Abstract
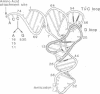
The CCA trinucleotide is a universally conserved feature of the 3' end of tRNAs, where it serves as the site of amino acid attachment. Despite this extreme conservation, we have isolated functional mutants of tRNA(His) and tRNA(Val1) with altered CCA ends. A mutant that leads to de-repression of the histidine biosynthetic operon in Salmonella typhimurium has been characterized and found to have the CCA end of the sole tRNA(His) species mutated to UCA. However, constructed mutants of tRNA(His) with ACA or GCA ends appeared to be nonfunctional in vivo. Mutants of Escherichia coli tRNA(Val1) with GCA or ACA ends were isolated on the basis of their ability to promote frameshifting at a specific sequence. These same tRNA(Val1) mutants also caused read-through of stop codons that were one, or in some instances two, codons downstream of the valine codon decoded by the mutant tRNA. A startling implication of these data is that disruption of interactions between the CCA end of the tRNA and the large ribosomal subunit promotes these aberrant codon-anticodon interactions on the small ribosomal subunit.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Atkins J. F., Nichols B. P., Thompson S. The nucleotide sequence of the first externally suppressible--1 frameshift mutant, and of some nearby leaky frameshift mutants. EMBO J. 1983;2(8):1345–1350. doi: 10.1002/j.1460-2075.1983.tb01590.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkins J. F., Ryce S. UGA and non-triplet suppressor reading of the genetic code. Nature. 1974 Jun 7;249(457):527–530. doi: 10.1038/249527a0. [DOI] [PubMed] [Google Scholar]
- Atkins J. F., Weiss R. B., Thompson S., Gesteland R. F. Towards a genetic dissection of the basis of triplet decoding, and its natural subversion: programmed reading frame shifts and hops. Annu Rev Genet. 1991;25:201–228. doi: 10.1146/annurev.ge.25.120191.001221. [DOI] [PubMed] [Google Scholar]
- Ayer D., Yarus M. The context effect does not require a fourth base pair. Science. 1986 Jan 24;231(4736):393–395. doi: 10.1126/science.3510456. [DOI] [PubMed] [Google Scholar]
- Bachmann B. J. Linkage map of Escherichia coli K-12, edition 8. Microbiol Rev. 1990 Jun;54(2):130–197. doi: 10.1128/mr.54.2.130-197.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergemann K., Nierhaus K. H. Codon-anticodon interaction at the ribosomal P site improves the accuracy of the decoding process. Biochem Int. 1984 Jan;8(1):121–126. [PubMed] [Google Scholar]
- Bitner R. M., Kuempel P. L. P1 transduction map spanning the replication terminus of Escherichia coli K12. Mol Gen Genet. 1981;184(2):208–212. doi: 10.1007/BF00272906. [DOI] [PubMed] [Google Scholar]
- Bossi L., Smith D. M. Conformational change in the DNA associated with an unusual promoter mutation in a tRNA operon of Salmonella. Cell. 1984 Dec;39(3 Pt 2):643–652. doi: 10.1016/0092-8674(84)90471-9. [DOI] [PubMed] [Google Scholar]
- Bossi L. The hisR locus of Salmonella: nucleotide sequence and expression. Mol Gen Genet. 1983;192(1-2):163–170. doi: 10.1007/BF00327662. [DOI] [PubMed] [Google Scholar]
- Brenner M., Ames B. N. Histidine regulation in Salmonella typhimurium. IX. Histidine transfer ribonucleic acid of the regulatory mutants. J Biol Chem. 1972 Feb 25;247(4):1080–1088. [PubMed] [Google Scholar]
