/nor209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nelson SJ, Kurhanewicz J, Vigneron DB, Larson PEZ, Harzstarck A, Ferrone M, et al. Metabolic Imaging of Patients with Prostate Cancer Using Hyperpolarized [1-13C]Pyruvate. Sci Transl Med. 2013 doi: 10.1126/scitranslmed.3006070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Andronesi OC, Kim GS, Gerstner E, Batchelor T, Tzika AA, Fantin VR, et al. Detection of 2-hydroxyglutarate in IDH-mutated glioma patients by in vivo spectral-editing and 2D correlation magnetic resonance spectroscopy. Sci Transl Med. 2012;4:116ra4. doi: 10.1126/scitranslmed.3002693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Choi C, Ganji SK, Deberardinis RJ, Hatanpaa KJ, Rakheja D, Kovacs Z, et al. 2-hydroxyglutarate detection by magnetic resonance spectroscopy in IDH-mutated patients with gliomas. Nat Med. 2012;18:624–9. doi: 10.1038/nm.2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lazovic J, Soto H, Piccioni D, Lou JR, Li S, Mirsadraei L, et al. Detection of 2-hydroxyglutaric acid in vivo by proton magnetic resonance spectroscopy in U87 glioma cells overexpressing isocitrate dehydrogenase-1 mutation. Neuro Oncol. 2012;14:1465–72. doi: 10.1093/neuonc/nos258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kalinina J, Carroll A, Wang L, Yu Q, Mancheno DE, Wu S, et al. Detection of “oncometabolite” 2-hydroxyglutarate by magnetic resonance analysis as a biomarker of IDH1/2 mutations in glioma. J Mol Med (Berl) 2012;90:1161–71. doi: 10.1007/s00109-012-0888-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chaumeil MM, Larson PE, Yoshihara HA, Danforth OM, Vigneron DB, Nelson SJ, et al. Non-invasive in vivo assessment of IDH1 mutational status in glioma. Nat Commun. 2013;4:2429. doi: 10.1038/ncomms3429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cooper AJ, Conway M, Hutson SM. A continuous 96-well plate spectrophotometric assay for branched-chain amino acid aminotransferases. Anal Biochem. 2002;308:100–5. doi: 10.1016/s0003-2697(02)00243-9. [DOI] [PubMed] [Google Scholar]
- 36.Brandes AH, Ward CS, Ronen SM. 17-allyamino-17-demethoxygeldanamycin treatment results in a magnetic resonance spectroscopy-detectable elevation in choline-containing metabolites associated with increased expression of choline transporter SLC44A1 and phospholipase A2. Breast Cancer Res. 2010;12:R84. doi: 10.1186/bcr2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lupo JM, Chen AP, Zierhut ML, Bok RA, Cunningham CH, Kurhanewicz J, et al. Analysis of hyperpolarized dynamic 13C lactate imaging in a transgenic mouse model of prostate cancer. Magn Reson Imaging. 2010;28:153–62. doi: 10.1016/j.mri.2009.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xing Y, Reed GD, Pauly JM, Kerr AB, Larson PE. Optimal variable flip angle schemes for dynamic acquisition of exchanging hyperpolarized substrates. J Magn Reson. 2013;234:75–81. doi: 10.1016/j.jmr.2013.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Larson PE, Bok R, Kerr AB, Lustig M, Hu S, Chen AP, et al. Investigation of tumor hyperpolarized [1-13C]-pyruvate dynamics using time-resolved multiband RF excitation echo-planar MRSI. Magnetic resonance in medicine. 2010;63:582–91. doi: 10.1002/mrm.22264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Larson PE, Kerr AB, Chen AP, Lustig MS, Zierhut ML, Hu S, et al. Multiband excitation pulses for hyperpolarized 13C dynamic chemical-shift imaging. J Magn Reson. 2008;194:121–7. doi: 10.1016/j.jmr.2008.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang H, Ye D, Guan KL, Xiong Y. IDH1 and IDH2 mutations in tumorigenesis: mechanistic insights and clinical perspectives. Clin Cancer Res. 2012;18:5562–71. doi: 10.1158/1078-0432.CCR-12-1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Popovici-Muller J, Saunders JO, Salituro FG, Travins JM, Yan SQ, Zhao F, et al. Discovery of the First Potent Inhibitors of Mutant IDH1 That Lower Tumor 2-HG in Vivo. Acs Med Chem Lett. 2012;3:850–5. doi: 10.1021/ml300225h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rohle D, Popovici-Muller J, Palaskas N, Turcan S, Grommes C, Campos C, et al. An Inhibitor of Mutant IDH1 Delays Growth and Promotes Differentiation of Glioma Cells. Science. 2013;340:626–30. doi: 10.1126/science.1236062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Haigis MC, Mostoslavsky R, Haigis KM, Fahie K, Christodoulou DC, Murphy AJ, et al. SIRT4 inhibits glutamate dehydrogenase and opposes the effects of calorie restriction in pancreatic beta cells. Cell. 2006;126:941–54. doi: 10.1016/j.cell.2006.06.057. [DOI] [PubMed] [Google Scholar]
- 45.Schlicker C, Gertz M, Papatheodorou P, Kachholz B, Becker CF, Steegborn C. Substrates and regulation mechanisms for the human mitochondrial sirtuins Sirt3 and Sirt5. J Mol Biol. 2008;382:790–801. doi: 10.1016/j.jmb.2008.07.048. [DOI] [PubMed] [Google Scholar]
- 46.Izquierdo Garcia JL, Eriksson P, Cai L, Chaumeil MM, Pieper RO, Phillips JJ, et al. Glutamine is the main source of 2-HG production in IDH1 mutant glioma cells. ISMRM; 2014; Milan, Italy. [Google Scholar]
- 47.Gallagher FA, Kettunen MI, Day SE, Hu DE, Karlsson M, Gisselsson A, et al. Detection of tumor glutamate metabolism in vivo using (13)C magnetic resonance spectroscopy and hyperpolarized [1-(13)C]glutamate. Magnetic resonance in medicine : official journal of the Society of Magnetic Resonance in Medicine/Society of Magnetic Resonance in Medicine. 2011;66:18–23. doi: 10.1002/mrm.22851. [DOI] [PubMed] [Google Scholar]
- 48.Karlsson M, Jensen PR, in’t Zandt R, Gisselsson A, Hansson G, Duus JO, et al. Imaging of branched chain amino acid metabolism in tumors with hyperpolarized 13C ketoisocaproate. Int J Cancer. 2010;127:729–36. doi: 10.1002/ijc.25072. [DOI] [PubMed] [Google Scholar]