SUMMARY
A 14-year-old boy with severe combined immunodeficiency presented three times to a medical facility over a period of 4 months with fever and headache that progressed to hydrocephalus and status epilepticus necessitating a medically induced coma. Diagnostic workup including brain biopsy was unrevealing. Unbiased next-generation sequencing of the cerebrospinal fluid identified 475 of 3,063,784 sequence reads (0.016%) corresponding to leptospira infection. Clinical assays for leptospirosis were negative. Targeted antimicrobial agents were administered, and the patient was discharged home 32 days later with a status close to his premorbid condition. Polymerase-chain-reaction (PCR) and serologic testing at the Centers for Disease Control and Prevention (CDC) subsequently confirmed evidence of Leptospira santarosai infection.
More than half the cases of meningoencephalitis remain undiagnosed, despite extensive clinical laboratory testing.1–4 Because more than 100 different infectious agents can cause encephalitis, establishing a diagnosis with the use of cultures, serologic tests, and pathogen-specific PCR assays can be difficult. Unbiased next-generation sequencing has the potential to revolutionize our ability to discover emerging pathogens, especially newly identified viruses.5–8 However, the usefulness of next-generation sequencing for the diagnosis of infectious diseases in a clinically relevant timeframe is largely unexplored.9 We used unbiased next-generation sequencing to identify a treatable, albeit rare, bacterial cause of meningoencephalitis. In this case, the results of next-generation sequencing contributed directly to a dramatic effect on the patient’s care, resulting ultimately in a favorable outcome.
CASE REPORT
A 14-year-old boy with severe combined immunodeficiency (SCID) caused by adenosine deaminase deficiency and partial immune reconstitution after he had undergone two haploidentical bone marrow transplantations initially presented to the emergency department in early April 2013 after having had headache and fevers, with temperatures up to 39.4°C, for 6 days (Fig. 1A). He was admitted to the hospital and discharged 1 day later after resolution of his fever and headache.
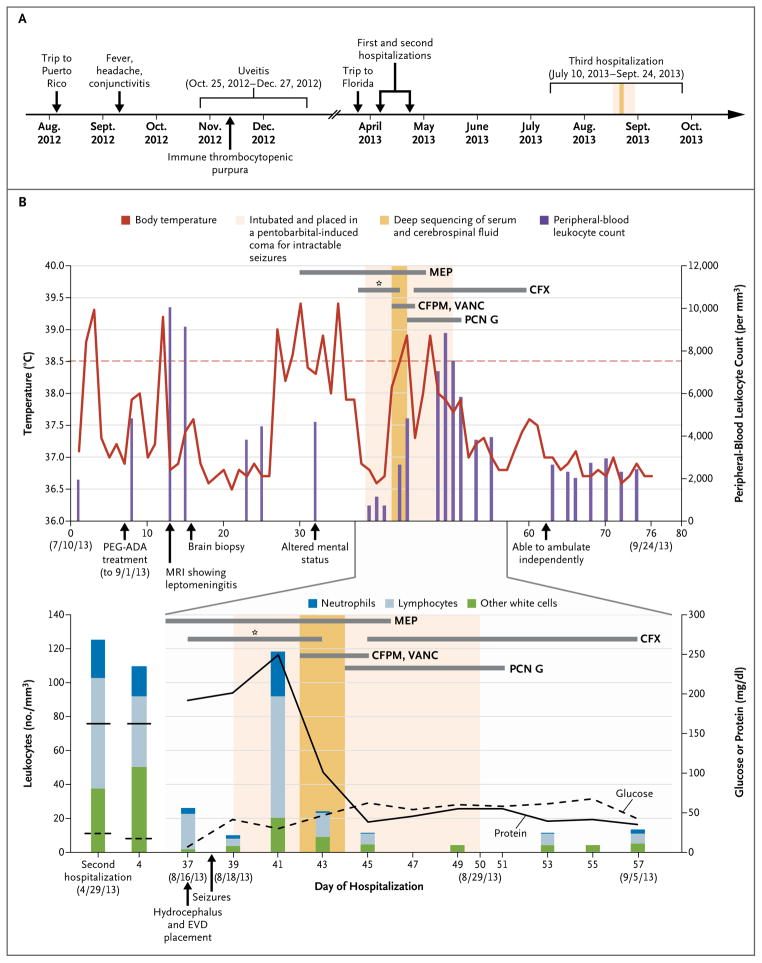
Figure 1. Clinical Course of the 14-Year-Old Patient with Fulminant Meningoencephalitis.
Panel A shows a timeline beginning with the patient’s trip to Puerto Rico in August 2012 and ending after his recovery in October 2013. Major events during the course of the patient’s illness are indicated by arrows. Panel B shows laboratory values obtained and pertinent medications administered during the patient’s third hospitalization. The upper graph shows the body-temperature curve (red line) and peripheral-blood leukocyte counts (purple bars). The lower graph shows the leukocyte count and differential (bars) and the glucose (dashed line) and protein (solid line) levels in serially collected cerebrospinal fluid samples. The horizontal thick gray lines show the medications administered. The asterisk denotes the first course of cefuroxime (CFX) given to the patient, which did not result in clinical improvement. CFPM denotes cefepime, EVD extraventricular drain, MEP methylprednisolone, MRI magnetic resonance imaging, PCN G penicillin G, PEG-ADA polyethylene glycol–modified adenosine deaminase, and VANC vancomycin.
