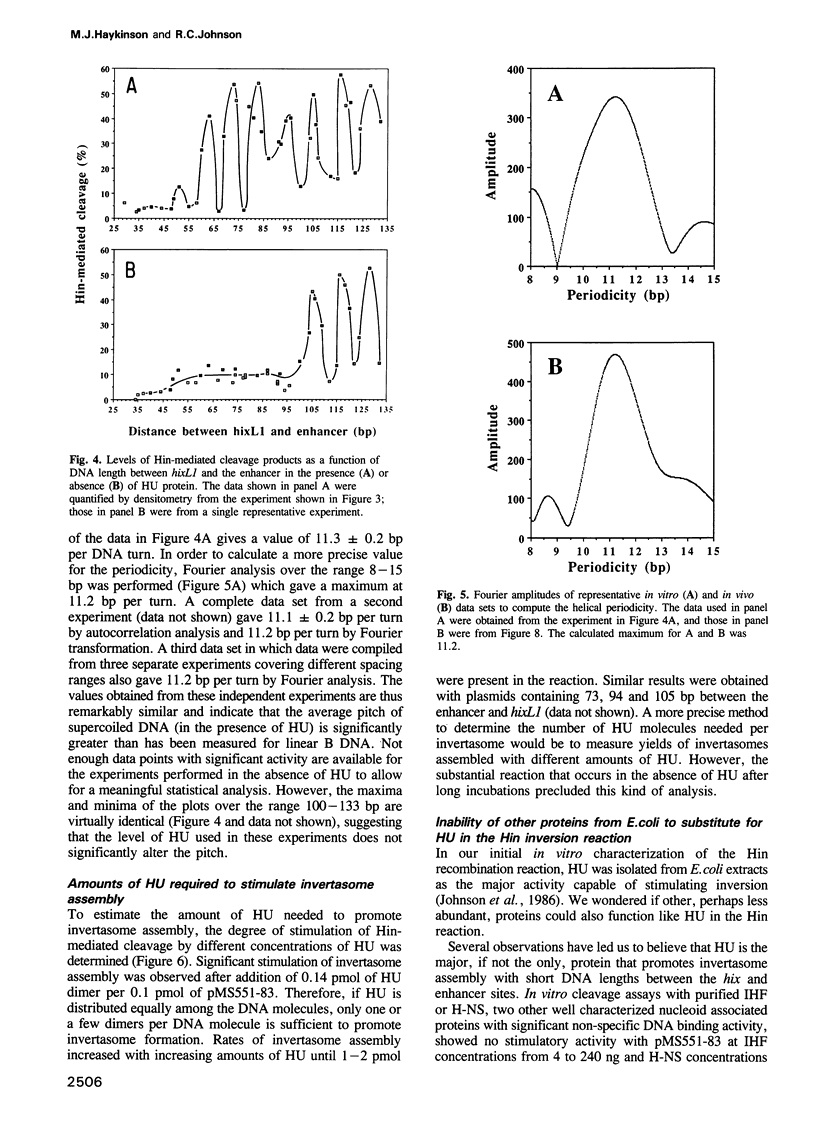
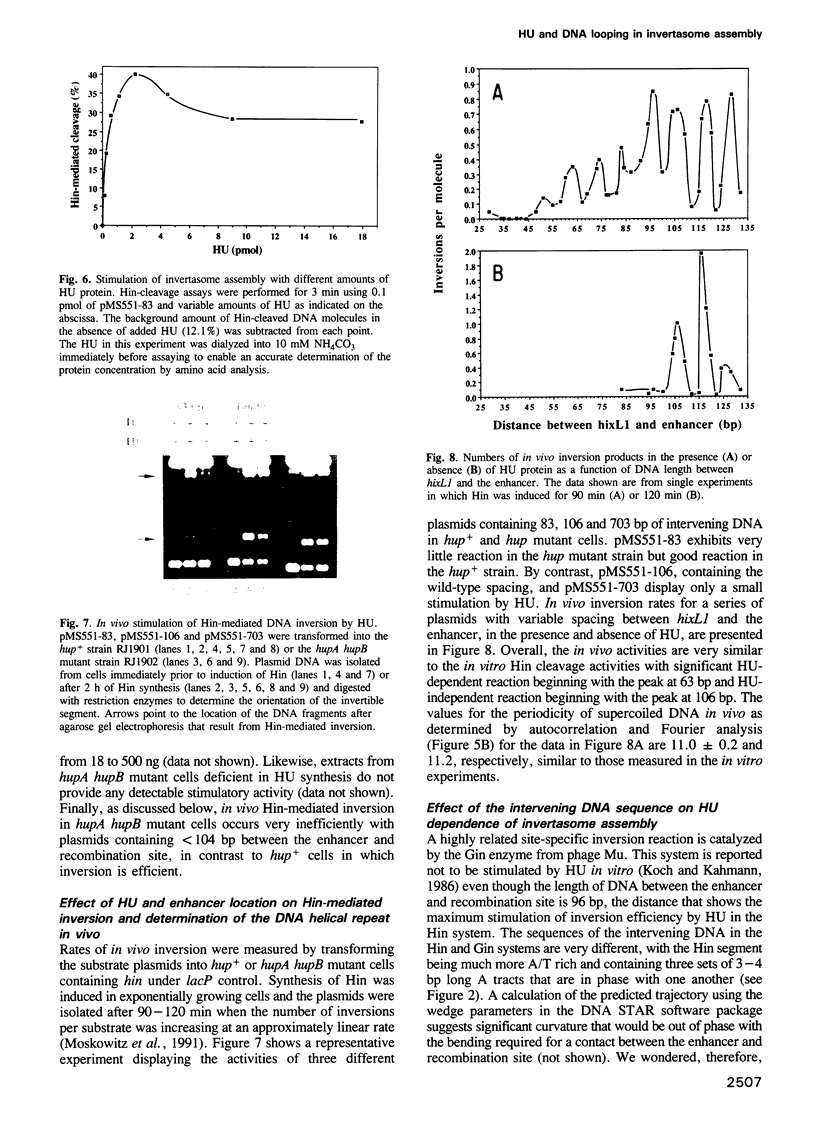
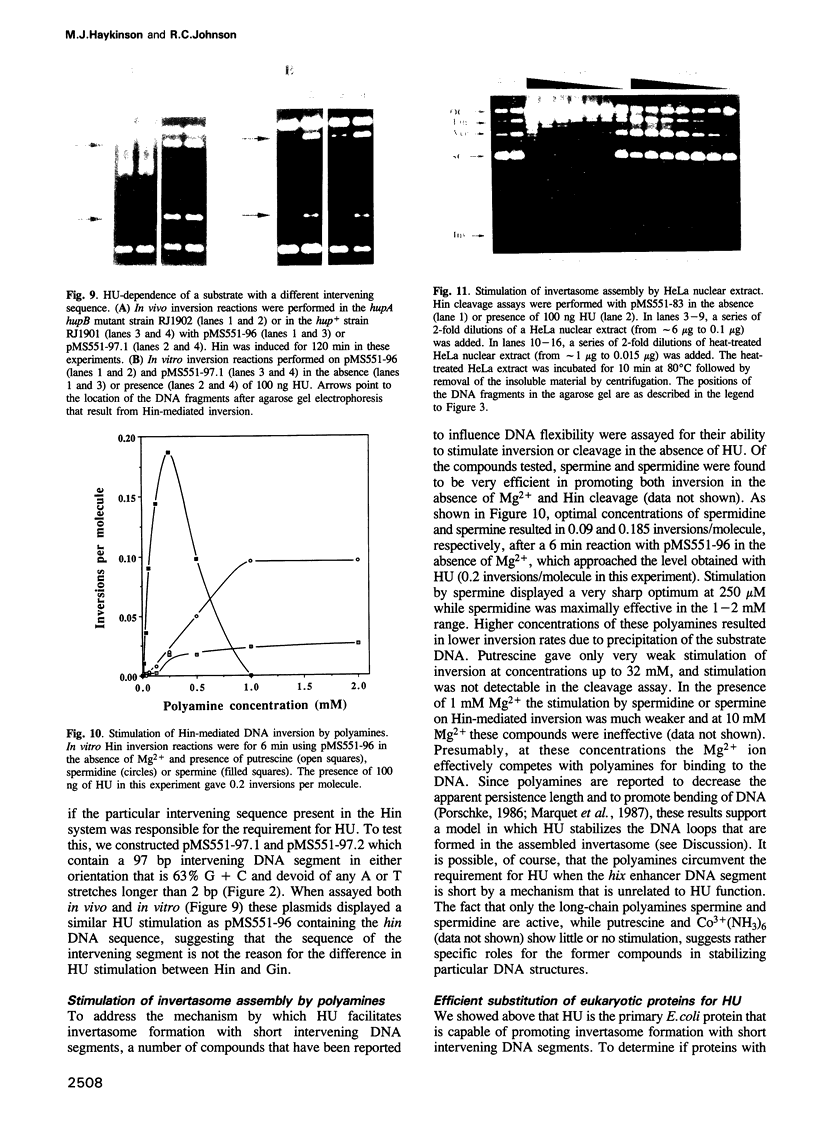
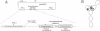Abstract
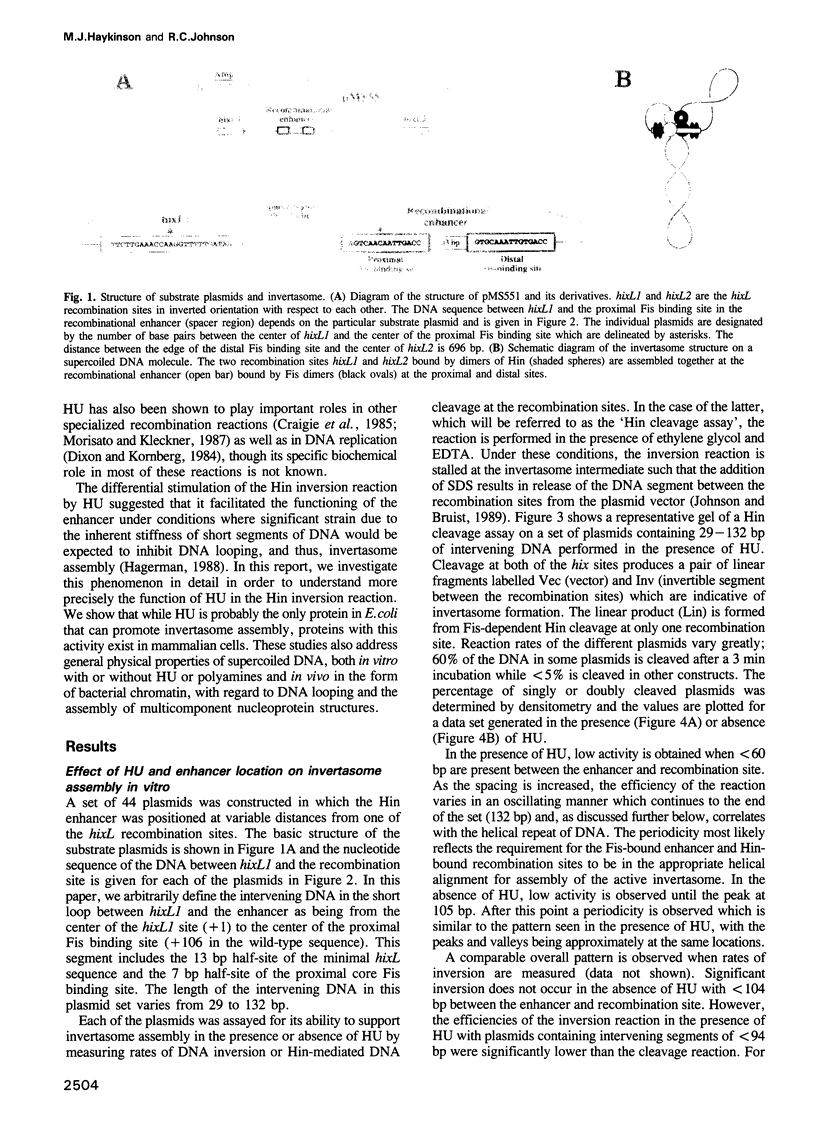
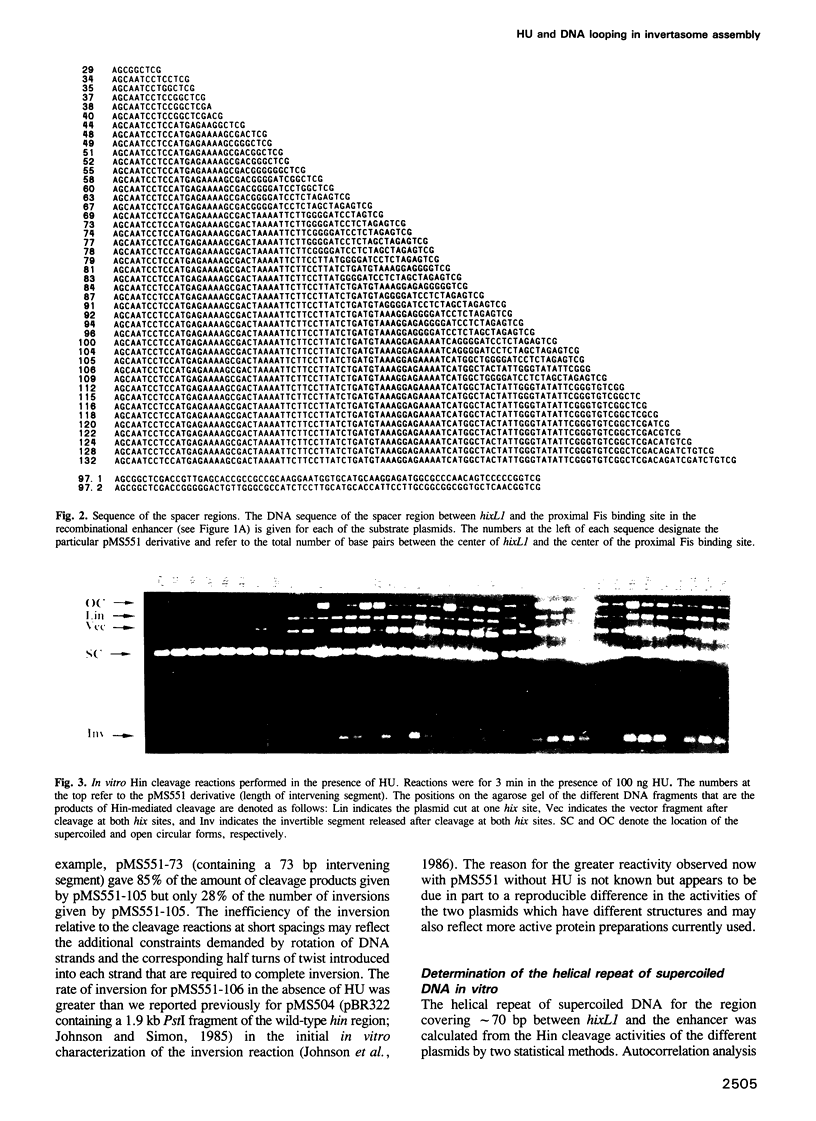
Site-specific DNA inversion by the Hin recombinase requires the formation of a multicomponent nucleo-protein structure called an invertasome. In this structure, the two recombination sites bound by Hin are assembled together at the Fis-bound recombinational enhancer with the requisite looping of the intervening DNA segments. We have analyzed the role of the HU protein in invertasome assembly when the enhancer is located at variable positions close to one of the recombination sites. In the absence of HU in vitro and in hupA hupB mutant cells in vivo, invertasome assembly is very inefficient when there is < 104 bp of DNA between the enhancer and recombination site. Invertasome assembly in the presence of HU in vitro or in vivo displayed a periodicity beginning with 60 bp of intervening DNA that reflected its helical repeat. The average helical repeat for this DNA region was calculated by autocorrelation and Fourier transformation to be 11.2 bp per turn for supercoiled DNA both in the presence of HU in vitro and in hup+ cells in vivo. HU is the only protein in Escherichia coli that can promote invertasome formation with short DNA lengths between the enhancer and recombination sites. However, the presence of certain polyamines and a protein activity present in HeLa nuclear extracts can efficiently substitute for HU in invertasome assembly. These data support a model in which HU binds non-specifically to the DNA between the enhancer and recombination site to facilitate DNA looping.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Abraham J. M., Freitag C. S., Clements J. R., Eisenstein B. I. An invertible element of DNA controls phase variation of type 1 fimbriae of Escherichia coli. Proc Natl Acad Sci U S A. 1985 Sep;82(17):5724–5727. doi: 10.1073/pnas.82.17.5724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adhya S. Multipartite genetic control elements: communication by DNA loop. Annu Rev Genet. 1989;23:227–250. doi: 10.1146/annurev.ge.23.120189.001303. [DOI] [PubMed] [Google Scholar]
