Abstract
repro27 mice exhibit fully penetrant male-specific infertility associated with a nonsense mutation in the golgin subfamily A member 3 gene (Golga3). GOLGA3 is a Golgi complex-associated protein implicated in protein trafficking, apoptosis, positioning of the Golgi, and spermatogenesis. In repro27 mutant mice, a point mutation in exon 18 of the Golga3 gene that inserts a premature termination codon leads to an absence of GOLGA3 protein expression. GOLGA3 protein was undetectable in the brain, heart, and liver in both mutant and control mice. While spermatogenesis in Golga3repro27 mutant mice appears to initiate normally, development is disrupted in late meiosis during the first wave of spermatogenesis, leading to significant germ cell loss between 15 and 18 days postpartum (dpp). Terminal deoxynucleotidyltransferase dUTP-mediated nick end labeling analysis showed elevated DNA fragmentation in meiotic germ cells by 12 dpp, suggesting apoptosis as a mechanism of germ cell loss. The few surviving postmeiotic round spermatids exhibited abnormal spermiogenesis with defects in acrosome formation, head and tail development, and extensive vacuolization in the seminiferous epithelium. Analysis of epididymal sperm showed significantly low sperm concentration and motility, and in vitro fertilization with mutant sperm was unsuccessful. Golga3repro27 mice lack GOLGA3 protein and thus provide an in vivo tool to aid in deciphering the role of GOLGA3 in Golgi complex positioning, cargo trafficking, and apoptosis signaling in male germ cells.
Introduction
Male infertility accounts for roughly half of the 15 percent of couples world-wide experiencing fertility problems (Matzuk & Lamb 2002). Genetic defects that result in abnormalities in sperm number, motility, and/or morphology may account for a significant percentage of idiopathic male infertility (Egozcue et al. 2000; Ferlin et al. 2007; Yan 2009). The Jackson Laboratory Reproductive Genomics mutagenesis program used a random N-ethyl-N-nitrosourea (ENU) mutagenesis scheme to create a panel of “repro” mutations that identify genes important for gametogenesis (Handel et al. 2006; Lessard et al. 2004). Similar large-scale ENU mutagenesis efforts have generated similar repositories, making mice with reproductive defects available to researchers worldwide (Clark et al. 2004; Kennedy et al. 2005). This unbiased, phenotype-driven approach identifies novel genes required for spermatogenesis and generates new alleles of known genes that may provide new insights and relevance into protein structure and function relationships.
We describe here a new allele of the Golga3 gene that causes male-specific infertility due to an ENU-induced nonsense mutation. Golga3 encodes a Golgi autoantigen that is a member of the golgin subfamily A. Golgins were initially identified in human patients with autoimmune disease (Fritzler et al. 1993) and represent a very diverse family of proteins including GOLGA1 (golgin-97), GOLGA2 (GM130), GOLGA3 (Golgin160, male-enhanced antigen two, MEA2), GOLGA4 (p230/golgin), and GOLGB1 (giantin)(Munro 2011). Proteins in this family associate on the cytoplasmic surface of the Golgi complex and contain a large coiled-coil domain that can form a rod-like structure (Barr 1999; Munro 2011; Short et al. 2005). Golgins have been implicated in a number of processes including tethering adjacent membranes during vesicular trafficking, Golgi positioning and cisternae stacking during mitosis and differentiation, and interacting with Golgi-associated Rab GTPases (Gleeson et al. 1996; Hennies et al. 2008; Linstedt et al. 2000; Short et al. 2005; Short et al. 2001; Yadav et al. 2009; Yadav et al. 2012). In the N-terminal protein region, GOLGA3 contains several phosphorylation sites on conserved serine residues, caspase cleavage sites, targeting signals to the Golgi complex and nucleus while the C-terminal end contains the characteristic golgin coiled-coil domain (Hicks & Machamer 2002). GOLGA3 specifically targets the β1 adrenergic receptor to the plasma membrane (Hicks et al. 2006; Williams et al. 2006), is required for the proper sequestration and sorting of GLUT4 glucose transporters (Williams et al. 2006), participates in protein degradation pathways (Dumin et al. 2006) and apoptosis pathways (Maag et al. 2005; Mancini et al. 2000; Mukherjee & Shields 2009), and most recently has been shown to recruit dyneins to the Golgi complex (Yadav et al. 2012). The GOLGA3-encoding gene has mammalian orthologs in human, rat, cattle, chimpanzee, and the domestic dog (Mouse Genome Database 2012).