The patient’s outpatient medications included monthly infusions of intravenous immune globulin for hypogammaglobulinemia and trimethoprim–sulfamethoxazole or atovaquone for prophylaxis against Pneumocystis jirovecii pneumonia. He had no known sick contacts but did have three pet cats. He had gone on a missionary trip to Puerto Rico during the first 2 weeks of August 2012 (Fig. 1A), where he swam in a river and the ocean. Notably, a 17-year-old fellow traveler had been hospitalized for 4 days with fever and hematuria. The patient had also vacationed in Florida in March 2013, where he swam in a pool at a resort where there were a number of feral cats.
In September 2012, the patient had presented to his primary care physician with fever, headache, and bilateral conjunctivitis that resolved spontaneously in 10 days (Fig. 1A). At the end of October 2012, he had had photophobia and pain with movement of his left eye. His ophthalmologist had prescribed eyedrops consisting of a combination of a glucocorticoid, a vasoconstrictor, and an antibiotic (ciprofloxacin) for uveitis. One week later, uveitis had developed in the contralateral eye and was treated in a similar manner. The ophthalmologic symptoms had resolved by December 2012. Thrombocytopenia had also developed in October 2012, and the patient was treated with rituximab for presumed immune thrombocytopenic purpura, with subsequent normalization of his platelet counts.
After the brief hospitalization in early April 2013, the patient was readmitted to the hospital at the end of April 2013 with fever, photophobia, and daily frontotemporal headaches (Fig. 1A). In addition, he reported increasing fatigue, abdominal pain, and a weight loss of 2.3 kg. On admission, he had normal vital signs, and the physical examination was unremarkable. The peripheral-blood leukocyte count was 3800 per cubic millimeter with 78% neutrophils. The erythrocyte sedimentation rate was 39 mm per hour (normal range, 0 to 20 mm per hour). The deoxyadenosine nucleotide percentage in the red cells, a measure of control of adenosine deaminase deficiency, was 5.9% (target range, <10%). Serum electrolyte, creatinine, liver-enzyme, and IgG values were within normal ranges. Analysis of the cerebrospinal fluid (CSF) showed 125 leukocytes per cubic millimeter (18% neutrophils and 52% lymphocytes), 0 red cells, a protein level of 97 mg per deciliter (normal range, 15 to 45 mg per deciliter), and a glucose level of 24 mg per deciliter (1.3 mmol per liter; normal range, 40 to 85 mg per deciliter [2.2 to 4.7 mmol per liter]) (Fig. 1B). The results of magnetic resonance imaging (MRI) of the head were unremarkable.
An extensive infectious disease workup was unrevealing (Table S1 in the Supplementary Appendix, available with the full text of this article at NEJM.org). Given the low suspicion for bacterial meningitis, the patient received no empirical antimicrobial agents. He was discharged with instructions to continue his routine prophylaxis and monthly intravenous immune globulin therapy. In May and June, his symptoms waxed and waned but were mild enough to allow him to return to school.
The patient was admitted in July for a third time with fever, headache, diffuse weakness, myalgias, nausea, and vomiting (Fig. 1A and 1B). He received two doses of rituximab for possible autoimmune involvement but had no reduction of symptoms. MRI of the head on admission showed patchy, nonenhancing, T2-weighted hyperintensities in the basal ganglia bilaterally. Analysis of CSF showed 109 leukocytes per cubic millimeter (16% neutrophils and 38% lymphocytes), 0 red cells, a protein level of 162 mg per deciliter, and a glucose level of 17 mg per deciliter (0.9 mmol per liter). The level of angiotensin-converting enzyme (ACE) in CSF was elevated at 7.2 U per liter (normal value, <2.5 U per milliliter).
A repeat MRI of the head at day 13 showed persistent hyperintensities in the basal ganglia and interval development of basilar leptomeningitis extending into the cerebral hemispheres (Fig. 2A, 2B, and 2C). Given these MRI findings and the lack of clinical improvement, a biopsy of the right frontal lobe was performed 2 weeks after admission. Histologic examination revealed inflamed leptomeninges with a granulomatous infiltrate (Fig. 2E). The cerebral cortex was normal with the exception of two microscopic foci of microglial proliferation. Immunohistochemical testing confirmed the presence of CD3-positive T cells (Fig. 2F) and CD68-positive histiocytes and the absence of CD20-positive B lymphocytes. Immunohistochemical testing and electron microscopy did not identify fungi, bacteria, or viruses (Fig. 2G through 2J).