- Bellomy G. R., Record M. T., Jr Stable DNA loops in vivo and in vitro: roles in gene regulation at a distance and in biophysical characterization of DNA. Prog Nucleic Acid Res Mol Biol. 1990;39:81–128. doi: 10.1016/s0079-6603(08)60624-8. [DOI] [PubMed] [Google Scholar]
- Bianchi M. E., Beltrame M., Paonessa G. Specific recognition of cruciform DNA by nuclear protein HMG1. Science. 1989 Feb 24;243(4894 Pt 1):1056–1059. doi: 10.1126/science.2922595. [DOI] [PubMed] [Google Scholar]
- Bliska J. B., Cozzarelli N. R. Use of site-specific recombination as a probe of DNA structure and metabolism in vivo. J Mol Biol. 1987 Mar 20;194(2):205–218. doi: 10.1016/0022-2836(87)90369-x. [DOI] [PubMed] [Google Scholar]
- Bonnefoy E., Rouvière-Yaniv J. HU, the major histone-like protein of E. coli, modulates the binding of IHF to oriC. EMBO J. 1992 Dec;11(12):4489–4496. doi: 10.1002/j.1460-2075.1992.tb05550.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broyles S. S., Pettijohn D. E. Interaction of the Escherichia coli HU protein with DNA. Evidence for formation of nucleosome-like structures with altered DNA helical pitch. J Mol Biol. 1986 Jan 5;187(1):47–60. doi: 10.1016/0022-2836(86)90405-5. [DOI] [PubMed] [Google Scholar]
- Bruist M. F., Glasgow A. C., Johnson R. C., Simon M. I. Fis binding to the recombinational enhancer of the Hin DNA inversion system. Genes Dev. 1987 Oct;1(8):762–772. doi: 10.1101/gad.1.8.762. [DOI] [PubMed] [Google Scholar]
- Bruist M. F., Simon M. I. Phase variation and the Hin protein: in vivo activity measurements, protein overproduction, and purification. J Bacteriol. 1984 Jul;159(1):71–79. doi: 10.1128/jb.159.1.71-79.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craigie R., Arndt-Jovin D. J., Mizuuchi K. A defined system for the DNA strand-transfer reaction at the initiation of bacteriophage Mu transposition: protein and DNA substrate requirements. Proc Natl Acad Sci U S A. 1985 Nov;82(22):7570–7574. doi: 10.1073/pnas.82.22.7570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dignam J. D., Martin P. L., Shastry B. S., Roeder R. G. Eukaryotic gene transcription with purified components. Methods Enzymol. 1983;101:582–598. doi: 10.1016/0076-6879(83)01039-3. [DOI] [PubMed] [Google Scholar]
