Abstract
The Aspergillus nidulans brlA locus controls conidiophore development in conjunction with the products of several other regulatory loci. In this paper, we show that the brlA locus consists of overlapping transcription units, designated alpha and beta, with alpha transcription initiating within beta intronic sequences. The predicted BrlA polypeptides differ by 23 amino acid residues at their N-termini. Targeted mutations specifically eliminating either the alpha or beta transcript led to developmental abnormalities similar to those produced by previously identified hypomorphic mutants, showing that both transcripts have essential functions for normal development. However, provision of additional doses of alpha in a beta- strain or of beta in an alpha- strain remediated the developmental defects, indicating that the polypeptides have redundant functions. It is likely that differential regulation of alpha and beta expression in the wild type is important for the initiation and temporal regulation of development.
Full text
PDF



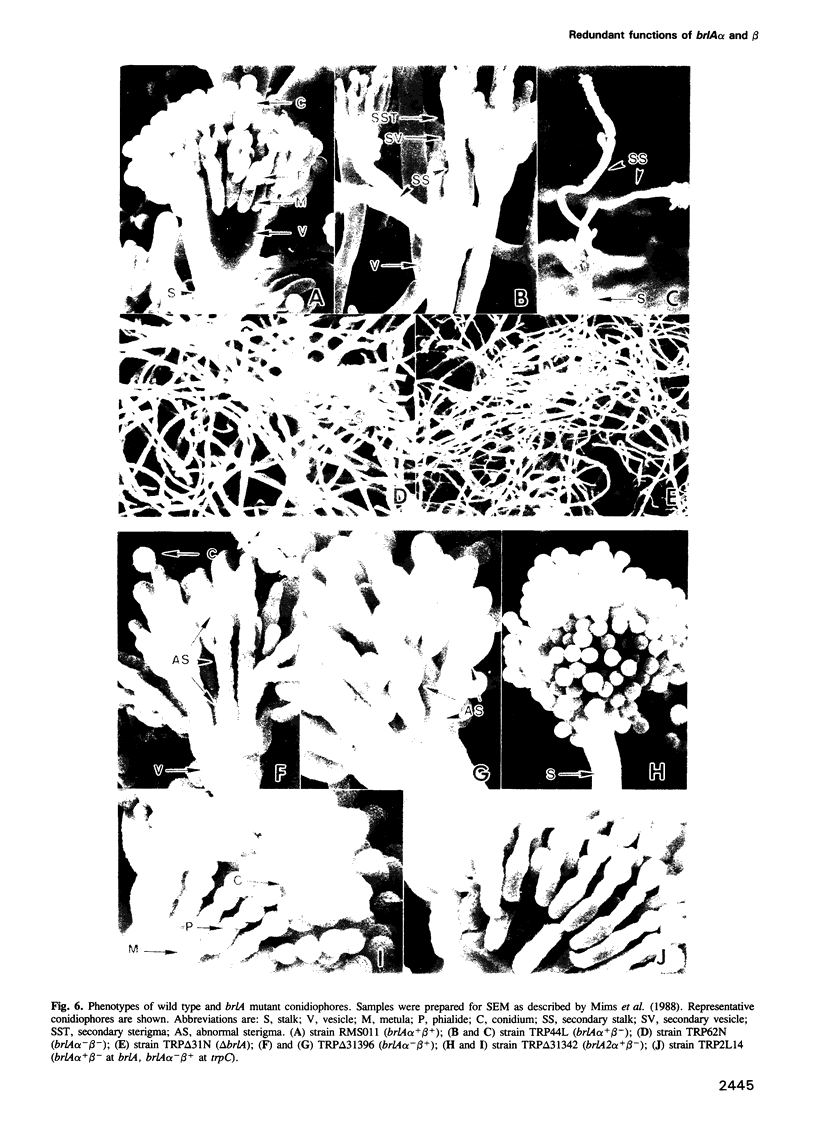


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adams T. H., Boylan M. T., Timberlake W. E. brlA is necessary and sufficient to direct conidiophore development in Aspergillus nidulans. Cell. 1988 Jul 29;54(3):353–362. doi: 10.1016/0092-8674(88)90198-5. [DOI] [PubMed] [Google Scholar]
- Adams T. H., Deising H., Timberlake W. E. brlA requires both zinc fingers to induce development. Mol Cell Biol. 1990 Apr;10(4):1815–1817. doi: 10.1128/mcb.10.4.1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams T. H., Timberlake W. E. Developmental repression of growth and gene expression in Aspergillus. Proc Natl Acad Sci U S A. 1990 Jul;87(14):5405–5409. doi: 10.1073/pnas.87.14.5405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams T. H., Timberlake W. E. Upstream elements repress premature expression of an Aspergillus developmental regulatory gene. Mol Cell Biol. 1990 Sep;10(9):4912–4919. doi: 10.1128/mcb.10.9.4912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aramayo R., Adams T. H., Timberlake W. E. A large cluster of highly expressed genes is dispensable for growth and development in Aspergillus nidulans. Genetics. 1989 May;122(1):65–71. doi: 10.1093/genetics/122.1.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aramayo R., Timberlake W. E. Sequence and molecular structure of the Aspergillus nidulans yA (laccase I) gene. Nucleic Acids Res. 1990 Jun 11;18(11):3415–3415. doi: 10.1093/nar/18.11.3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birse C. E., Clutterbuck A. J. Isolation and developmentally regulated expression of an Aspergillus nidulans phenol oxidase-encoding gene, ivoB. Gene. 1991 Feb 1;98(1):69–76. doi: 10.1016/0378-1119(91)90105-k. [DOI] [PubMed] [Google Scholar]
- Birse C. E., Clutterbuck A. J. N-acetyl-6-hydroxytryptophan oxidase, a developmentally controlled phenol oxidase from Aspergillus nidulans. J Gen Microbiol. 1990 Sep;136(9):1725–1730. doi: 10.1099/00221287-136-9-1725. [DOI] [PubMed] [Google Scholar]