GOLGA3 was previously demonstrated to be necessary for normal spermatogenesis. The Golga3Tg(06MGMT)T604Kccri allele, commonly referred to as ΔMea2, is the result of a spontaneous chromosomal translocation that eliminated the first seven exons in a transgenic mouse line carrying the E. coli 06-methylguanine-DNA methyltransferase gene (Banu et al. 2002; Matsukuma et al. 1999). This translocation led to the expression of a truncated GOLGA3 protein, defects in spermatogenesis, and male infertility with variable penetrance that was partially rescued in the ΔMea2H line (Banu et al. 2002). Translin-associated factor X (TSNAX) protein, identified by yeast two-hybrid and glutathione S-transferase pull-down experiments, associates with GOLGA3 (Bray et al. 2002) and colocalized with GOLGA3 in pachytene spermatocytes (Banu et al. 2002; Matsuda et al. 2004). However, the observation that GOLGA3 and TSNAX were localized to separate regions of the Golgi in both the ΔMea2 line and ΔMea2H lines suggests that protein-protein interactions were not necessary for the rescue of spermatogenesis (Matsuda et al. 2004).
Here we report that homozygosity for the new Golga3repro27 allele completely eliminates GOLGA3 protein expression and causes severe spermatogenic defects. We also report development of a C3HeB/FeJ congenic strain, providing a resource to facilitate future analyses of male-specific infertility due to an absence of GOLGA3.
Materials and Methods
Animals and Gene Mapping
All animal procedures were approved by the New Mexico Highlands University and the Jackson Laboratory Animal Care and Use Committees. C3Fe;B6-repro27 were generated by the Jackson Laboratory Reproductive Genomics Program (http://reproductivegenomics.jax.org) using ethylnitrosouria (ENU) chemical mutagenesis, and a three-generation breeding strategy and fertility screening protocol (Handel et al. 2006; Lessard et al. 2004). Mice are a mixture of C3HeB/FeJ (C3Fe) and C57BL/6J (B6) with the mutated DNA being of B6 origin. Regional genetic mapping was conducted at the Jackson Laboratory by analyzing meiotic recombination using microsatellite markers as described previously (Dietrich et al. 1996). Briefly, the repro27 mutation was initially mapped to Chr 5 by genome scanning of DNA from affected and unaffected mice using 2-3 single sequence length polymorphic (SSLP) markers per autosomal chromosome. The single B6 region identified as homozygous in all affected mice but not in unaffected mice was considered the candidate gene region. Polymorphic DNA markers flanking this genomic interval were used for genotyping subsequent progeny and colony maintenance. Intercrosses of heterozygous mice were used to maintain the colony. Narrowing the candidate gene region was accomplished by testing all meiotic recombinant progeny for fertility and the spermatogenesis defect and selecting mice carrying the mutation for future breeding. Once the gene mutation was identified, “mixed” C3Fe;B6-Golga3repro27 mice were maintained by mating either heterozygous or homozygous females with heterozygous males.
A C3Fe congenic strain was generated by backcrossing the mutation 10 generations to C3Fe mice (Jackson Laboratory, C3Fe.B6-Golga3repro27). Congenic mice were maintained by mating homozygous females with heterozygous males to produce 50% mutant and 50% heterozygotes while C3Fe mice were used as wildtype controls.
Mutant and control pups were identified at 8-10 days post-partum (dpp) using a polymerase chain reaction (PCR)-based genotyping protocol. DNA was isolated from tail snips using the sodium hydroxide hot shot protocol as previously described elsewhere (Truett et al. 2000). PCR using flanking SSLP DNA markers (D5Mit317 or D5Mit24, and D5Mit338) were then used to identify mice carrying the repro27 genetic mutation using the following conditions: 94°C for 30 sec, 55°C for 35 sec, 72°C for 30 seconds (39 cycles), and a final extension at 72°C for 7 minutes. PCR products were separated on a 3% Metaphor agarose gel (Cambrex, Rockland, MD). Genotyping results were confirmed phenotypically by the presence of very small testes in mutant mice. In addition, an allele-specific assay, designed by Dr. Matsukuma's laboratory using loop hybrid mobility shift assay (Matsukuma et al. 2006), was used regularly within the colony to confirm breeders and for routine quality control.
RNA Isolation and cDNA synthesis
Total RNA from adult mixed C3Fe;B6-Golga3repro27 and B6 testes were extracted using TRIzol reagent (Invitrogen Corp., Carlsbad, CA) according to the manufacturer's guidelines. Reverse-transcription PCR was used to synthesize single-stranded cDNA from total RNA (1-2 μg) with MMLV-reverse transcriptase (Promega Corp., Madison, WI). Synthesized cDNA was then used for Golga3 transcript sequencing.