- Dixon N. E., Kornberg A. Protein HU in the enzymatic replication of the chromosomal origin of Escherichia coli. Proc Natl Acad Sci U S A. 1984 Jan;81(2):424–428. doi: 10.1073/pnas.81.2.424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drlica K., Rouviere-Yaniv J. Histonelike proteins of bacteria. Microbiol Rev. 1987 Sep;51(3):301–319. doi: 10.1128/mr.51.3.301-319.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Echols H. Multiple DNA-protein interactions governing high-precision DNA transactions. Science. 1986 Sep 5;233(4768):1050–1056. doi: 10.1126/science.2943018. [DOI] [PubMed] [Google Scholar]
- Echols H. Nucleoprotein structures initiating DNA replication, transcription, and site-specific recombination. J Biol Chem. 1990 Sep 5;265(25):14697–14700. [PubMed] [Google Scholar]
- Feuerstein B. G., Pattabiraman N., Marton L. J. Molecular mechanics of the interactions of spermine with DNA: DNA bending as a result of ligand binding. Nucleic Acids Res. 1990 Mar 11;18(5):1271–1282. doi: 10.1093/nar/18.5.1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flashner Y., Gralla J. D. DNA dynamic flexibility and protein recognition: differential stimulation by bacterial histone-like protein HU. Cell. 1988 Aug 26;54(5):713–721. doi: 10.1016/s0092-8674(88)80016-3. [DOI] [PubMed] [Google Scholar]
- Gellert M., Nash H. Communication between segments of DNA during site-specific recombination. 1987 Jan 29-Feb 4Nature. 325(6103):401–404. doi: 10.1038/325401a0. [DOI] [PubMed] [Google Scholar]
- Glasgow A. C., Bruist M. F., Simon M. I. DNA-binding properties of the Hin recombinase. J Biol Chem. 1989 Jun 15;264(17):10072–10082. [PubMed] [Google Scholar]
- Gober J. W., Shapiro L. Integration host factor is required for the activation of developmentally regulated genes in Caulobacter. Genes Dev. 1990 Sep;4(9):1494–1504. doi: 10.1101/gad.4.9.1494. [DOI] [PubMed] [Google Scholar]