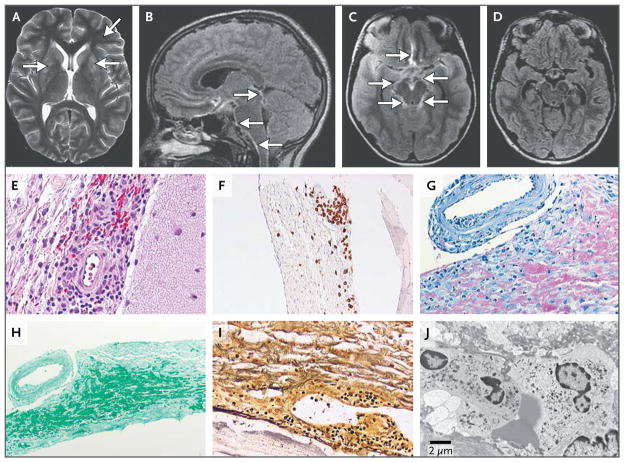
Figure 2. Neuroradiologic MRI and Brain-Biopsy Findings.
The images shown in Panels A, B, and C were acquired on day 13 of the patient’s third hospitalization, whereas the image shown in Panel D was acquired on day 55, after the patient had completed a 7-day course of intravenous penicillin G. An axial T2-weighted image of the head revealed persistent hyperintensities in the basal ganglia and deep frontal white matter (Panel A, arrows). Sagittal and axial T2-weighted fluid-attenuated inversion recovery (FLAIR) images (Panels B and C, respectively) showed thickening in and around the basilar meninges (arrows). An axial T2-weighted FLAIR image (Panel D) depicts near resolution of the previously seen (Panels B and C) basilar meningitis. Results of a biopsy of the right frontal lobe performed 2 weeks after the third hospital admission showed infiltration by lymphocytes and epithelioid histiocytes in the subarachnoid space with a perivascular predilection (Panel E, hematoxylin and eosin). T lymphocytes were visualized by means of immunolabeling with anti-CD3 antibody (Panel F). Various stains also showed the absence of mycobacteria (Panel G, acid-fast), fungi (Panel H, Gomori methenamine silver), and leptospira or other spirochetes (Panel I, Warthin–Starry silver). Electron microscopy revealed the presence of an inflammatory infiltrate and the absence of inclusion bodies, viral particles, or other evidence of microorganisms (Panel J).
Given the negative results of the infectious disease workup (Table S1 in the Supplementary Appendix), the elevated ACE level in CSF, the patient’s history of presumed autoimmune disease, and the presence of granulomatous leptomeningitis, the patient was treated with intravenous glucocorticoids for possible neurosarcoidosis. Treatment with polyethylene glycol–modified adenosine deaminase (PEG-ADA) was also administered in an attempt to boost his immune system.10 Despite these interventions, the patient’s condition continued to decline, with new-onset psychiatric symptoms. An extraventricular drain was placed because of worsening hydrocephalus, and the patient was placed in a medically induced coma to control new-onset status epilepticus. Cefuroxime, administered as prophylaxis against infection at the site of the extraventricular drain, had no effect on his CSF values or clinical status (Fig. 1B).
After written informed consent was obtained from a parent on behalf of the patient, he was enrolled in a research study for pathogen detection and discovery in hospitalized patients with the use of unbiased next-generation sequencing. Coincident with sending CSF and serum samples for next-generation sequencing, broad-spectrum antimicrobial coverage with cefepime and vancomycin was started empirically for fever suggestive of a potential nosocomial infection (Fig. 1B). Within 48 hours after receipt of the samples (Fig. 3A), next-generation sequencing analysis of more than 8 million sequences with the use of a bioinformatics pipeline for the detection of all known pathogens detected sequence reads corresponding to leptospira infection in the patient’s CSF but not in the serum. Although the identification of leptospira in the CSF was later confirmed with the use of targeted PCR and Sanger sequencing by two independent laboratories (Table 1), a decision was made to treat the patient immediately for neuroleptospirosis before confirmatory testing could be obtained. Therefore, glucocorticoids were tapered, and antibiotic coverage was narrowed to high-dose intravenous penicillin G (13 million units daily).