- Boylan M. T., Mirabito P. M., Willett C. E., Zimmerman C. R., Timberlake W. E. Isolation and physical characterization of three essential conidiation genes from Aspergillus nidulans. Mol Cell Biol. 1987 Sep;7(9):3113–3118. doi: 10.1128/mcb.7.9.3113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brody H., Griffith J., Cuticchia A. J., Arnold J., Timberlake W. E. Chromosome-specific recombinant DNA libraries from the fungus Aspergillus nidulans. Nucleic Acids Res. 1991 Jun 11;19(11):3105–3109. doi: 10.1093/nar/19.11.3105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang Y. C., Timberlake W. E. Identification of Aspergillus brlA response elements (BREs) by genetic selection in yeast. Genetics. 1993 Jan;133(1):29–38. doi: 10.1093/genetics/133.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clutterbuck A. J. A mutational analysis of conidial development in Aspergillus nidulans. Genetics. 1969 Oct;63(2):317–327. doi: 10.1093/genetics/63.2.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clutterbuck A. J., Stark M. S., Gupta G. An intragenic map of the brlA locus of Aspergillus nidulans. Mol Gen Genet. 1992 Jan;231(2):212–216. doi: 10.1007/BF00279793. [DOI] [PubMed] [Google Scholar]
- Clutterbuck A. J. The genetics of conidiophore pigmentation in Aspergillus nidulans. J Gen Microbiol. 1990 Sep;136(9):1731–1738. doi: 10.1099/00221287-136-9-1731. [DOI] [PubMed] [Google Scholar]
- Johnstone I. L., Hughes S. G., Clutterbuck A. J. Cloning an Aspergillus nidulans developmental gene by transformation. EMBO J. 1985 May;4(5):1307–1311. doi: 10.1002/j.1460-2075.1985.tb03777.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel T. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc Natl Acad Sci U S A. 1985 Jan;82(2):488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Käfer E. Meiotic and mitotic recombination in Aspergillus and its chromosomal aberrations. Adv Genet. 1977;19:33–131. doi: 10.1016/s0065-2660(08)60245-x. [DOI] [PubMed] [Google Scholar]
- Law D. J., Timberlake W. E. Developmental regulation of laccase levels in Aspergillus nidulans. J Bacteriol. 1980 Nov;144(2):509–517. doi: 10.1128/jb.144.2.509-517.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall M. A., Timberlake W. E. Aspergillus nidulans wetA activates spore-specific gene expression. Mol Cell Biol. 1991 Jan;11(1):55–62. doi: 10.1128/mcb.11.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinelli S. D. Phenotypes of double conidiation mutants of Aspergillus nidulans. J Gen Microbiol. 1979 Oct;114(2):277–287. doi: 10.1099/00221287-114-2-277. [DOI] [PubMed] [Google Scholar]
- Mayorga M. E., Timberlake W. E. Isolation and molecular characterization of the Aspergillus nidulans wA gene. Genetics. 1990 Sep;126(1):73–79. doi: 10.1093/genetics/126.1.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller K. Y., Toennis T. M., Adams T. H., Miller B. L. Isolation and transcriptional characterization of a morphological modifier: the Aspergillus nidulans stunted (stuA) gene. Mol Gen Genet. 1991 Jun;227(2):285–292. doi: 10.1007/BF00259682. [DOI] [PubMed] [Google Scholar]