- Goodman S. D., Nash H. A. Functional replacement of a protein-induced bend in a DNA recombination site. Nature. 1989 Sep 21;341(6239):251–254. doi: 10.1038/341251a0. [DOI] [PubMed] [Google Scholar]
- Goodman S. D., Nicholson S. C., Nash H. A. Deformation of DNA during site-specific recombination of bacteriophage lambda: replacement of IHF protein by HU protein or sequence-directed bends. Proc Natl Acad Sci U S A. 1992 Dec 15;89(24):11910–11914. doi: 10.1073/pnas.89.24.11910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagerman P. J. Flexibility of DNA. Annu Rev Biophys Biophys Chem. 1988;17:265–286. doi: 10.1146/annurev.bb.17.060188.001405. [DOI] [PubMed] [Google Scholar]
- Heichman K. A., Johnson R. C. The Hin invertasome: protein-mediated joining of distant recombination sites at the enhancer. Science. 1990 Aug 3;249(4968):511–517. doi: 10.1126/science.2166334. [DOI] [PubMed] [Google Scholar]
- Heichman K. A., Moskowitz I. P., Johnson R. C. Configuration of DNA strands and mechanism of strand exchange in the Hin invertasome as revealed by analysis of recombinant knots. Genes Dev. 1991 Sep;5(9):1622–1634. doi: 10.1101/gad.5.9.1622. [DOI] [PubMed] [Google Scholar]
- Hillyard D. R., Edlund M., Hughes K. T., Marsh M., Higgins N. P. Subunit-specific phenotypes of Salmonella typhimurium HU mutants. J Bacteriol. 1990 Sep;172(9):5402–5407. doi: 10.1128/jb.172.9.5402-5407.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodges-Garcia Y., Hagerman P. J., Pettijohn D. E. DNA ring closure mediated by protein HU. J Biol Chem. 1989 Sep 5;264(25):14621–14623. [PubMed] [Google Scholar]
- Hoover T. R., Santero E., Porter S., Kustu S. The integration host factor stimulates interaction of RNA polymerase with NIFA, the transcriptional activator for nitrogen fixation operons. Cell. 1990 Oct 5;63(1):11–22. doi: 10.1016/0092-8674(90)90284-l. [DOI] [PubMed] [Google Scholar]
