Abstract
The serotonin 2A receptor gene (HTR2A) harbors two functional single nucleotide polymorphisms (SNPs) that are frequent in populations of African and European descent; rs6311, which affects mRNA expression, and rs6314, which changes the amino acid sequence of the encoded protein and affects the signaling properties of the receptor. Multiple clinical associations support a role for these SNPs in cognitive and neuropsychiatric phenotypes, although studies in autism spectrum disorder (ASD) remain equivocal. Here, we tested transmission disequilibrium of rs6311 and rs6314 in a cohort of 158 ASD trios (simplex and multiplex), observing significant under-transmission of the minor “A” allele of rs6311 to offspring with ASD (permuted p=0.0004). Consistent with our previous findings in the dorsolateral prefrontal cortex of unaffected individuals, rs6311/A decreases expression of HTR2A mRNA with an extended 5′ untranslated region in the frontopolar cortex in brain samples from 54 ASD patients and controls. Interpreting the clinical results in the context of our mRNA expression analysis, we speculate that any risk associated with rs6311 is conferred by greater expression of the long 5′UTR mRNA isoform. The current study corroborates earlier associations between rs6311 and ASD in a family study, supporting the hypothesis that rs6311 plays a modulatory role in ASD risk.
Keywords: Autism, serotonin, gene expression, HTR2A, rs6311, monoamine
Introduction
Autism spectrum disorders (ASD) are a highly heterogeneous group of developmental disorders. Genetic factors, including chromosome abnormalities and copy number variation (CNV), as well as inherited and de novo mutations in highly penetrant single risk genes can now be identified in approximately 20-30% of cases (Abrahams & Geschwind, 2008; Benvenuto et al., 2009; Marshall & Scherer, 2012; Matsunami et al., 2013; Pinto et al., 2010; Zafeiriou et al., 2007). However, the underlying genetic factors contributing to the remaining 70-80% of cases remain unexplained (Benvenuto et al., 2009). Possible explanations to account for the considerable portion of remaining genetic risk include highly penetrant single gene mutations in previously unrecognized risk genes, polygenic inheritance of multiple low penetrance risk factors, and combined genetic/epigenetic/environmental factors, among others (Devlin & Scherer, 2012).
Appreciation for common synaptic pathophysiology in syndromic and non-syndromic ASD (Auerbach et al., 2011; Baudouin et al., 2012) bolsters the argument for highly penetrant rare variants in genes crucial for synapse formation and maturation contributing some of the missing genetic risk, assuming functional consequences of these rare variants converges on a common synaptic pathophysiology. However, the rarity of these putative causative variants, often arising de novo (O’Roak et al., 2011, 2012) or demonstrating incomplete penetrance (Morrow et al., 2008; Yu et al., 2013), requires additional sources of genetic risk to account for ASD etiology. One potential source for this risk are common functional genetic variants (present in the general population at greater than 1% minor allele frequency) amenable to risk estimation and identification by genome-wide association studies (GWAS). However, this genome-wide approach for uncovering genetic risk in ASD has provided few replicable loci, each contributing low-to-modest genetic risk (Anney et al., 2010, 2012; Connolly et al., 2013; Ma et al., 2009; Wang et al., 2009; Weiss et al., 2009).
By first identifying polymorphisms with known biological effects – or closely linked surrogates – we can take a more targeted approach than GWAS, asking whether polymorphisms in plausible candidate genes with strong evidence for biological function have any impact on ASD etiology or behavior. In addition, by excluding likely syndromic causes through careful clinical phenotyping, we may enable detection of distinct genetic factors conferring risk, such as common functional variants. Given our a priori knowledge of the polymorphism’s function, we can further characterize implicated polymorphisms in the context of ASD using affected brain tissues.
The two polymorphisms chosen for this study reside in the genomic region encoding the serotonin 2A receptor (HTR2A). The first polymorphism, rs6311 (G/A), modulates expression of HTR2A mRNA in the human dorsolateral prefrontal cortex, affecting the usage of a newly identified transcription start site, which encodes a longer 5′UTR with increased protein translation efficiency (Smith et al., 2013). The second polymorphism, rs6314 (G/A), changes the encoded amino acid at residue 452 from a histidine to a tyrosine (His452Tyr), affecting ligand binding and second-messenger signaling in vitro (Davies et al., 2006; Hazelwood & Sanders-Bush, 2004). The minor alleles for both of these polymorphisms correspond to a loss-of-function, exhibiting reduced long 5′UTR mRNA expression and reduced second messenger signaling and ligand binding, respectively. Both rs6311 and rs6314 have numerous clinical implications, including meta-analytic associations with fibromyalgia (Lee et al., 2012), rheumatoid arthritis (Kling et al., 2008), psychosis in Alzheimer’s disease (Ramanathan & Glatt, 2009), anorexia (Gorwood et al., 2003; Martaskova et al., 2009), and multiple replicated pharmacogenetic associations with response or side-effects to atypical antipsychotics and selective serotonin reuptake inhibitors (Arranz et al., 1998; Lerer et al., 2005; Kato & Serretti, 2010), often used to treat these disorders. With respect to ASD, previous studies have found equivocal support for rs6311 and none for rs6314 (Cho et al., 2007; Guhathakurta et al., 2009; Hranilovic et al., 2010; Veenstra-VanderWeele et al., 2002). Thus, it is clear that these HTR2A variants impact biological function to modulate risk for an assortment of clinical disorders, but further study is necessary to resolve a role for HTR2A in ASD.
Here, we test whether genetic variants in HTR2A with identified biological functions that influence risk for a variety of clinical disorders are associated with ASD in a cohort of 158 simplex and multiplex ASD trios. Given our previous studies of rs6311 on HTR2A expression in the prefrontal cortex of typically-developed individuals, we asked how this polymorphism affected mRNA expression in ASD brain tissue to address the most likely mechanism(s) by which rs6311 confers risk when implicated in ASD.