- Brosius J., Holy A. Regulation of ribosomal RNA promoters with a synthetic lac operator. Proc Natl Acad Sci U S A. 1984 Nov;81(22):6929–6933. doi: 10.1073/pnas.81.22.6929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang A. C., Cohen S. N. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J Bacteriol. 1978 Jun;134(3):1141–1156. doi: 10.1128/jb.134.3.1141-1156.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chopra I., Shales S. W., Ward J. M., Wallace L. J. Reduced expression of Tn 10-mediated tetracycline resistance in Escherichia coli containing more than one copy of the transposon. J Gen Microbiol. 1981 Sep;126(1):45–54. doi: 10.1099/00221287-126-1-45. [DOI] [PubMed] [Google Scholar]
- Church G. M., Gilbert W. Genomic sequencing. Proc Natl Acad Sci U S A. 1984 Apr;81(7):1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies J., Jones D. S., Khorana H. G. A further study of misreading of codons induced by streptomycin and neomycin using ribopolynucleotides containing two nucleotides in alternating sequence as templates. J Mol Biol. 1966 Jun;18(1):48–57. doi: 10.1016/s0022-2836(66)80075-x. [DOI] [PubMed] [Google Scholar]
- Deutscher M. P. Ribonucleases, tRNA nucleotidyltransferase, and the 3' processing of tRNA. Prog Nucleic Acid Res Mol Biol. 1990;39:209–240. doi: 10.1016/s0079-6603(08)60628-5. [DOI] [PubMed] [Google Scholar]
- Eriani G., Delarue M., Poch O., Gangloff J., Moras D. Partition of tRNA synthetases into two classes based on mutually exclusive sets of sequence motifs. Nature. 1990 Sep 13;347(6289):203–206. doi: 10.1038/347203a0. [DOI] [PubMed] [Google Scholar]
- Francklyn C., Musier-Forsyth K., Schimmel P. Small RNA helices as substrates for aminoacylation and their relationship to charging of transfer RNAs. Eur J Biochem. 1992 Jun 1;206(2):315–321. doi: 10.1111/j.1432-1033.1992.tb16929.x. [DOI] [PubMed] [Google Scholar]
- Geigenmüller U., Nierhaus K. H. Significance of the third tRNA binding site, the E site, on E. coli ribosomes for the accuracy of translation: an occupied E site prevents the binding of non-cognate aminoacyl-tRNA to the A site. EMBO J. 1990 Dec;9(13):4527–4533. doi: 10.1002/j.1460-2075.1990.tb07904.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazelbauer G. L., Engström P., Harayama S. Methyl-accepting chemotaxis protein III and transducer gene trg. J Bacteriol. 1981 Jan;145(1):43–49. doi: 10.1128/jb.145.1.43-49.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henson J. M., Kopp B., Kuempel P. L. Deletion of 60 kilobase pairs of DNA from the terC region of the chromosome of Escherichia coli. Mol Gen Genet. 1984;193(2):263–268. doi: 10.1007/BF00330678. [DOI] [PubMed] [Google Scholar]
- Hirsh D., Gold L. Translation of the UGA triplet in vitro by tryptophan transfer RNA's. J Mol Biol. 1971 Jun 14;58(2):459–468. doi: 10.1016/0022-2836(71)90363-9. [DOI] [PubMed] [Google Scholar]
- Hughes D., Atkins J. F., Thompson S. Mutants of elongation factor Tu promote ribosomal frameshifting and nonsense readthrough. EMBO J. 1987 Dec 20;6(13):4235–4239. doi: 10.1002/j.1460-2075.1987.tb02772.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes D., Thompson S., O'Connor M., Tuohy T., Nichols B. P., Atkins J. F. Genetic characterization of frameshift suppressors with new decoding properties. J Bacteriol. 1989 Feb;171(2):1028–1034. doi: 10.1128/jb.171.2.1028-1034.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes K. T., Roth J. R. Transitory cis complementation: a method for providing transposition functions to defective transposons. Genetics. 1988 May;119(1):9–12. doi: 10.1093/genetics/119.1.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston H. M., Barnes W. M., Chumley F. G., Bossi L., Roth J. R. Model for regulation of the histidine operon of Salmonella. Proc Natl Acad Sci U S A. 1980 Jan;77(1):508–512. doi: 10.1073/pnas.77.1.508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato M., Nishikawa K., Uritani M., Miyazaki M., Takemura S. The difference in the type of codon-anticodon base pairing at the ribosomal P-site is one of the determinants of the translational rate. J Biochem. 1990 Feb;107(2):242–247. doi: 10.1093/oxfordjournals.jbchem.a123033. [DOI] [PubMed] [Google Scholar]