- Johnson R. C., Bruist M. F. Intermediates in Hin-mediated DNA inversion: a role for Fis and the recombinational enhancer in the strand exchange reaction. EMBO J. 1989 May;8(5):1581–1590. doi: 10.1002/j.1460-2075.1989.tb03542.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson R. C., Bruist M. F., Simon M. I. Host protein requirements for in vitro site-specific DNA inversion. Cell. 1986 Aug 15;46(4):531–539. doi: 10.1016/0092-8674(86)90878-0. [DOI] [PubMed] [Google Scholar]
- Johnson R. C., Glasgow A. C., Simon M. I. Spatial relationship of the Fis binding sites for Hin recombinational enhancer activity. Nature. 1987 Oct 1;329(6138):462–465. doi: 10.1038/329462a0. [DOI] [PubMed] [Google Scholar]
- Johnson R. C. Mechanism of site-specific DNA inversion in bacteria. Curr Opin Genet Dev. 1991 Oct;1(3):404–411. doi: 10.1016/s0959-437x(05)80307-7. [DOI] [PubMed] [Google Scholar]
- Johnson R. C., Simon M. I. Hin-mediated site-specific recombination requires two 26 bp recombination sites and a 60 bp recombinational enhancer. Cell. 1985 Jul;41(3):781–791. doi: 10.1016/s0092-8674(85)80059-3. [DOI] [PubMed] [Google Scholar]
- Kanaar R., Klippel A., Shekhtman E., Dungan J. M., Kahmann R., Cozzarelli N. R. Processive recombination by the phage Mu Gin system: implications for the mechanisms of DNA strand exchange, DNA site alignment, and enhancer action. Cell. 1990 Jul 27;62(2):353–366. doi: 10.1016/0092-8674(90)90372-l. [DOI] [PubMed] [Google Scholar]
- Kanaar R., van de Putte P., Cozzarelli N. R. Gin-mediated DNA inversion: product structure and the mechanism of strand exchange. Proc Natl Acad Sci U S A. 1988 Feb;85(3):752–756. doi: 10.1073/pnas.85.3.752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S., Moitoso de Vargas L., Nunes-Düby S. E., Landy A. Mapping of a higher order protein-DNA complex: two kinds of long-range interactions in lambda attL. Cell. 1990 Nov 16;63(4):773–781. doi: 10.1016/0092-8674(90)90143-3. [DOI] [PubMed] [Google Scholar]
- Koch C., Kahmann R. Purification and properties of the Escherichia coli host factor required for inversion of the G segment in bacteriophage Mu. J Biol Chem. 1986 Nov 25;261(33):15673–15678. [PubMed] [Google Scholar]
- Landy A. Dynamic, structural, and regulatory aspects of lambda site-specific recombination. Annu Rev Biochem. 1989;58:913–949. doi: 10.1146/annurev.bi.58.070189.004405. [DOI] [PubMed] [Google Scholar]