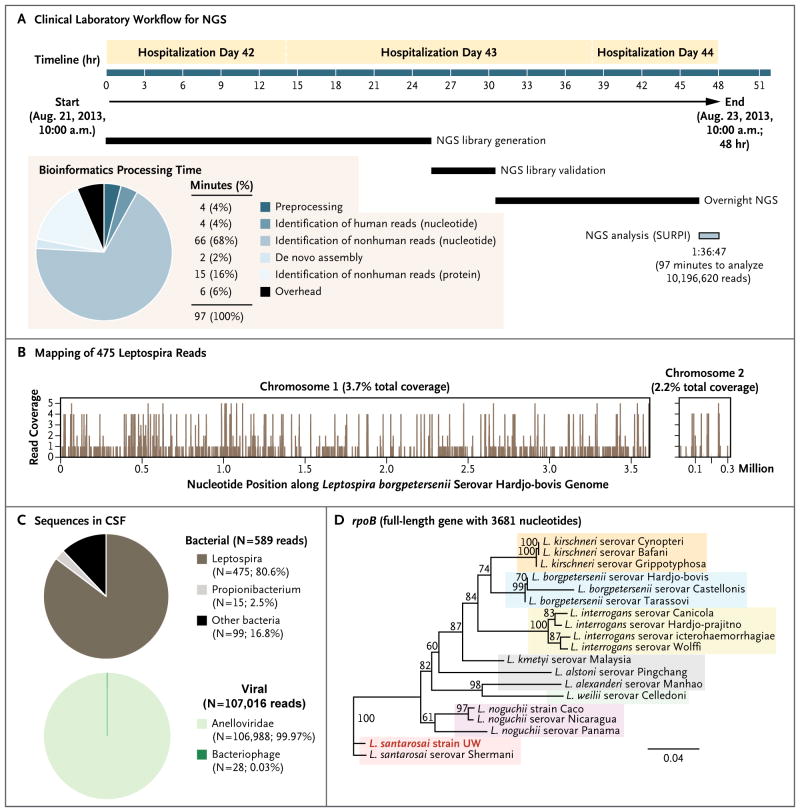
Figure 3. Diagnosis of Leptospira Infection by Means of Unbiased Next-Generation Sequencing (NGS).
A clinical laboratory workflow for a comprehensive pathogen-detection assay with the use of unbiased NGS is shown (Panel A). The total time required for the bioinformatics analysis of NGS data, 97 minutes (pie chart, inset), was only 3.4% of the 48-hour turnaround time for the assay. Sequence-based ultra-rapid pathogen identification (SURPI) is a bioinformatics pipeline for pathogen identification from data obtained by means of next-generation sequencing of clinical samples. In de novo assembly, overlapping sequence reads are joined together in order to create longer contiguous sequences. The overhead time refers to computational time necessary for file-format conversion, sequence retrieval of identified matches in the database, and taxonomic classification of aligned reads. A total of 475 sequence reads derived from the patient’s cerebrospinal fluid (CSF) sample were mapped to the closest matched leptospira genome in the reference database, Leptospira borgpetersenii (Panel B). The distribution of bacterial and viral sequences identified in the patient’s CSF included propionibacteria, anelloviruses, and bacteriophages, which are generally considered to be nonpathogenic body flora (Panel C). Phylogenetic analysis of the full-length gene rpoB showed sequences from representative serovars, strains, and species of leptospira, highlighting the strain identified in the patient with fulminant meningoencephalitis (provisionally named L. santarosai strain UW [University of Wisconsin]) (Panel D). The scale bar denotes the number of nucleotide substitutions per site. GenBank accession numbers are provided in the Supplementary Appendix.
Table 1.
Confirmatory Diagnostic Testing for Neuroleptospirosis.*
| Assay† | Testing Site | Sample Type | Before Diagnosis‡ | After Diagnosis‡ | Date of Test Result |
|---|---|---|---|---|---|
| 16S rRNA bacterial PCR assay | UW | CSF | Negative | July 14, 2013 | |
| 16S rRNA bacterial PCR assay | UW | CSF | Negative | Aug. 12, 2013 | |
| 16S rRNA bacterial PCR assay | UW | Serum | Negative | Aug. 24, 2013 | |
| Leptospira PCR assays targeting lipL32, lipL41, ompA, rpoB, and secY | UCSF | CSF | Positive | Aug. 28, 2013 | |
| Leptospira PCR assays targeting lipL32, ompA, and secY | UCSF | Serum | Negative | Aug. 28, 2013 | |
| Leptospira culture | CDC | CSF | Negative | Oct. 15, 2013 | |
| Leptospira PCR assay targeting lipL32 with the use of a clinically validated assay11 | CDC | CSF | Negative | Oct. 15, 2013 | |
| 16S rRNA bacterial PCR assay | CDC | CSF | Negative | Oct. 15, 2013 | |
| Leptospira PCR assay targeting lipL32 with the use of a clinically validated assay11 | CDC | Serum | Negative | Oct. 15, 2013 | |
| Leptospira IgM antibody with the use of dot blot ELISA | CDC | Serum | Negative | Oct. 15, 2013 | |
| Leptospira IgM antibody with the use of dot blot ELISA | CDC | Serum | Negative (sample obtained on Oct. 9, 2013) | Oct. 17, 2013 | |
| Leptospira PCR assays targeting lipL32, ompA, and secY | UCSF | Brain | Negative | Oct. 17, 2013 | |
| Leptospira PCR assays targeting lipL32, ompA, and secY | UCSF | Serum | Negative | Oct. 31, 2013 | |
| Leptospira PCR assay targeting lipL32 with the use of a clinically validated assay and a change in the amplification mix11 | CDC | CSF | Positive | Jan. 16, 2014 | |
| Leptospira IgM antibody with the use of latex agglutination ELISA12 | CDC | Serum | Positive (sample obtained on Oct. 9, 2013) | Feb. 6, 2014 | |
| Leptospira PCR assay targeting lipL32 with the use of a clinically validated assay and a change in the amplification mix11 | CDC | CSF | Negative (sample obtained on Feb. 5, 2014) | Feb. 24, 2014 |
CDC denotes Centers for Disease Control and Prevention, CSF cerebrospinal fluid, ELISA enzyme-linked immunosorbent assay, PCR polymerase chain reaction, rRNA ribosomal RNA, UCSF University of California, San Francisco, and UW University of Wisconsin.