- Komine Y., Adachi T., Inokuchi H., Ozeki H. Genomic organization and physical mapping of the transfer RNA genes in Escherichia coli K12. J Mol Biol. 1990 Apr 20;212(4):579–598. doi: 10.1016/0022-2836(90)90224-A. [DOI] [PubMed] [Google Scholar]
- Kopelowitz J., Hampe C., Goldman R., Reches M., Engelberg-Kulka H. Influence of codon context on UGA suppression and readthrough. J Mol Biol. 1992 May 20;225(2):261–269. doi: 10.1016/0022-2836(92)90920-f. [DOI] [PubMed] [Google Scholar]
- Kunkel T. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc Natl Acad Sci U S A. 1985 Jan;82(2):488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lill R., Lepier A., Schwägele F., Sprinzl M., Vogt H., Wintermeyer W. Specific recognition of the 3'-terminal adenosine of tRNAPhe in the exit site of Escherichia coli ribosomes. J Mol Biol. 1988 Oct 5;203(3):699–705. doi: 10.1016/0022-2836(88)90203-3. [DOI] [PubMed] [Google Scholar]
- Lill R., Robertson J. M., Wintermeyer W. Binding of the 3' terminus of tRNA to 23S rRNA in the ribosomal exit site actively promotes translocation. EMBO J. 1989 Dec 1;8(12):3933–3938. doi: 10.1002/j.1460-2075.1989.tb08574.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moazed D., Noller H. F. Intermediate states in the movement of transfer RNA in the ribosome. Nature. 1989 Nov 9;342(6246):142–148. doi: 10.1038/342142a0. [DOI] [PubMed] [Google Scholar]
- Moazed D., Noller H. F. Sites of interaction of the CCA end of peptidyl-tRNA with 23S rRNA. Proc Natl Acad Sci U S A. 1991 May 1;88(9):3725–3728. doi: 10.1073/pnas.88.9.3725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moir P. D., Spiegelberg R., Oliver I. R., Pringle J. H., Masters M. Proteins encoded by the Escherichia coli replication terminus region. J Bacteriol. 1992 Apr;174(7):2102–2110. doi: 10.1128/jb.174.7.2102-2110.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monro R. E., Cerná J., Marcker K. A. Ribosome-catalyzed peptidyl transfer: substrate specificity at the P-site. Proc Natl Acad Sci U S A. 1968 Nov;61(3):1042–1049. doi: 10.1073/pnas.61.3.1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monro R. E., Marcker K. A. Ribosome-catalysed reaction of puromycin with a formylmethionine-containing oligonucleotide. J Mol Biol. 1967 Apr 28;25(2):347–350. doi: 10.1016/0022-2836(67)90146-5. [DOI] [PubMed] [Google Scholar]
- Nierhaus K. H. The allosteric three-site model for the ribosomal elongation cycle: features and future. Biochemistry. 1990 May 29;29(21):4997–5008. doi: 10.1021/bi00473a001. [DOI] [PubMed] [Google Scholar]
- Noller H. F. Ribosomal RNA and translation. Annu Rev Biochem. 1991;60:191–227. doi: 10.1146/annurev.bi.60.070191.001203. [DOI] [PubMed] [Google Scholar]