- Lavoie B. D., Chaconas G. Immunoelectron microscopic analysis of the A, B, and HU protein content of bacteriophage Mu transpososomes. J Biol Chem. 1990 Jan 25;265(3):1623–1627. [PubMed] [Google Scholar]
- Lee D. H., Schleif R. F. In vivo DNA loops in araCBAD: size limits and helical repeat. Proc Natl Acad Sci U S A. 1989 Jan;86(2):476–480. doi: 10.1073/pnas.86.2.476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marquet R., Wyart A., Houssier C. Influence of DNA length on spermine-induced condensation. Importance of the bending and stiffening of DNA. Biochim Biophys Acta. 1987 Aug 25;909(3):165–172. doi: 10.1016/0167-4781(87)90074-1. [DOI] [PubMed] [Google Scholar]
- Morisato D., Kleckner N. Tn10 transposition and circle formation in vitro. Cell. 1987 Oct 9;51(1):101–111. doi: 10.1016/0092-8674(87)90014-6. [DOI] [PubMed] [Google Scholar]
- Moskowitz I. P., Heichman K. A., Johnson R. C. Alignment of recombination sites in Hin-mediated site-specific DNA recombination. Genes Dev. 1991 Sep;5(9):1635–1645. doi: 10.1101/gad.5.9.1635. [DOI] [PubMed] [Google Scholar]
- Osuna R., Finkel S. E., Johnson R. C. Identification of two functional regions in Fis: the N-terminus is required to promote Hin-mediated DNA inversion but not lambda excision. EMBO J. 1991 Jun;10(6):1593–1603. doi: 10.1002/j.1460-2075.1991.tb07680.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettijohn D. E., Pfenninger O. Supercoils in prokaryotic DNA restrained in vivo. Proc Natl Acad Sci U S A. 1980 Mar;77(3):1331–1335. doi: 10.1073/pnas.77.3.1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porschke D. Structure and dynamics of double helices in solution: modes of DNA bending. J Biomol Struct Dyn. 1986 Dec;4(3):373–389. doi: 10.1080/07391102.1986.10506356. [DOI] [PubMed] [Google Scholar]
- Ptashne M. Gene regulation by proteins acting nearby and at a distance. Nature. 1986 Aug 21;322(6081):697–701. doi: 10.1038/322697a0. [DOI] [PubMed] [Google Scholar]
- Rhodes D., Klug A. Helical periodicity of DNA determined by enzyme digestion. Nature. 1980 Aug 7;286(5773):573–578. doi: 10.1038/286573a0. [DOI] [PubMed] [Google Scholar]