Sequencing Golga3
The Golga3 cDNA was sequenced using published primers 15L, 19R, Ex23L and Ex24R (Banu et al. 2002), and additional primers designed to span the expressed transcript. PCR product sizes ranged between approximately 300-750 bp. PCR products were purified using a PCR purification kit (QIAGEN, Inc., Valencia, CA). Forward and reverse primers were then used in BigDye® Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems, Foster City, CA). The reaction was purified with a DyeEx 2.0 Spin Column (QIAGEN Inc.) and lyophilized before shipment to University of New Mexico for analysis on an ABI 3130 DNA Sequencer. Sequence data were analyzed using Vector NTI 10.1.1 (Invitrogen Corp., Carlsbad, CA) to compare repro27 mutant DNA with B6 and published sequence. The point mutation identified in exon 18 was confirmed by sequencing exon 18 from Golga3repro27 mutant (n=3) and B6 genomic DNA.
Quantitative Real Time PCR
Real time validation and reactions were carried out essentially as described (Gifford et al. 2007) using a Corbett RG-6000 real time analyzer (Qiagen Inc.). Testes for RNA isolation were collected from three to twelve mice from congenic C3Fe.B6-Golga3repro27 mutant (Golga3repro27/Golga3repro27, or m/m) and C3HeB/FeJ wildtype (+/+) at 4, 5, 7, 9, and 14 dpp. First-strand cDNA were synthesized as described previously using MMLV reverse transcriptase, oligo dT, DNaseI, and RNAse H (Invitrogen Corp.). A 1:10 dilution of cDNA was used as the template in a 25 μl reaction with SYBR SensiMix™ (Bioline, Taunton, MA). The following primers (300 nM) were used to amplify Golga3 (forward primer: 5′- ACAGGTGGAGGCTTGTCAT -3′ and reverse primer: 5′- TTCTAGCATGCTCTGCAGGT-3′, these primers span an intron) and the housekeeping gene β-Actin (forward primer: 5′-AGCCATGTACGTAGCCATCC-3′and reverse primer: 5′-TTTGATGTCACGCACGATTT-3′). β-actin Ct values were used to calculate ΔCt values for each sample. β-Actin Ct values did not vary across genotype and the melting curves for β-Actin and Golga3 at each time point gave only a single peak unique to each primer set. A cDNA serial dilution standard curve was included in each real time PCR run. The primer efficiency averaged 92%. β-Actin Ct threshold values ranged from 13-17 cycles while Golga3 Ct threshold values ranged from 18-28 cycles. Samples, no reverse transcriptase controls, and no template controls were run in duplicate. Golga3 (forward primer: 5′- GAACAGCCTGAAGGAACAGA-3′and reverse primer: 5′-GAACAGCCTGAAGGAACAGA-3′) and Gapdh (forward primer: 5′-CTGGAGAAACCTGCCAAGTA-3′ and reverse primer: 5′- TGTTGCTGTAGCCGTATTCA-3′) primers from RealTimePrimers.com (Elkins Park, PA) were used according to recommended conditions to validate 14 day data in five mice per genotype. ΔΔCt values were calculated by subtracting the mean ΔCt of the 4 dpp wildtype from the ΔCt at each time point. The fold change compared to the 4 dpp wildtype was calculated using the 2-ΔΔCt method (Livak & Schmittgen 2001).
Northern Blotting
Ten μg of total RNA isolated from 14 dpp (n=3/genotype) and adult testes (n=3) from congenic Golga3repro27 mutant and control testes were run on a 1% denaturing agarose gel, transferred to nylon membrane, and hybridized with 32P endlabeled DNA oligo probes for Golga3 (642 bp corresponding to a transcript located between ∼603 and 1245 bp) and to Gapdh. Golga3 transcript sizes were determined from the migration of ribosomal RNAs.