Materials and Methods
Clinical Sample
Families were recruited through the Central Ohio Registry for Autism (CORA) at Nationwide Children’s Hospital as previously reported (Cottrell et al., 2011). The cohort included 158 unrelated trios (affected child and parents); 78% (124/158) were simplex families and 22% were multiplex (more than 1 affected child) families. In multiplex families, only the identified proband was analyzed. Sixty-three percent of the families self-identified as Caucasian, 28% as Caucasian/Other, 4% as African American, and 4% as Other. ASD diagnosis was made by a qualified clinical psychologist or developmental pediatrician using DSM-IV diagnostic criteria. The Autism Diagnostic Observation Schedule (ADOS) was available on 97% (153/158) of the children, and IQ testing results were available on 53% (83/158) assessed by either the Stanford-Binet Intelligence Scales or the Leiter International Performance Scale, depending on the verbal ability of the child.
A clinical genetics evaluation was completed on 72% (113/158) of the children. Two unrelated affected probands each had a pathogenic de novo PTEN gene mutation, and 1 child was identified with a pathogenic 22q11.2 deletion of unknown parental origin. Three probands with significant intellectual disability who were found to have pathogenic chromosomal rearrangements following enrollment in the study (2 with large unbalanced autosomal translocations and one with a mosaic ring chromosome) were excluded from any molecular studies. Chromosomal microarray (CMA) was completed on 98% (155/158) of the children, with 9 “likely pathogenic” or susceptibility copy number variations identified (Supplemental Table 1). The individuals with the PTEN mutations, the 22q11.2 deletion and the CNVs listed in Supplemental Table 1 were included since penetrance for these entities is variable, and we felt that additional loci analyzed in our studies might influence the phenotype.
Clinical sample genotyping and analysis
Genomic DNA (gDNA) was isolated from whole blood or lymphoblastoid cell lines derived from the patients and their parents and rs6311 and rs6314 were genotyped using TaqMan probes (Life Technologies, Carlsbad, CA) on an ABI 7500 Real-Time PCR System (Life Technologies), according to manufacturer’s instructions. Transmission disequilibrium tests (TDT) for each SNP were performed using PLINK (Purcell et al., 2007). Permuted p-values are reported for each SNP, calculated from 10,000 family-wise permutations. For post-hoc genotype-phenotype comparisons for rs6311 (Supplemental Table 2), categorical measures were analyzed using Chi-square tests, while quantitative measures were analyzed with univariate analysis of variance (ANOVA) tests in SPSS v19.0 (IBM Corporation, Armonk, NY), subsequently Bonferroni-corrected for multiple hypothesis testing.
Brain samples
54 total frontopolar cortex samples (Brodmann Area 10) obtained from the Autism Tissue Program (Autism Speaks) were used in this study. These included 20 samples from individuals diagnosed with ASD, 14 first-degree relatives of individuals with ASD (but unrelated to our autism cohort), and 20 control brain tissues matched to the ASD samples by sex and age. Demographic characteristics are available in Table 1.
Table 1. Demographic characteristics for BA10 brain tissue.
| Diagnosis (n) | Sex (M:F) | Age [Avg.±S.D. (Range)] | PMIa [Avg.±S.D. (Range)] | Race |
|---|---|---|---|---|
| ASD (20) | 15:5 | 24.7±20.1 (2-82) | 17.1±6.2 (4-25) | Cauc: 15, AA: 3, Unk: 2 |
| First-degree relative (14) | 6:8 | 65.5±26.7 (0-90) b | 16.3±3.6 (10-22) | Cauc: 8, AA: 0, Unk: 6 |
| Control (20) | 18:2 | 41.9±23.5 (7-88) | 21.4±5.0 (11-33) b | Cauc: 1, AA: 1, Unk: 18 |
Post-mortem interval (hours)
p<0.05 versus other diagnosis groups
Abbreviations – AA: African-American; ASD: autism spectrum disorder; Avg.: average; BA10: Brodmann Area 10; Cauc: Caucasian;
S.D.: standard deviation; Unk: unknown
Allelic mRNA expression and quantitative PCR (qPCR) measurement in brain
gDNA and RNA were isolated from each brain tissue, as previously described (Smith et al., 2013). RNA was reverse-transcribed into complementary DNA (cDNA) with SuperScript III using gene-specific primers supplemented with oligo-dT.
Allelic mRNA expression measurements were performed in duplicate or triplicate in samples heterozygous for rs6311. A region surrounding rs6311 was PCR-amplified in gDNA (25ng) or cDNA (12.5ng) for 30 cycles using a standard 3-step protocol. Following amplification, unincorporated primers were digested by simultaneous incubation with exonuclease I (New England Biolabs; NEB, Ipswich, MA) and Antarctic phosphatase (NEB). In a single-nucleotide extension reaction using SNaPshot (Life Technologies), primers immediately adjacent to the SNP direct the incorporation of a fluorescent dideoxynucleotide (ddNTP) at the SNP position, using the PCR product as a template. Unincorporated ddNTPs are then digested with calf intestinal phosphatase (NEB). The fluorescently-labeled product is resolved in an ABI 3730 (Life Technologies) and the fluorescent peaks for each allele, represented by different fluorophores, are detected by GeneMapper 4.0 (Life Technologies) and peak heights are used to calculate a ratio for cDNA and gDNA by dividing the major ancestral allele by the variant allele. cDNA ratios are then normalized to gDNA ratios, which represent a 1:1 relationship, to yield the relative difference in expression across the two alleles. Samples with an average allelic ratio more than two times greater than the overall average within-sample standard deviation are designated as having significant allelic expression imbalance (AEI).
Quantitative PCR was performed using an ABI 7500 Fast Real-Time PCR System (Life Technologies). For each sample, HTR2A expression was measured in duplicate at two different sites; in the constitutively-expressed exon 4 and in the extended 5′ untranslated region (UTR) we previously found to be modulated by rs6311 (Smith et al., 2013). HTR2A expression was normalized to β-actin (ACTB) expression, measured in duplicate for each sample. Group-wise ANOVAs comparing HTR2A mRNA expression across genotype and diagnosis was performed using SPSS. One first-degree relative (sibling) was a statistical outlier (>3 standard deviations from the total cohort mean), expressing approximately 6-fold less total HTR2A mRNA. This subject died at 7 weeks of age and had no confirmed ASD diagnosis, although three older siblings were diagnosed on the ASD spectrum. Consequently, group-wise comparisons reported here exclude this sample.