- Rouvière-Yaniv J., Gros F. Characterization of a novel, low-molecular-weight DNA-binding protein from Escherichia coli. Proc Natl Acad Sci U S A. 1975 Sep;72(9):3428–3432. doi: 10.1073/pnas.72.9.3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouvière-Yaniv J., Yaniv M., Germond J. E. E. coli DNA binding protein HU forms nucleosomelike structure with circular double-stranded DNA. Cell. 1979 Jun;17(2):265–274. doi: 10.1016/0092-8674(79)90152-1. [DOI] [PubMed] [Google Scholar]
- Santero E., Hoover T. R., North A. K., Berger D. K., Porter S. C., Kustu S. Role of integration host factor in stimulating transcription from the sigma 54-dependent nifH promoter. J Mol Biol. 1992 Oct 5;227(3):602–620. doi: 10.1016/0022-2836(92)90211-2. [DOI] [PubMed] [Google Scholar]
- Schleif R. DNA looping. Annu Rev Biochem. 1992;61:199–223. doi: 10.1146/annurev.bi.61.070192.001215. [DOI] [PubMed] [Google Scholar]
- Schmid M. B. Structure and function of the bacterial chromosome. Trends Biochem Sci. 1988 Apr;13(4):131–135. doi: 10.1016/0968-0004(88)90069-2. [DOI] [PubMed] [Google Scholar]
- Schneider G. J., Sayre M. H., Geiduschek E. P. DNA-bending properties of TF1. J Mol Biol. 1991 Oct 5;221(3):777–794. doi: 10.1016/0022-2836(91)80175-t. [DOI] [PubMed] [Google Scholar]
- Shore D., Baldwin R. L. Energetics of DNA twisting. I. Relation between twist and cyclization probability. J Mol Biol. 1983 Nov 15;170(4):957–981. doi: 10.1016/s0022-2836(83)80198-3. [DOI] [PubMed] [Google Scholar]
- Skarstad K., Baker T. A., Kornberg A. Strand separation required for initiation of replication at the chromosomal origin of E.coli is facilitated by a distant RNA--DNA hybrid. EMBO J. 1990 Jul;9(7):2341–2348. doi: 10.1002/j.1460-2075.1990.tb07406.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenzel T. T., MacAllister T., Bastia D. Cooperativity at a distance promoted by the combined action of two replication initiator proteins and a DNA bending protein at the replication origin of pSC101. Genes Dev. 1991 Aug;5(8):1453–1463. doi: 10.1101/gad.5.8.1453. [DOI] [PubMed] [Google Scholar]