All the tests were performed on clinical samples obtained during the patient’s third hospitalization and collected before treatment with intravenous penicillin G on August 23, 2013, with the exception of serum and CSF collected during the convalescent phase on October 9, 2013, and February 5, 2014, respectively.
The diagnosis was made on the basis of unbiased next-generation sequencing on August 23, 2013.
The patient gradually recovered over the next 7 days, with resolution of his status epilepticus, normalization of his CSF, and resolution of leptomeningitis as observed on serial MRI scans (Fig. 1A, 1B, and 2D). He was discharged to inpatient rehabilitation 14 days after completing a 7-day course of intravenous penicillin G. At discharge, the patient had largely recovered but still had occasional right facial twitching from simple partial seizures. After 11 days of rehabilitation, he returned home near the end of September (76 days after hospital admission) close to his premorbid functional status. CSF collected 4 months later revealed 0 leukocytes and red cells, a protein level of 49 mg per deciliter, and a glucose level of 47 mg per deciliter (2.6 mmol per liter); the CSF was negative for leptospira on repeat PCR testing (Table 1).
METHODS
Analysis of the patient’s clinical samples for the identification of potential pathogens was approved by the institutional review board at the University of Wisconsin and at the University of California, San Francisco (UCSF). Amplified DNA libraries for next-generation sequencing were constructed from extracted nucleic acid derived from clinical samples as previously described,13,14 followed by library validation and sequencing on an Illumina MiSeq instrument. Reads were analyzed with the use of sequence-based ultra-rapid pathogen identification (SURPI), a bioinformatics pipeline developed at UCSF to rapidly classify next-generation sequencing reads according to their origin.15 The SURPI pipeline first identifies and subtracts human host sequences, followed by alignment of reads to reference sequences in National Center for Biotechnology Information (NCBI) databases, including all of GenBank, for the comprehensive identification of bacteria, viruses, fungi, and parasites. Full details regarding sample processing, next-generation sequencing analysis, PCR confirmation, and phylogenetic analysis are provided in the Supplementary Appendix.
RESULTS
RAPID IDENTIFICATION OF LEPTOSPIRA SEQUENCES IN CSF
CSF and serum samples were processed in a clinical laboratory with the use of an unbiased next-generation sequencing assay protocol with a sample-to-answer turnaround time of 48 hours7,16,17 (Fig. 3A). Approximately half the total CSF volume (750 μl) was pretreated with DNase before nucleic acid extraction to enrich the sample for viral sequencing; the remaining half was not pretreated (i.e., untreated CSF), and the nucleic acid was directly extracted for the detection of bacteria, fungi, and parasites. The next-generation sequencing run of four individually indexed samples, including a serum sample from an unrelated patient that served as a negative control, yielded 10,196,620 raw reads. A total of 8,187,737 reads were derived from the patient’s serum, untreated CSF, and DNase-treated CSF, and the remaining 2,008,883 reads corresponded to the negative control sample. The full data set of 10,196,620 raw single-end reads was analyzed in approximately 100 minutes with the use of SURPI.
In the untreated CSF, the majority of bacterial reads (475 of 589 reads; 80.6%) corresponded to the Leptospiraceae family (Fig. 3B and 3C, and Table S2 in the Supplementary Appendix), with the mapped reads spanning the closest matched full leptospira genome deposited in the NCBI nucleotide reference database at the time, L. borgpetersenii,18 at 3.7% and 2.2% coverage for chromosomes 1 and 2, respectively. No convincing hits to other bacteria or viruses were found in the next-generation sequencing data (Tables S3 and S4 in the Supplementary Appendix), and no reads corresponding to leptospira, either single-end or paired-end, were detected in serum samples from the patient or the negative control (Table S3 in the Supplementary Appendix).
CONFIRMATORY TESTING FOR LEPTOSPIROSIS
Primers targeting the genes ompA and secY were designed directly from the mapped next-generation sequencing reads, and the identification of leptospira was confirmed 5 days after the next-generation sequencing analysis with the use of PCR analysis and Sanger sequencing (Table 1, and Fig. S1 and Table S5 in the Supplementary Appendix). This approach was also used to sequence the partial-length gene lipL32 and the full-length genes lipL41, rpoB, and secY. Phylogenetic analysis revealed that the CSF sample harbored L. santarosai (Fig. 3D, and Fig. S2 in the Supplementary Appendix), a pathogenic species whose genome has been sequenced19 but was not deposited in the NCBI nucleotide reference database as of August 2013 and thus was not identified by the SURPI pipeline. The CSF titer, calculated from a sample collected 1 day before the initiation of vancomycin and cefepime (Fig. S3 in the Supplementary Appendix), was 958 genome copies per milliliter.