Results and Discussion
Clinical association
The results of the TDT-based clinical association are presented in Table 2. rs6311 was significantly associated with ASD, exhibiting under-transmission of the minor “A” allele (rs6311/A) to affected probands. If we excluded the 12 probands with additional risk factors (pathogenic PTEN mutation, 22q11.2 deletion, CNVs listed in Supplemental Table 1), our results remain statistically significant for rs6311. Several previous studies have examined rs6311 in ASD. In a case-control association study, Hranilovic et al. (2010) found that rs6311/A was underrepresented in ASD relative to controls, consistent with our findings. However, two other case-control studies and three transmission-based examinations of HTR2A found no significant association between rs6311 or rs6313, as a surrogate marker for rs6311, and ASD (Cho et al., 2007; Guhathakurta et al., 2009; Herault, et al., 1996; Veenstra-VanderWeele et al., 2002). One explanation for discrepant findings is that each of these previous studies examined fewer affected probands than the current study, generating less statistical power to detect the genetic effect reported here. Moreover, our ASD population excludes individuals with identifiable common causes of “syndromic autism”, such as Fragile X syndrome, large chromosomal rearrangements, or those with significant intellectual disability (IQ<40). Thus, regulatory variants in key candidate genes could exert a larger influence in the context of affected subjects who do not carry known highly penetrant chromosomal abnormalities. We must also consider the ethnic make-up of our population, relative to these other studies. Allele frequencies for rs6311 are similar across all 1000 Genomes populations, ranging from 30-50% minor allele frequency (MAF), and all studies reported here are within this range, including ours (Table 2). Therefore, population stratification is unlikely to account for differences in findings related to rs6311 here. In contrast, MAF for rs6314 varies across populations; as high as 20% in individuals of African descent and absent in Asian descent. Our study reports slightly higher MAF for rs6314 relative to the other studies (11% versus 7-9%), but similar to previous reports, we find no association between rs6314 and ASD in our cohort (Guhathakurta et al., 2009; Hranilovic et al., 2010; Veenstra-VanderWeele et al., 2002). Thus, the ethnic make-up of our population is unlikely to account for discrepancies between studies, and our results provide further support for a lack of association between rs6314 and ASD.
Table 2. Transmission disequilibrium test results for HTR2A polymorphisms in 158 CORA Trios.
| Gene | dbSNP IDa | Minor Allele |
Major Allele |
HapMap CEU MAF |
ASD MAF |
Parent MAF |
Total Trios |
TDT Allele |
Trans | Untrans | Chi Sq | p-value | Permuted p-valueb |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| HTR2A | rs6311 | A | G | 0.465 | 0.364 | 0.435 | 158 | A | 58 | 103 | 12.58 | 0.0004 | 0.0004 |
| Excluding samples with pathogenic or likely pathogenic mutationsc | 146 | A | 54 | 95 | 11.28 | 0.0008 | 0.0011 | ||||||
| HTR2A | rs6314 | T | C | 0.062 | 0.117 | 0.106 | 158 | T | 34 | 27 | 0.80 | 0.3701 | 0.6417 |
| Excluding samples with pathogenic or likely pathogenic mutationsc | 146 | T | 32 | 25 | 0.86 | 0.3538 | 0.6174 | ||||||
National Center for Biotechnology Information (NCBI) dbSNP build 137
corrected p-value after 10,000 permutations
includes PTEN and 22q11.2 mutations and samples listed in Supplemental Table 1.
Abbreviations – CEU: HapMap CEPH (Utah Residents with Northern and Western European Ancestry) population; CORA: Central Ohio Registry for Autism; MAF: minor allele frequency; TDT: transmission disequilibrium test; Trans: transmitted; Untrans: untransmitted
We examined the relationship between proband rs6311 genotype and several phenotypes, including diagnostic testing scores, prenatal complications and environmental exposures, and proband IQ (Supplemental Table 2). After multiple hypothesis correction, rs6311 was not significantly associated with any phenotypic measures examined. The most significant associations included IQ (F=4.00, uncorrected p=0.02, corrected p=0.29), the ADOS stereotyped behaviors subscore (F=4.51, uncorrected p=0.02, corrected p=0.27), and concurrent ADHD diagnosis (χ2=6.73, uncorrected p=0.04, corrected p=0.53). For the exploratory analysis of measures reported in Supplemental Table 2, we urge caution when interpreting these associations, as these phenotypes are not likely independent and have varying degrees of genetic complexity and environmental contributions that underlie their manifestation.
Although both rs6311 and rs6314 can be classified as loss-of-function variants, they impact HTR2A function at different levels – rs6311 through RNA and rs6314 through protein. In addition, we presume that rs6314 impacts HTR2A function everywhere it is expressed by altering the encoded protein, while it is possible that rs6311 has varying degrees of influence on HTR2A mRNA expression, depending on the amount and constellation of trans-acting factors available to locally bind to the regulatory element(s) in which rs6311 is encoded. Based on this observation, we deduce that global reductions in HTR2A signaling are not necessarily a risk factor for ASD and other clinical phenotypes associated with rs6311 but not rs6314.
HTR2A mRNA expression in ASD and control brain tissues
Considering the significant clinical association found here, we examined the functional effects of rs6311 specifically in postmortem brain tissues from ASD patients, first-degree relatives of affected individuals, and matched controls. Since this tissue was dissected from the frontopolar cortex (BA10), as compared to the dorsolateral prefrontal cortex in our earlier study (BA46; Smith et al., 2013), we tested the pervasiveness of rs6311 function in this second cortical area.
All qPCR expression analyses are summarized in Table 3. The three cohorts (ASD, relatives, and controls) significantly differed in age (F=12.76, p<0.001) and post-mortem interval (PMI) (F=5.07, p=0.01) but not race or sex (Table 1). Bonferroni-corrected pairwise comparisons indicated that for age, first-degree relatives were significantly older than either ASD or control cohorts, which did not significantly differ from each other. For PMI, the control cohort had significantly higher PMI than either the ASD or first-degree relative cohorts, which did not significantly differ from each other. Consequently, both age and PMI were included as covariates in all between-group comparisons. Total HTR2A mRNA expression trended towards significance when compared across the three diagnosis groups (F=2.68, p=0.079), with ASD samples expressing 1.6-fold less total HTR2A mRNA than control samples, while first degree relatives expressed intermediate levels. We found no significant differences across the groups for extended 5′UTR mRNA expression or the percent of 5′UTR mRNA relative to total HTR2A mRNA.