Western Blotting
Paired testes were collected from 15 dpp mice (n=3 per genotype). Testes, brain, liver, and heart tissues were collected from adult animals (4 months old, n=3 per genotype) and either processed immediately or flash frozen in liquid nitrogen and stored at -80°C. Tissues were homogenized in RIPA lysis buffer (150 mM sodium chloride; 1 mM EDTA; 20 mM Tris HCl pH 7.6; 1% IGEPAL (NP-40); 1% sodium deoxycholate; 5 mM sodium fluoride and 0.1% SDS) containing 10% PMSF, 0.1% DDT, and 1% Protease Inhibitor Cocktail (Sigma, St. Louis, MO). Homogenates were incubated on ice for 30 minutes, and then centrifuged at 12,000 g for 5 minutes at 4°C. The protein concentration of the supernatant was determined using the BCA protein assay (Pierce Biotechnology, Rockford, IL). Forty μg of protein extracts were separated by 7% SDS- PAGE, transferred overnight (30V at 4°C) to nitrocellulose membrane, and blocked for one hour with 5% Carnation® non-fat dried milk in TBS-T (Tris-buffered saline with 0.1% Tween 20). Kaleideoscope Precision Plus Protein Standards (Bio-Rad, Hercules, CA) were run on each gel. Custom GOLGA3 peptide antibodies were produced to the N- terminal (EGSVRKEALQSLRLSL, in exon 3) and C-terminal (EELLRPPPAVSKEPLK, in exon 23) domains (ThermoScientific Open Biosystems). Crude sera from 131-day bleeds were used at a dilution of 1:1500 for all western blots. A monoclonal antibody clone C4 recognizing β-Actin (Millipore, Billerica, MA) diluted 1:1500 (Immuno-Pure goat anti-mouse (H+L) peroxidase conjugated secondary antibody, 1:40,000, ThermoScientific) was used as a protein loading control as it did not vary across Golga3repro27 genotypes. Prior to obtaining the custom antibodies, GOLGA3 antibodies (rabbit anti-N terminal GOLGA3/MEA2 and rabbit anti-C terminal GOLGA3/MEA2, both used 1:500 with 100 μg total protein) provided by Dr. Matsukuma, Kanagawa Cancer Center Research Institute, Nakao, Japan, were used in western blot analysis with 100 μg total protein. Custom GOLGA3 antibodies recognized the same molecular weight protein as the Matsukuma antibodies but were more sensitive.
Membranes were incubated in primary antibodies overnight at 4°C, washed in TBS-T, and then incubated for 1 hour in goat anti-rabbit horseradish peroxidase secondary antibody (0.4 mg/ml, Pierce Biotechnology) diluted to 1:40,000. All antibody incubations were carried out in TBS-T with 5% non-fat dried milk. SuperSignal West Dura Extended Duration Substrate (Pierce Biotechnology) and Hyperfilm™ ECL film (VWR, Radner, PA) were used according to manufacturer's recommendations. GOLGA3 antibodies were validated by western blotting with immunoabsorbed and unabsorbed N- and C-terminal antiserum following a peptide pre-absorption protocol supplied by Open Biosystems. Antiserum was spiked with peptide 1:1 and incubated overnight at 4°C (Supplemental Fig. 1).
Developmental Profiling
Reproductive organs were collected from C3Fe;B6- Golga3repro27 and C3Fe.B6- Golga3repro27 mutant and control mice. Mice were euthanized by CO2 anesthesia followed by cervical dislocation. Developmental profiles of mice were established based on body weight and the paired testes surgically removed from at least six male mice per genotype, mutant, heterozygous (Golga3repro27/+, or m/+), and wildtype mice from 12 to 70 dpp. Paired seminal vesicle weights were measured in mutant, heterozygous, and wildtype mice from 2 to 20 weeks of age. Epididymal sperm were collected and sperm smears were stained with 5% eosin Y (Sigma, St. Louis, MO). Fertilization potential of epididymal sperm was tested from three B6 and C3Fe;B6-Golga3repro27 adult male mice using a high-throughput diagnostic in vitro fertilization (IVF) assay developed by the Reproductive Genomics Program according to the protocol described at the following url: http://reproductivegenomics.jax.org/maleprotocol.html.
Histology
Histological analysis was used to verify the germ cell population and the developmental stage from 15 to 21 dpp and at 10 weeks of age (Russell et al. 1990). Testes were fixed by immersion in Bouins, cut sagittally, and paraffin embedded. 4-5 μm sections were cut and stained with periodic acid Schiffs (PAS reagent) and counterstained with hematoxylin. Total numbers of germ cells were counted in 20 round tubules per cross-section. Two sections were analyzed for each sample (n ≥ 3/genotype) by light microscopy. The average numbers of total germ cells per tubule cross-sectional area were determined. The percentage of post-meiotic germ cells per total number of germ cells per tubule in mixed C3Fe;B6-Golga3repro27 mutant and control mice was determined in 20 tubule cross sections (n ≥ 3/genotype).