Clinical diagnostic tests of CSF and serum samples obtained during the acute and convalescent periods were negative for leptospirosis; testing included IgM and IgG antibody testing by means of dot-blot enzyme-linked immunosorbent assay, direct culture, and a Clinical Laboratory Improvement Amendments–validated PCR assay targeting the gene lipL3211 (Table 1). These results suggest that the diagnosis could not have been made routinely with the use of available clinically validated assays for leptospirosis. The CDC eventually detected leptospira in the CSF 5 months after the next-generation sequencing analysis by repeating the PCR assay targeting lipL32 with the use of a different amplification mix (cycle threshold of approximately 35; Fig. S4 in the Supplementary Appendix). This yielded an amplicon that was shown by means of direct Sanger sequencing to correspond to L. santarosai. Separate confirmation of leptospira infection from a serum sample drawn in October 2013 (during the convalescent period) was also obtained with the use of a qualitative IgM latex agglutination assay targeting pathogenic leptospira that had been newly approved by the Food and Drug Administration.12
DISCUSSION
Leptospirosis is a worldwide zoonotic disease caused by a spirochete.20 Bacteria are shed in urine from reservoir animals and transmitted to humans by means of mucosal exposure or breaks in the skin.21–25 The disease phenotype varies from subclinical infection to severe illness with multiorgan involvement. The acute stage of the disease is characterized by fever, myalgias, headache, and conjunctivitis. The chronic stage of the disease can include meningitis, encephalitis, or both (predominantly in male patients),26 nephritis, cholecystitis, uveitis, and thrombocytopenia.
The standard diagnostic assay for leptospirosis detects the host’s serologic response.27 The degree of immunosuppression resulting from SCID and rituximab therapy in the case patient made it unlikely that he would have had a detectable antibody response even if the diagnosis had been considered a priori. Furthermore, his treatment with intravenous immune globulin may have clouded the interpretation of a positive result.28 Leptospires, like other spirochetes, are typically not detected by means of routine microscopy and cannot be cultured with the use of standard media.20 PCR-based testing is available but has limited sensitivity given the wide diversity of leptospira species and the lack of standardization among assays.29
We suspect that the patient most likely contracted leptospirosis during his trip to Puerto Rico in August 2012, during which he swam in freshwater and during which fever and hematuria suggestive of leptospira-associated nephritis developed in a fellow traveler.30,31 The diagnosis of L. santarosai infection is also consistent with the geographic distribution of this bacterial species, which includes Latin America, the Caribbean, and Taiwan.32,33 The patient’s fever, headache, thrombocytopenia, uveitis, and conjunctivitis within 3 months after visiting Puerto Rico were, in hindsight, probable clinical manifestations of leptospirosis.30,31 Although the neurotropism and persistence of leptospira in the brains of animal reservoir hosts are well described,34–36 this case is a rare instance of chronic neuroleptospirosis in a human lasting for months after the acute presentation.37,38
The decision to treat this patient before confirmatory tests could be completed was the result of a 2-hour multidisciplinary discussion during which the risk that the leptospirosis result was a spurious finding was weighed against the potential benefit of treatment and the low toxicity of penicillin G. The concern that the finding was spurious was considered to be negligible given that a large number of reads spanning the leptospira genome were detected in the patient’s CSF (Fig. 3B), leptospira reads were absent in the patient’s serum and in a control sample from an unrelated patient processed concomitantly (Tables S2, S3, and S4 in the Supplementary Appendix), and leptospira sequences have hitherto never been detected in the next-generation sequencing laboratory. In addition, the patient’s travel history, clinical presentation, and CSF values were all consistent with a diagnosis of neuroleptospirosis, and no other pathogens associated with the clinical syndrome were identified in the next-generation sequencing data or by means of conventional microbiologic testing.
In summary, unbiased next-generation sequencing coupled with a rapid bioinformatics pipeline provided a clinically actionable diagnosis of a specific infectious disease from an uncommon pathogen that eluded conventional testing for months after the initial presentation. This approach thus facilitated the use of targeted and efficacious antimicrobial therapy.
Supplementary Material
Acknowledgments
Supported by the American Brain Foundation Clinical Research Training Fellowship (to Dr. Wilson), a National Human Genome Research Institute Intramural Research Program appointment (to Dr. Sokolic, Ms. Garabedian, and Dr. Candotti), a Howard Hughes Medical Institute appointment (to Dr. DeRisi), a grant from the National Institutes of Health (R01-HL105704, to Dr. Chiu), a University of California Discovery Grant (to Dr. Chiu), an Amazon Web Services in Education Research Grant (to Dr. Chiu), and an Abbott Viral Discovery Award (to Dr. Chiu).