- Miller K. Y., Wu J., Miller B. L. StuA is required for cell pattern formation in Aspergillus. Genes Dev. 1992 Sep;6(9):1770–1782. doi: 10.1101/gad.6.9.1770. [DOI] [PubMed] [Google Scholar]
- Mirabito P. M., Adams T. H., Timberlake W. E. Interactions of three sequentially expressed genes control temporal and spatial specificity in Aspergillus development. Cell. 1989 Jun 2;57(5):859–868. doi: 10.1016/0092-8674(89)90800-3. [DOI] [PubMed] [Google Scholar]
- O'Hara E. B., Timberlake W. E. Molecular characterization of the Aspergillus nidulans yA locus. Genetics. 1989 Feb;121(2):249–254. doi: 10.1093/genetics/121.2.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver P. T. Conidiophore and spore development in Aspergillus nidulans. J Gen Microbiol. 1972 Nov;73(1):45–54. doi: 10.1099/00221287-73-1-45. [DOI] [PubMed] [Google Scholar]
- PONTECORVO G., ROPER J. A., HEMMONS L. M., MACDONALD K. D., BUFTON A. W. J. The genetics of Aspergillus nidulans. Adv Genet. 1953;5:141–238. doi: 10.1016/s0065-2660(08)60408-3. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sewall T. C., Mims C. W., Timberlake W. E. Conidium differentiation in Aspergillus nidulans wild-type and wet-white (wetA) mutant strains. Dev Biol. 1990 Apr;138(2):499–508. doi: 10.1016/0012-1606(90)90215-5. [DOI] [PubMed] [Google Scholar]
- Stringer M. A., Dean R. A., Sewall T. C., Timberlake W. E. Rodletless, a new Aspergillus developmental mutant induced by directed gene inactivation. Genes Dev. 1991 Jul;5(7):1161–1171. doi: 10.1101/gad.5.7.1161. [DOI] [PubMed] [Google Scholar]
- Tilburn J., Roussel F., Scazzocchio C. Insertional inactivation and cloning of the wA gene of Aspergillus nidulans. Genetics. 1990 Sep;126(1):81–90. doi: 10.1093/genetics/126.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timberlake W. E. Developmental gene regulation in Aspergillus nidulans. Dev Biol. 1980 Aug;78(2):497–510. doi: 10.1016/0012-1606(80)90349-8. [DOI] [PubMed] [Google Scholar]
- Timberlake W. E. Molecular genetics of Aspergillus development. Annu Rev Genet. 1990;24:5–36. doi: 10.1146/annurev.ge.24.120190.000253. [DOI] [PubMed] [Google Scholar]
- Timberlake W. E. Temporal and spatial controls of Aspergillus development. Curr Opin Genet Dev. 1991 Oct;1(3):351–357. doi: 10.1016/s0959-437x(05)80299-0. [DOI] [PubMed] [Google Scholar]
- Yelton M. M., Hamer J. E., Timberlake W. E. Transformation of Aspergillus nidulans by using a trpC plasmid. Proc Natl Acad Sci U S A. 1984 Mar;81(5):1470–1474. doi: 10.1073/pnas.81.5.1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann C. R., Orr W. C., Leclerc R. F., Barnard E. C., Timberlake W. E. Molecular cloning and selection of genes regulated in Aspergillus development. Cell. 1980 Oct;21(3):709–715. doi: 10.1016/0092-8674(80)90434-1. [DOI] [PubMed] [Google Scholar]