- Surette M. G., Chaconas G. A protein factor which reduces the negative supercoiling requirement in the Mu DNA strand transfer reaction is Escherichia coli integration host factor. J Biol Chem. 1989 Feb 15;264(5):3028–3034. [PubMed] [Google Scholar]
- Tabor C. W., Tabor H. Polyamines in microorganisms. Microbiol Rev. 1985 Mar;49(1):81–99. doi: 10.1128/mr.49.1.81-99.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tullius T. D., Dombroski B. A. Iron(II) EDTA used to measure the helical twist along any DNA molecule. Science. 1985 Nov 8;230(4726):679–681. doi: 10.1126/science.2996145. [DOI] [PubMed] [Google Scholar]
- Wada M., Kutsukake K., Komano T., Imamoto F., Kano Y. Participation of the hup gene product in site-specific DNA inversion in Escherichia coli. Gene. 1989;76(2):345–352. doi: 10.1016/0378-1119(89)90174-1. [DOI] [PubMed] [Google Scholar]
- Wang J. C. Helical repeat of DNA in solution. Proc Natl Acad Sci U S A. 1979 Jan;76(1):200–203. doi: 10.1073/pnas.76.1.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasserman S. A., White J. H., Cozzarelli N. R. The helical repeat of double-stranded DNA varies as a function of catenation and supercoiling. Nature. 1988 Aug 4;334(6181):448–450. doi: 10.1038/334448a0. [DOI] [PubMed] [Google Scholar]
- White S. W., Appelt K., Wilson K. S., Tanaka I. A protein structural motif that bends DNA. Proteins. 1989;5(4):281–288. doi: 10.1002/prot.340050405. [DOI] [PubMed] [Google Scholar]
- Yang C. C., Nash H. A. The interaction of E. coli IHF protein with its specific binding sites. Cell. 1989 Jun 2;57(5):869–880. doi: 10.1016/0092-8674(89)90801-5. [DOI] [PubMed] [Google Scholar]
- Zacharias W., Jaworski A., Larson J. E., Wells R. D. The B- to Z-DNA equilibrium in vivo is perturbed by biological processes. Proc Natl Acad Sci U S A. 1988 Oct;85(19):7069–7073. doi: 10.1073/pnas.85.19.7069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zieg J., Simon M. Analysis of the nucleotide sequence of an invertible controlling element. Proc Natl Acad Sci U S A. 1980 Jul;77(7):4196–4200. doi: 10.1073/pnas.77.7.4196. [DOI] [PMC free article] [PubMed] [Google Scholar]