We thank Nicholas Marinelli, M.D., in the Department of Radiology at the University of Wisconsin, for providing the magnetic resonance images and associated text; and Jerome LeGoff for editorial comments on an earlier version of the manuscript.
Footnotes
Disclosure forms provided by the authors are available with the full text of this article at NEJM.org.
References
- 1.Glaser CA, Gilliam S, Schnurr D, et al. In search of encephalitis etiologies: diagnostic challenges in the California Encephalitis Project, 1998–2000. Clin Infect Dis. 2003;36:731–42. doi: 10.1086/367841. [DOI] [PubMed] [Google Scholar]
- 2.Glaser CA, Honarmand S, Anderson LJ, et al. Beyond viruses: clinical profiles and etiologies associated with encephalitis. Clin Infect Dis. 2006;43:1565–77. doi: 10.1086/509330. [DOI] [PubMed] [Google Scholar]
- 3.Granerod J, Tam CC, Crowcroft NS, Davies NW, Borchert M, Thomas SL. Challenge of the unknown: a systematic review of acute encephalitis in non-outbreak situations. Neurology. 2010;75:924–32. doi: 10.1212/WNL.0b013e3181f11d65. [DOI] [PubMed] [Google Scholar]
- 4.Mailles A, Stahl JP. Infectious encephalitis in France in 2007: a national prospective study. Clin Infect Dis. 2009;49:1838–47. doi: 10.1086/648419. [DOI] [PubMed] [Google Scholar]
- 5.Briese T, Paweska JT, McMullan LK, et al. Genetic detection and characterization of Lujo virus, a new hemorrhagic fever-associated arenavirus from southern Africa. PLoS Pathog. 2009;5(5):e1000455. doi: 10.1371/journal.ppat.1000455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chiu CY. Viral pathogen discovery. Curr Opin Microbiol. 2013;16:468–78. doi: 10.1016/j.mib.2013.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grard G, Fair JN, Lee D, et al. A novel rhabdovirus associated with acute hemorrhagic fever in central Africa. PLoS Pathog. 2012;8(9):e1002924. doi: 10.1371/journal.ppat.1002924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Palacios G, Druce J, Du L, et al. A new arenavirus in a cluster of fatal transplant-associated diseases. N Engl J Med. 2008;358:991–8. doi: 10.1056/NEJMoa073785. Erratum, N Engl J Med 2008; 358:1204. [DOI] [PubMed] [Google Scholar]
- 9.Dunne WM, Jr, Westblade LF, Ford B. Next-generation and whole-genome sequencing in the diagnostic clinical microbiology laboratory. Eur J Clin Microbiol Infect Dis. 2012;31:1719–26. doi: 10.1007/s10096-012-1641-7. [DOI] [PubMed] [Google Scholar]
- 10.Gaspar HB, Aiuti A, Porta F, Candotti F, Hershfield MS, Notarangelo LD. How I treat ADA deficiency. Blood. 2009;114:3524–32. doi: 10.1182/blood-2009-06-189209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stoddard RA, Gee JE, Wilkins PP, Mc-Caustland K, Hoffmaster AR. Detection of pathogenic Leptospira spp. through TaqMan polymerase chain reaction targeting the LipL32 gene. Diagn Microbiol Infect Dis. 2009;64:247–55. doi: 10.1016/j.diagmicrobio.2009.03.014. [DOI] [PubMed] [Google Scholar]
- 12.Brownlow T, Kavanagh OV, Logan EF, et al. ‘Leptorapide’ — a one-step assay for rapid diagnosis of human leptospirosis. Epidemiol Infect. 2013 Sep 19; doi: 10.1017/S0950268813002112. (Epub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Naccache SN, Federman S, Veeraraghavan N, et al. A Cloud-compatible bioinformatics pipeline for ultra-rapid pathogen identification from next-generation sequencing of clinical samples. Genome Res. doi: 10.1101/gr.171934.113. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Naccache SN, Greninger AL, Lee D, et al. The perils of pathogen discovery: origin of a novel parvovirus-like hybrid genome traced to nucleic acid extraction spin columns. J Virol. 2013;87:11966–77. doi: 10.1128/JVI.02323-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Swei A, Russell BJ, Naccache SN, et al. The genome sequence of Lone Star virus, a highly divergent bunyavirus found in the Amblyomma americanum tick. PLoS One. 2013;8(4):e62083. doi: 10.1371/journal.pone.0062083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Greninger AL, Chen EC, Sittler T, et al. A metagenomic analysis of pandemic influenza A (2009 H1N1) infection in patients from North America. PLoS One. 2010;5(10):e13381. doi: 10.1371/journal.pone.0013381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yu G, Greninger AL, Isa P, et al. Discovery of a novel polyomavirus in acute diarrheal samples from children. PLoS One. 2012;7(11):e49449. doi: 10.1371/journal.pone.0049449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bulach DM, Zuerner RL, Wilson P, et al. Genome reduction in Leptospira borgpetersenii reflects limited transmission potential. Proc Natl Acad Sci U S A. 2006;103:14560–5. doi: 10.1073/pnas.0603979103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chou LF, Chen YT, Lu CW, et al. Sequence of Leptospira santarosai serovar Shermani genome and prediction of virulence-associated genes. Gene. 2012;511:364–70. doi: 10.1016/j.gene.2012.09.074. [DOI] [PubMed] [Google Scholar]