Table 3. HTR2A mRNA expression across diagnostic groups and rs6311 genotypes (group mean ± SEM [Fold Changea]).
| Comparison | Controls (n=20) | Relatives (n=13) | Autism (n=20) | p-value |
|---|---|---|---|---|
| Total HTR2A Ctb,c | 5.79 ± 0.20 | 6.08 ± 0.29 [-1.22] | 6.46 ± 0.21 [-1.59] | 0.079 |
| 5′UTR HTR2A Ctb,c | 9.87 ± 0.17 | 9.95 ± 0.25 [-1.05] | 10.06 ± 0.18 [-1.14] | 0.761 |
| 5′UTR % of Totald,e | 6.7 ± 0.9 | 8.3 ± 1.4 [+1.24] | 9.3 ± 1.0 [+1.38] | 0.166 |
| Comparison | rs6311 – G/G (n=15) | rs6311 – G/A (n=30) | rs6311 – A/A (n=8) | p-value |
|---|---|---|---|---|
| Total HTR2A Ctb,f | 6.10 ± 0.20 | 6.28 ± 0.14 [+1.13] | 5.51 ± 0.28 [+1.51] | 0.059 |
| 5′UTR HTR2A Ctb | 9.72 ± 0.18 | 9.96 ± 0.13 [+1.18] | 10.41 ± 0.25 [-1.61] | 0.084 g |
| 5′UTR % of Totald,e | 8.7 ± 1.0 | 9.0 ± 0.7 [+1.03] | 3.3 ± 1.3 [-2.63] | 0.001 |
Fold change relative to Controls (top table) or Ancestral G/G genotype (bottom table)
Normalized to within-sample ACTB (β-actin) cycle threshold
Corrected for significant covariates: age and post-mortem interval
Percentage calculated based on cycle threshold values
Corrected for significant covariates: age
Corrected for significant covariates: diagnosis and age
p-value = 0.061 when including diagnosis and age as covariates
Abbreviations: Ct = cycle threshold
Genotypic comparisons of HTR2A mRNA expression
To examine only the genetic effects of rs6311 on HTR2A mRNA expression, we first performed step-wise linear regression comparing demographic variables (diagnosis, age, sex, race, and post-mortem interval) against total HTR2A mRNA expression, extended 5′UTR mRNA expression, and percent of extended 5′UTR mRNA relative to total HTR2A mRNA; significant covariates were included in subsequent ANOVAs comparing expression values across rs6311 genotype. Total HTR2A mRNA expression, when examined across rs6311 genotype trended towards significance (F=3.00, p=0.059; diagnosis and age included as significant covariates). However the largest difference (1.7-fold) was observed between lower expressing heterozygotes and higher expressing homozygous minor allele rs6311/A samples and not homozygous major allele rs6311/G versus rs6311/A samples. While previous studies have found higher total HTR2A expression corresponding to rs6311/A (Polesskaya et al., 2002), the lack of a significant difference between homozygous G and A allele samples is consistent with our previous study and others (Bray et al., 2004; Fukuda et al., 2006; Smith et al., 2013) that suggest rs6311 does not directly influence total HTR2A mRNA expression..
Instead, rs6311 acts in cis to modulate use of an upstream transcription start site that encodes an alternative HTR2A transcript with an extended 5′UTR (Smith et al., 2013). Analysis of extended 5′UTR expression across rs6311 genotype yielded a statistical trend towards significance (F=2.60, p=0.084), but in contrast to total expression, rs6311/A displays an allele-dose dependent reduction in expression, with rs6311/A homozygotes expressing 1.6-fold lower extended 5′UTR mRNA than major allele rs6311/G homozygotes. Including diagnosis and age as covariates, as done for total HTR2A expression, improves this genotype-expression association (F=2.96, p=0.061). The relative percent of extended 5′UTR mRNA expression in the total HTR2A mRNA pool calculated from normalized expression via qPCR is significantly lower for rs6311/A homozygotes versus rs6311/G homozygotes or heterozygotes (3.3% versus 8.7% and 9.0%, respectively; F=7.88, p=0.001; age included as a significant covariate). Interestingly, the sample identified as an outlier was heterozygous for rs6311 and exhibited a 120-fold reduction in extended 5′UTR expression when compared to the total group mean, constituting only 0.4% of total HTR2A mRNA in this sample (group mean = 8%). Sequencing a contiguous 6.5kb region of this sample (chr13:47,469,225-47,475,723 in GRCh37/hg19), including the promoter and first two exons, revealed no polymorphisms that are likely to account for this decreased expression.
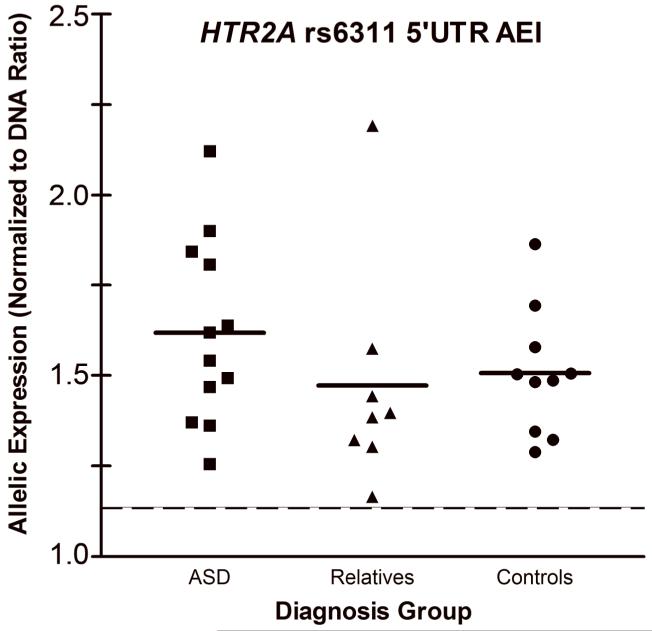
To further address the nature of cis-versus trans-acting factors affecting expression, we measured allelic expression imbalance in samples heterozygous for rs6311 in the extended 5′UTR or rs6313 in exon 2. Allelic expression measured at rs6311 is indicative only of cis-acting influence on the extended 5′UTR, while allelic expression measured at rs6313 reveals cis-acting influence on total HTR2A mRNA expression. Thirty-one of the 54 brain samples were heterozygous for rs6311, including 12 of 20 (60%) autistic samples, 9 of 14 (64%) first-degree relatives of autistic individuals, and 10 of 20 (50%) control samples. Only the outlying sample failed to yield reliable allelic expression ratios, due to low expression, as noted above. The average normalized allelic ratio for gDNA was 1.0±0.059 (mean±s.d.), while the within-sample SEM for cDNA allelic ratios ranged from 0.0006-0.097, both indicating high assay consistency. Every sample heterozygous for rs6311 displayed significant AEI, ranging from 1.16 to 2.2-fold greater expression of rs6311/G relative to rs6311/A alleles (Figure 1), consistent with the relationship of absolute expression for the extended 5′UTR transcripts measured via qPCR. The magnitude of allelic differences did not differ across diagnosis groups (Figure 1), suggesting that rs6311 behaves in a similar fashion in the context of ASD. As with previously published reports of allelic expression in HTR2A (Bray et al., 2004; Fukuda et al., 2006; Smith et al., 2013), measuring AEI at rs6313 did not reveal evidence for a common cis-acting functional variant affecting total HTR2A mRNA expression. This strongly suggests that rs6314 also does not act in cis to significantly influence transcription.