- O'Connor M., Gesteland R. F., Atkins J. F. Sequence of E.coli K12 valU operon which contains 3 genes for tRNA(Val1) and 1 gene for tRNA(Lys). Nucleic Acids Res. 1990 Feb 11;18(3):672–672. doi: 10.1093/nar/18.3.672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connor M., Gesteland R. F., Atkins J. F. tRNA hopping: enhancement by an expanded anticodon. EMBO J. 1989 Dec 20;8(13):4315–4323. doi: 10.1002/j.1460-2075.1989.tb08618.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Mahony D. J., Hughes D., Thompson S., Atkins J. F. Suppression of a -1 frameshift mutation by a recessive tRNA suppressor which causes doublet decoding. J Bacteriol. 1989 Jul;171(7):3824–3830. doi: 10.1128/jb.171.7.3824-3830.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagel F. T., Tuohy T. M., Atkins J. F., Murgola E. J. Doublet translocation at GGA is mediated directly by mutant tRNA(2Gly). J Bacteriol. 1992 Jun;174(12):4179–4182. doi: 10.1128/jb.174.12.4179-4182.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quiggle K., Kumar G., Ott T. W., Ryu E. K., Chládek S. Donor site of ribosomal peptidyltransferase: investigation of substrate specificity using 2'(3')-O-(N-acylaminoacyl)dinucleoside phosphates as models of the 3' terminus of N-acylaminoacyl transfer ribonucleic acid. Biochemistry. 1981 Jun 9;20(12):3480–3485. doi: 10.1021/bi00515a027. [DOI] [PubMed] [Google Scholar]
- Roth J. R., Antón D. N., Hartman P. E. Histidine regulatory mutants in Salmonella typhimurium. I. Isolation and general properties. J Mol Biol. 1966 Dec 28;22(2):305–323. doi: 10.1016/0022-2836(66)90134-3. [DOI] [PubMed] [Google Scholar]
- Stoker N. G., Fairweather N. F., Spratt B. G. Versatile low-copy-number plasmid vectors for cloning in Escherichia coli. Gene. 1982 Jun;18(3):335–341. doi: 10.1016/0378-1119(82)90172-x. [DOI] [PubMed] [Google Scholar]
- Tezuka M., Chládek S. Effect of nucleotide substitution on the peptidyltransferase activity of 2'(3')-O-(aminoacyl) oligonucleotides. Biochemistry. 1990 Jan 23;29(3):667–670. doi: 10.1021/bi00455a011. [DOI] [PubMed] [Google Scholar]
- Tuohy T. M., Thompson S., Gesteland R. F., Hughes D., Atkins J. F. The role of EF-Tu and other translation components in determining translocation step size. Biochim Biophys Acta. 1990 Aug 27;1050(1-3):274–278. doi: 10.1016/0167-4781(90)90180-a. [DOI] [PubMed] [Google Scholar]
- VOGEL H. J., BONNER D. M. Acetylornithinase of Escherichia coli: partial purification and some properties. J Biol Chem. 1956 Jan;218(1):97–106. [PubMed] [Google Scholar]
- Wanner B. L. Novel regulatory mutants of the phosphate regulon in Escherichia coli K-12. J Mol Biol. 1986 Sep 5;191(1):39–58. doi: 10.1016/0022-2836(86)90421-3. [DOI] [PubMed] [Google Scholar]
- Weiner A. M., Maizels N. tRNA-like structures tag the 3' ends of genomic RNA molecules for replication: implications for the origin of protein synthesis. Proc Natl Acad Sci U S A. 1987 Nov;84(21):7383–7387. doi: 10.1073/pnas.84.21.7383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss R. B., Dunn D. M., Atkins J. F., Gesteland R. F. Slippery runs, shifty stops, backward steps, and forward hops: -2, -1, +1, +2, +5, and +6 ribosomal frameshifting. Cold Spring Harb Symp Quant Biol. 1987;52:687–693. doi: 10.1101/sqb.1987.052.01.078. [DOI] [PubMed] [Google Scholar]
- Weiss R., Gallant J. Mechanism of ribosome frameshifting during translation of the genetic code. 1983 Mar 31-Apr 6Nature. 302(5907):389–393. doi: 10.1038/302389a0. [DOI] [PubMed] [Google Scholar]
- Weiss W. A., Edelman I., Culbertson M. R., Friedberg E. C. Physiological levels of normal tRNA(CAGGln) can effect partial suppression of amber mutations in the yeast Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1987 Nov;84(22):8031–8034. doi: 10.1073/pnas.84.22.8031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wower J., Scheffer P., Sylvers L. A., Wintermeyer W., Zimmermann R. A. Topography of the E site on the Escherichia coli ribosome. EMBO J. 1993 Feb;12(2):617–623. doi: 10.1002/j.1460-2075.1993.tb05694.x. [DOI] [PMC free article] [PubMed] [Google Scholar]