- 20.Levett PN. Leptospirosis. Clin Microbiol Rev. 2001;14:296–326. doi: 10.1128/CMR.14.2.296-326.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brockmann S, Piechotowski I, Bock-Hensley O, et al. Outbreak of leptospirosis among triathlon participants in Germany, 2006. BMC Infect Dis. 2010;10:91. doi: 10.1186/1471-2334-10-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Haake DA, Dundoo M, Cader R, et al. Leptospirosis, water sports, and chemoprophylaxis. Clin Infect Dis. 2002;34(9):e40–e43. doi: 10.1086/339942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Morgan J, Bornstein SL, Karpati AM, et al. Outbreak of leptospirosis among triathlon participants and community residents in Springfield, Illinois, 1998. Clin Infect Dis. 2002;34:1593–9. doi: 10.1086/340615. [DOI] [PubMed] [Google Scholar]
- 24.Sejvar J, Bancroft E, Winthrop K, et al. Leptospirosis in “Eco-Challenge” athletes, Malaysian Borneo, 2000. Emerg Infect Dis. 2003;9:702–7. doi: 10.3201/eid0906.020751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stern EJ, Galloway R, Shadomy SV, et al. Outbreak of leptospirosis among Adventure Race participants in Florida, 2005. Clin Infect Dis. 2010;50:843–9. doi: 10.1086/650578. [DOI] [PubMed] [Google Scholar]
- 26.Mathew T, Satishchandra P, Mahadevan A, et al. Neuroleptospirosis — revisited: experience from a tertiary care neurological centre from south India. Indian J Med Res. 2006;124:155–62. [PubMed] [Google Scholar]
- 27.Levett PN, Branch SL, Whittington CU, Edwards CN, Paxton H. Two methods for rapid serological diagnosis of acute leptospirosis. Clin Diagn Lab Immunol. 2001;8:349–51. doi: 10.1128/CDLI.8.2.349-351.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lichtiger B, Rogge K. Spurious serologic test results in patients receiving infusions of intravenous immune gammaglobulin. Arch Pathol Lab Med. 1991;115:467–9. [PubMed] [Google Scholar]
- 29.Bourhy P, Bremont S, Zinini F, Giry C, Picardeau M. Comparison of real-time PCR assays for detection of pathogenic Leptospira spp. in blood and identification of variations in target sequences. J Clin Microbiol. 2011;49:2154–60. doi: 10.1128/JCM.02452-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sharma J, Suryavanshi M. Thrombocytopenia in leptospirosis and role of platelet transfusion. Asian J Transfus Sci. 2007;1:52–5. doi: 10.4103/0973-6247.33447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rathinam SR. Ocular manifestations of leptospirosis. J Postgrad Med. 2005;51:189–94. [PubMed] [Google Scholar]
- 32.Lin PC, Chi CY, Ho MW, Chen CM, Ho CM, Wang JH. Demographic and clinical features of leptospirosis: three-year experience in central Taiwan. J Microbiol Immunol Infect. 2008;41:145–50. [PubMed] [Google Scholar]
- 33.Nalam K, Ahmed A, Devi SM, et al. Genetic affinities within a large global collection of pathogenic Leptospira: implications for strain identification and molecular epidemiology. PLoS One. 2010;5(8):e12637. doi: 10.1371/journal.pone.0012637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Garcia-Monco JC, Benach JL. A disconnect between the neurospirochetoses in humans and rodent models of disease. PLoS Pathog. 2013;9(4):e1003288. doi: 10.1371/journal.ppat.1003288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sunbul M, Esen S, Leblebicioglu H, Hokelek M, Pekbay A, Eroglu C. Rattus norvegicus acting as reservoir of Leptospira interrogans in the Middle Black Sea region of Turkey, as evidenced by PCR and presence of serum antibodies to Leptospira strain. Scand J Infect Dis. 2001;33:896–8. doi: 10.1080/00365540110076796. [DOI] [PubMed] [Google Scholar]
- 36.Vinetz JM, Glass GE, Flexner CE, Mueller P, Kaslow DC. Sporadic urban leptospirosis. Ann Intern Med. 1996;125:794–8. doi: 10.7326/0003-4819-125-10-199611150-00002. [DOI] [PubMed] [Google Scholar]
- 37.Murgatroyd F. Chronic meningitis in Weil’s disease. Br Med J. 1937;1:7–11. doi: 10.1136/bmj.1.3965.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sakellaridis N, Panagopoulos D, Androulis A. Neuroleptospirosis with hydrocephalus and very elevated cerebrospinal fluid protein. South Med J. 2009;102:549–50. doi: 10.1097/SMJ.0b013e3181a0ae80. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.