Figure 1.
Allelic expression in brain samples heterozygous for rs6311 in the extended 5′UTR. All brain samples heterozygous for rs6311 display significant allelic expression imbalance (AEI), with the major rs6311/G allele expressing 1.16 to 2.2-fold more mRNA relative to the minor rs6311/A allele. The magnitude of allelic expression is not significantly different across diagnosis groups. Note: dashed line represents 2 standard deviations of the within-sample assay error, indicative of significant AEI.
We speculate that the function of the extended 5′UTR of HTR2A mRNA is responsible for modulating clinical associations with rs6311. To date, the functional consequences of differential HTR2A 5′UTR length have only been apparent for protein translation efficiency (Smith et al., 2013), with the rs6311/A reducing expression of the longer 5′UTRs that have greater translational efficiency, thereby also indirectly reducing HTR2A protein. Although both rs6311 and rs6314 are loss-of-function variants according to our analyses, they do not confer risk uniformly across all clinical association studies (i.e. when one is implicated, often the other is not). Therefore, we find it less likely that a simple reduction in protein expression resulting from shorter 5′UTR length would be responsible for risk conferred by rs6311, as this would be similar to the net reduction of 5-HT2A receptor (5-HT2AR) signaling produced by rs6314. Instead, we speculate that the 5′UTR has additional functions yet to be uncovered. Prominent among the possible biological functions of the 5′UTR include regulation of the spatial or cellular distribution of HTR2A mRNAs for local translation. In situ hybridization studies of HTR2A mRNA in primates clearly show the strongest labeling in apical dendrites of pyramidal neurons (Jakab & Goldman-Rakic, 1998). However, HTR2A mRNA is also present to a lesser extent at dendritic spines and in other cell types (Jakab & Goldman-Rakic, 1998). It is plausible that the longer 5′UTR transcripts, constituting approximately 10% of all HTR2A mRNAs in the cortex (Smith et al., 2013), correspond to one or more of these less-dense populations of HTR2A mRNAs.
Serotonergic signaling in ASD
Apart from genetic association studies, there is evidence for general serotonergic and specific 5-HT2AR dysfunction in ASD. Reduced cortical 5-HT2AR density has been observed in adult men with Asperger’s syndrome using single-photon emission computed tomography (SPECT) (Murphy et al., 2006) and in first-degree relatives of autistic individuals using positron emission tomography (PET) (Goldberg et al., 2009), while subcortical thalamic areas also show decreased 5-HT2AR density by PET in high-functional adults with autism (Beversdorf et al., 2012). However, although another PET study did not find differences in 5-HT2AR density between ASD patients and controls (Girgis et al., 2011). Additional changes in 5-HT signaling identified through neuroimaging studies include a reduction in medial frontal cortex serotonin transporter density (Makkonen et al., 2008) and a reduced 5-HT synthesis capacity during childhood in autistic children (Chugani et al., 1999). Taken together, these neuroimaging studies lend support for disrupted central 5-HT signaling, in which the 5-HT2AR is a major component.
Peripherally, platelet hyperserotonemia is estimated to occur in up to one-third of ASD patients (Cook & Leventhal, 1996). 5-HT2AR expressed on platelets promotes aggregation, and 5-HT2AR ligand binding affinity or the ability to promote aggregation can be measured as an indication of 5-HT2AR function. Elevated HTR2A mRNA has been observed in platelets from ASD patients (Kazek et al., 2010) although physiological studies find reduced 5-HT2AR function or binding in platelets from ASD patients (Hranilovic et al., 2009; McBride et al., 1989) and first degree-relatives (Cook et al., 1993). Given the multiple observations of altered 5-HT2AR function in ASD by various methods, delineating molecular and genetic factors modulating HTR2A expression is critical to better understanding how polymorphisms in this gene contribute to the disorder and can be used to guide therapeutics. The latter point is critical, considering behaviors inherent to ASD are treated with atypical antipsychotics that directly antagonize 5-HT2AR (Scott & Dhillon, 2008).
Conclusions
The etiologies of ASD are likely a complex mixture of highly penetrant rare mutations and less penetrant but more common modifying variants, both subject to varying degrees of environmental influence for the manifestation of the disorder. Here, instead of extensively searching for variants likely causing ASD, we asked whether common polymorphisms in HTR2A with known biological function are contributing to ASD, finding rs6311/A was significantly under-transmitted to affected offspring in our study population. Our examination of the frontopolar cortex from affected individuals, first-degree relatives, and matched controls, found that the biological function of rs6311 in ASD was similar to that observed in unaffected frontopolar cortex. The combination of these two results allows for multiple complementary interpretations. It is possible that increased expression of the longer 5′UTR form of HTR2A transcripts confers risk, reduced expression of the short 5′UTR transcripts confers protection, or a reduction in the ratio of the short relative to the long 5′UTR confers risk.
Our study lends support to the hypothesis that regulatory variants, observed at an appreciable frequency across the human population and affecting mRNA expression, can contribute to complex genetic disorders, even when acting in a biological manner consistent with that observed in typically-developed individuals. Considering inconsistent clinical associations between rs6311 and ASD and the fact that rs6311 is a common regulatory variant, we emphasize the importance of context, genetic and environmental, for such a variant to impact risk for complex genetic disorders. The biological role of the extended 5′UTR in HTR2A mRNA requires further study. Already, we have determined that it promotes greater translational efficiency in vitro (Smith et al., 2013). Our previous study leads us to suggest that a decrease in short 5′UTR and/or increase in long 5′UTR transcripts, as observed by clinical and molecular results in the ASD samples here, would result in more 5-HT2AR protein. However, it remains to be seen how the different UTR isoforms affect subcellular HTR2A mRNA expression in neurons or in the periphery, especially in platelets or with respect to hyperserotonemia. A major goal of our future research is to delineate the circumstances (genetic background, epistasis, environment, etc.) under which frequent variants such as rs6311 contribute to ASD.
Supplementary Material
Acknowledgements
The authors declare no competing interests. This work was supported by the National Institute of General Medical Sciences [U01GM092655 to W.S.] and the United States Air Force Department of Defense [FA7014-09-2-0004 and FA8650-12-2-6359 to Gail E. Herman]. We are grateful to all the families participating in the CORA registry. We thankfully acknowledge Harvard Brain Tissue Resource Center and the NICHD Brain and Tissue Bank for providing brain tissues, granted to Ryan M. Smith and Wolfgang Sadee from the Autism Tissue Program, made possible by the donations of generous family members.
The views and opinions expressed in this article are those of the author(s) and do not reflect official policy or position of the United State Air Force, Department of Defense, or US Government. The use of product(s) and/or manufacturer name(s) is added for clarification only; in no way implies endorsement by the authors, USAF, or DoD of the product(s) or manufacturer(s).
Grant Sponsor: National Institute of General Medical Sciences; Grant number: U01GM092655
Grant Sponsor: United States Air Force Department of Defense; Grant number: FA7014-09-2-0004
Grant Sponsor: United States Air Force Department of Defense; Grant number: FA8650-12-2-6359
Literature Cited
- Abrahams BS, Geschwind DH. Advances in autism genetics: on the threshold of a new neurobiology. Nature Reviews Genetics. 2008;9:341–355. doi: 10.1038/nrg2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anney R, Klei L, Pinto D, Almeida J, Bacchelli E, Baird G, et al. Individual common variants exert weak effects on the risk for autism spectrum disorderspi. Human Molecular Genetics. 2012;21:4781–4792. doi: 10.1093/hmg/dds301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anney R, Klei L, Pinto D, Regan R, Conroy J, Magalhaes TR, et al. A genomewide scan for common alleles affecting risk for autism. Human Molecular Genetics. 2010;19:4072–4082. doi: 10.1093/hmg/ddq307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arranz MJ, Munro J, Sham P, Kirov G, Murray RM, Collier DA, et al. Metaanalysis of studies on genetic variation in 5-HT2A receptors and clozapine response. Schizophrenia Research. 1998;32:93–99. doi: 10.1016/s0920-9964(98)00032-2. [DOI] [PubMed] [Google Scholar]
- Auerbach BD, Osterweil EK, Bear MF. Mutations causing syndromic autism define an axis of synaptic pathophysiology. Nature. 2011;480:63–68. doi: 10.1038/nature10658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baudouin SJ, Gaudias J, Gerharz S, Hatstatt L, Zhou K, Punnakkal P, et al. Shared synaptic pathophysiology in syndromic and nonsyndromic rodent models of autism. Science. 2012;338:128–132. doi: 10.1126/science.1224159. [DOI] [PubMed] [Google Scholar]
- Benvenuto A, Moavero R, Alessandrelli R, Manzi B, Curatolo P. Syndromic autism: causes and pathogenetic pathways. World Journal of Pediatrics. 2009;5:169–176. doi: 10.1007/s12519-009-0033-2. [DOI] [PubMed] [Google Scholar]
- Beversdorf DQ, Nordgren RE, Bonab AA, Fischman AJ, Weise SB, Dougherty DD, et al. 5-HT2 receptor distribution shown by [18F] setoperone PET in highfunctioning autistic adults. The Journal of Neuropsychiatry and Clinical Neurosciences. 2012;24:191–197. doi: 10.1176/appi.neuropsych.11080202. [DOI] [PubMed] [Google Scholar]
- Bray NJ, Buckland PR, Hall H, Owen MJ, O’Donovan MC. The serotonin-2A receptor gene locus does not contain common polymorphism affecting mRNA levels in adult brain. Molecular Psychiatry. 2004;9:109–114. doi: 10.1038/sj.mp.4001366. [DOI] [PubMed] [Google Scholar]
- Cho IH, Yoo HJ, Park M, Lee YS, Kim SA. Family-based association study of 5-HTTLPR and the 5-HT2A receptor gene polymorphisms with autism spectrum disorder in Korean trios. Brain Research. 2007;1139:34–41. doi: 10.1016/j.brainres.2007.01.002. [DOI] [PubMed] [Google Scholar]
- Chugani DC, Muzik O, Behen M, Rothermel R, Janisse JJ, Lee J, et al. Developmental changes in brain serotonin synthesis capacity in autistic and nonautistic children. Annals of Neurology. 1999;45:287–295. doi: 10.1002/1531-8249(199903)45:3<287::aid-ana3>3.0.co;2-9. [DOI] [PubMed] [Google Scholar]
- Connolly JJ, Glessner JT, Hakonarson H. A genome-wide association study of autism incorporating autism diagnostic interview-revised, autism diagnostic observation schedule, and social responsiveness scale. Child Development. 2013;84:17–33. doi: 10.1111/j.1467-8624.2012.01838.x. [DOI] [PubMed] [Google Scholar]
- Cook EH, Jr., Arora RC, Anderson GM, Berry-Kravis EM, Yan SY, Yeoh HC, et al. Platelet serotonin studies in hyperserotonemic relatives of children with autistic disorder. Life Sciences. 1993;52:2005–2015. doi: 10.1016/0024-3205(93)90685-v. [DOI] [PubMed] [Google Scholar]
- Cook EH, Leventhal BL. The serotonin system in autism. Current Opinion in Pediatrics. 1996;8:348–354. doi: 10.1097/00008480-199608000-00008. [DOI] [PubMed] [Google Scholar]
- Cottrell CE, Bir N, Varga E, Alvarez CE, Bouyain S, Zernzach R, et al. Contactin 4 as an autism susceptibility locus. Autism Research. 2011;4:189–199. doi: 10.1002/aur.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies MA, Setola V, Strachan RT, Sheffler DJ, Salay E, Hufeisen SJ, et al. Pharmacologic analysis of non-synonymous coding h5-HT2A SNPs reveals alterations in atypical antipsychotic and agonist efficacies. Pharmacogenomics Journal. 2006;6:42–51. doi: 10.1038/sj.tpj.6500342. [DOI] [PubMed] [Google Scholar]
- Devlin B, Scherer SW. Genetic architecture in autism spectrum disorder. Current Opinion in Genetics and Development. 2012;22:229–237. doi: 10.1016/j.gde.2012.03.002. [DOI] [PubMed] [Google Scholar]
- Fukuda Y, Koga M, Arai M, Noguchi E, Ohtsuki T, Horiuchi Y, et al. Monoallelic and unequal allelic expression of the HTR2A gene in human brain and peripheral lymphocytes. Biological Psychiatry. 2006;60:1331–1335. doi: 10.1016/j.biopsych.2006.06.024. [DOI] [PubMed] [Google Scholar]
- Girgis RR, Slifstein M, Xu X, Frankle WG, Anagnostou E, Wasserman S, et al. The 5-HT(2A) receptor and serotonin transporter in Asperger’s disorder: A PET study with [(1)(1)C]MDL 100907 and [(1)(1)C]DASB. Psychiatry Research. 2011;194:230–234. doi: 10.1016/j.pscychresns.2011.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg J, Anderson GM, Zwaigenbaum L, Hall GB, Nahmias C, Thompson A, et al. Cortical serotonin type-2 receptor density in parents of children with autism spectrum disorders. Journal of Autism and Developmental Disorders. 2009;39:97–104. doi: 10.1007/s10803-008-0604-4. [DOI] [PubMed] [Google Scholar]
- Gorwood P, Kipman A, Foulon C. The human genetics of anorexia nervosa. European Journal of Pharmacology. 2003;480:163–170. doi: 10.1016/j.ejphar.2003.08.103. [DOI] [PubMed] [Google Scholar]
- Gotham K, Risi S, Pickles A, Lord C. The Autism Diagnostic Observation Schedule: Revised algorithms for improved diagnostic validity. Journal of Autism and Developmental Disorders. 2007;37:613–627. doi: 10.1007/s10803-006-0280-1. [DOI] [PubMed] [Google Scholar]
- Guhathakurta S, Singh AS, Sinha S, Chatterjee A, Ahmed S, Ghosh S, et al. Analysis of serotonin receptor 2A gene (HTR2A): association study with autism spectrum disorder in the Indian population and investigation of the gene expression in peripheral blood leukocytes. Neurochemistry International. 2009;55:754–759. doi: 10.1016/j.neuint.2009.07.008. [DOI] [PubMed] [Google Scholar]
- Hazelwood LA, Sanders-Bush E. His452Tyr polymorphism in the human 5-HT2A receptor destabilizes the signaling conformation. Molecular Pharmacology. 2004;66:1293–1300. [PubMed] [Google Scholar]
- Herault J, Petit E, Martineau J, Cherpi C, Perrot A, Barthelemy C, et al. Serotonin and autism: biochemical and molecular biology features. Psychiatry Research. 1996;65:33–43. doi: 10.1016/0165-1781(96)02882-x. [DOI] [PubMed] [Google Scholar]
- Hranilovic D, Blazevic S, Babic M, Smurinic M, Bujas-Petkovic Z, Jernej B. 5- HT2A receptor gene polymorphisms in Croatian subjects with autistic disorder. Psychiatry Research. 2010;178:556–558. doi: 10.1016/j.psychres.2010.04.007. [DOI] [PubMed] [Google Scholar]
- Hranilovic D, Bujas-Petkovic Z, Tomicic M, Bordukalo-Niksic T, Blazevic S, Cicin-Sain L. Hyperserotonemia in autism: activity of 5HT-associated platelet proteins. Journal of Neural Transmission. 2009;116:493–501. doi: 10.1007/s00702-009-0192-2. [DOI] [PubMed] [Google Scholar]
- Jakab RL, Goldman-Rakic PS. 5-Hydroxytryptamine2A serotonin receptors in the primate cerebral cortex: possible site of action of hallucinogenic and antipsychotic drugs in pyramidal cell apical dendrites. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:735–740. doi: 10.1073/pnas.95.2.735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato M, Serretti A. Review and meta-analysis of antidepressant pharmacogenetic findings in major depressive disorder. Molecular Psychiatry. 2010;15:473–500. doi: 10.1038/mp.2008.116. [DOI] [PubMed] [Google Scholar]
- Kazek B, Huzarska M, Grzybowska-Chlebowczyk U, Kajor M, Ciupinska-Kajor M, Wos H, et al. Platelet and intestinal 5-HT2A receptor mRNA in autistic spectrum disorders - results of a pilot study. Acta Neurobiologiae Experimentalis. 2010;70:232–238. doi: 10.55782/ane-2010-1794. [DOI] [PubMed] [Google Scholar]
- Kling A, Seddighzadeh M, Arlestig L, Alfredsson L, Rantapaa-Dahlqvist S, Padyukov L. Genetic variations in the serotonin 5-HT2A receptor gene (HTR2A) are associated with rheumatoid arthritis. Annals of the Rheumatic Diseases. 2008;67:1111–1115. doi: 10.1136/ard.2007.074948. [DOI] [PubMed] [Google Scholar]
- Lee YH, Choi SJ, Ji JD, Song GG. Candidate gene studies of fibromyalgia: a systematic review and meta-analysis. Rheumatology International. 2012;32:417–426. doi: 10.1007/s00296-010-1678-9. [DOI] [PubMed] [Google Scholar]
- Lerer B, Segman RH, Tan EC, Basile VS, Cavallaro R, Aschauer HN, et al. Combined analysis of 635 patients confirms an age-related association of the serotonin 2A receptor gene with tardive dyskinesia and specificity for the non-orofacial subtype. International Journal of Neuropsychopharmacology. 2005;8:411–425. doi: 10.1017/S1461145705005389. [DOI] [PubMed] [Google Scholar]
- Lord C, Risi S, Lambrecht L, Cook EH, Jr., Leventhal BL, DiLavore PC, et al. The Autism Diagnostic Observation Schedule-Generic: a standard measure of social and communication deficits associated with the spectrum of autism. Journal of Autism and Developmental Disorders. 2000;30:205–223. [PubMed] [Google Scholar]
- Ma D, Salyakina D, Jaworski JM, Konidari I, Whitehead PL, Andersen AN, et al. A genome-wide association study of autism reveals a common novel risk locus at 5p14.1. Annals of Human Genetics. 2009;73:263–273. doi: 10.1111/j.1469-1809.2009.00523.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makkonen I, Riikonen R, Kokki H, Airaksinen MM, Kuikka JT. Serotonin and dopamine transporter binding in children with autism determined by SPECT. Developmental Medicine & Child Neurology. 2008;50:593–597. doi: 10.1111/j.1469-8749.2008.03027.x. [DOI] [PubMed] [Google Scholar]
- Marshall CR, Scherer SW. Detection and characterization of copy number variation in autism spectrum disorder. Methods in Molecular Biology. 2012;838:115–135. doi: 10.1007/978-1-61779-507-7_5. [DOI] [PubMed] [Google Scholar]
- Martaskova D, Slachtova L, Kemlink D, Zahorakova D, Papezova H. Polymorphisms in serotonin-related genes in anorexia nervosa. The first study in Czech population and metaanalyses with previously performed studies. Folia Biologica. 2009;55:192–197. doi: 10.14712/fb2009055050192. [DOI] [PubMed] [Google Scholar]
- Matsunami N, Hadley D, Hensel CH, Christensen GB, Kim C, Frackelton E, et al. Identification of rare recurrent copy number variants in high-risk autism families and their prevalence in a large ASD population. PLoS One. 2013;8:e52239. doi: 10.1371/journal.pone.0052239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride PA, Anderson GM, Hertzig ME, Sweeney JA, Kream J, Cohen DJ, et al. Serotonergic responsivity in male young adults with autistic disorder. Results of a pilot study. Archives of General Psychiatry. 1989;46:213–221. doi: 10.1001/archpsyc.1989.01810030019003. [DOI] [PubMed] [Google Scholar]
- Morrow EM, Yoo SY, Flavell SW, Kim TK, Lin Y, Hill RS, et al. Identifying autism loci and genes by tracing recent shared ancestry. Science. 2008;321:218–223. doi: 10.1126/science.1157657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy DG, Daly E, Schmitz N, Toal F, Murphy K, Curran S, et al. Cortical serotonin 5-HT2A receptor binding and social communication in adults with Asperger’s syndrome: an in vivo SPECT study. American Journal of Psychiatry. 2006;163:934–936. doi: 10.1176/ajp.2006.163.5.934. [DOI] [PubMed] [Google Scholar]
- O’Roak BJ, Deriziotis P, Lee C, Vives L, Schwartz JJ, Girirajan S, et al. Exome sequencing in sporadic autism spectrum disorders identifies severe de novo mutations. Nature Genetics. 2011;43:585–589. doi: 10.1038/ng.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Roak BJ, Vives L, Girirajan S, Karakoc E, Krumm N, Coe BP, et al. Sporadic autism exomes reveal a highly interconnected protein network of de novo mutations. Nature. 2012;485:246–250. doi: 10.1038/nature10989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto D, Pagnamenta AT, Klei L, Anney R, Merico D, Regan R, et al. Functional impact of global rare copy number variation in autism spectrum disorders. Nature. 2010;466:368–372. doi: 10.1038/nature09146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polesskaya OO, Sokolov BP. Differential expression of the "C" and "T" alleles of the 5-HT2A receptor gene in the temporal cortex of normal individuals and schizophrenics. Journal of Neuroscience Research. 2002;67:812–822. doi: 10.1002/jnr.10173. [DOI] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MAR, Bender D, et al. PLINK: a toolset for whole-genome association and population-based linkage analysis. American Journal of Human Genetics. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramanathan S, Glatt SJ. Serotonergic system genes in psychosis of Alzheimer dementia: meta-analysis. American Journal of Geriatric Psychiatry. 2009;17:839–846. doi: 10.1097/JGP.0b013e3181ab8c3f. [DOI] [PubMed] [Google Scholar]
- Scott LJ, Dhillon S. Spotlight on risperidone in irritability associated with autistic disorder in children and adolescents. CNS Drugs. 2008;22:259–262. doi: 10.2165/00023210-200822030-00006. [DOI] [PubMed] [Google Scholar]
- Smith RM, Papp AC, Webb A, Ruble CL, Munsie LM, Nisenbaum LK, et al. Multiple regulatory variants modulate expression of 5-hydroxytryptamine 2A receptors in human cortex. Biological Psychiatry. 2013;73:546–554. doi: 10.1016/j.biopsych.2012.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veenstra-VanderWeele J, Kim SJ, Lord C, Courchesne R, Akshoomoff N, Leventhal BL, et al. Transmission disequilibrium studies of the serotonin 5-HT2A receptor gene (HTR2A) in autism. American Journal of Medical Genetics. 2002;114:277–283. doi: 10.1002/ajmg.10192. [DOI] [PubMed] [Google Scholar]
- Wang K, Zhang H, Ma D, Bucan M, Glessner JT, Abrahams BS, et al. Common genetic variants on 5p14.1 associate with autism spectrum disorders. Nature. 2009;459:528–533. doi: 10.1038/nature07999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss LA, Arking DE, Daly MJ, Chakravarti A. A genome-wide linkage and association scan reveals novel loci for autism. Nature. 2009;461:802–808. doi: 10.1038/nature08490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu TW, Chahrour MH, Coulter ME, Jiralerspong S, Okamura-Ikeda K, Ataman B, et al. Using whole-exome sequencing to identify inherited causes of autism. Neuron. 2013;77:259–273. doi: 10.1016/j.neuron.2012.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zafeiriou DI, Ververi A, Vargiami E. Childhood autism and associated comorbidities. Brain and Development. 2007;29:257–272. doi: 10.1016/j.braindev.2006.09.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.