Transmission electron microscopy
Paired testes were collected from 12 week old mixed C3Fe;B6-Golga3repro27 mutant and control mice (n = 3 per genotype), sectioned, pre-fixed in 2.5 % glutaraldehyde (Electron Microscopy Sciences, Hatfield, PA) overnight at 4°C, and then washed in 0.1 M sodium cacodylate buffer. Tissue sections were stained with toluidine blue and analyzed under a light microscope to guide observation under the transmission electron microscope [Hitachi (H-7500 TEM)].
DNA Fragmentation Assay (TUNEL)
DNA fragmentation was assessed by terminal deoxynucleotidyl transferase (Tdt) dUTP-mediated Nick End Labeling (TUNEL assay) using the DeadEnd™ Fluorometric TUNEL System (Promega Corp.) following manufacturer's instructions. Three unstained paraffin-embedded histological testes sections per genotype from mice ages 12 to 21 dpp were used in the assay. Slides were deparaffinized, rehydrated, and washed sequentially in 0.85% NaCl and 1× PBS, fixed in 4% PFA in 1× PBS and incubated in 20 μg/mL proteinase K for antigen retrieval. Sections were equilibrated for 10 minutes and labeled in a dark humidified chamber with recombinant Tdt incubation buffer at 37°C for 1 hour. The tailing reaction was terminated in 2× salt sodium citrate (SSC) for 15 minutes at room temperature. Slides were washed in 1× PBS and Aqueous SlowFade Gold Antifade reagent (Invitrogen Corp.) added to treated sections prior to mounting. Negative controls included incubation without recombinant Tdt enzyme in the incubation buffer while positive controls were pretreated with 10 U/mL DNase I enzyme prior to incubation with recombinant Tdt enzyme. All slides were kept in the dark and stored at 4°C until microscopic evaluations at 100× and 400×.
The numbers of germ cells per testis for each genotype from 12 to 21 dpp were determined after scoring tissue sections following a protocol previously described (Nandi et al. 1999). TUNEL-stained germ cells in 5 μm whole testicular cross-sections were counted under a 40× objective by moving an ocular field across the section in a non-overlapping fashion and counting cells within its boundaries. The numbers of ocular fields were estimated using the 10× objective and ranged between 4 and 15 fields per section. The average number of stained cells per ocular field was determined. The numbers of TUNEL-stained germ cells per testis were calculated as follows: average numbers of stained cells per ocular volume multiplied by ocular volume per testis equals the number of stained cells per testis. The ocular volume was defined by the product of the thickness of section and the area of a 10× objective field. The number of ocular volumes per testis was obtained by dividing the ocular volume by the volume of the testis, essentially the testis weight. It is important to note that this quantification technique may not represent the true absolute numbers of cells, since larger cells span more sections than smaller ones. However, this method is useful for comparison between genotypes, which was our goal.
Statistical analysis
Mean values were obtained from at least three parallel specimens after quantification. All statistical evaluations were by ANOVA and multiple comparison test (Tukey's HSD) using SPSS 13.0 software. Regression analysis was performed using Microsoft® Excel. Significant statistical difference was p < 0.05 and a highly significant statistical difference was p < 0.01.
Results
repro27 congenic line
The repro27 mutation was induced by ENU in B6 male mice that were subsequently mated for several generations to C3Fe mice. The original strain was thus mixed and designated C3Fe;B6-Golga3repro27. This mixed strain was used for genetic mapping and characterization of the spermatogenic defects. A congenic strain, C3Fe.B6-Golga3repro27, was developed by backcrossing. The only difference between the mixed and congenic nomenclature is the replacement of the semicolon with a period in the strain name. The congenic strain was developed to standardize the genetic background, minimize introduction of background modifier genes, and permit the use of C3Fe inbred mice as controls. Gene expression was analyzed in the congenic strain.
repro27 mice have a point mutation in Golga3
Mapping of the infertility phenotype by meiotic recombination narrowed the repro27 candidate gene region to a region of ∼2 megabases (Mb) on Chr 5. Data mining of known genes in the region identified Golga3 as the strongest candidate gene. Sequence analysis of the 4,805 bp Golga3-002 cDNA identified a point mutation in exon 18 in repro27 mutants not present in B6 cDNA. This mutation converted a cytosine to thymine and replaced glutamine with a stop codon (Fig. 1). Genomic sequencing of exon 18 further confirmed this result. This nonsense mutation is unique to repro27 and is not a common single nucleotide polymorphism (SNP). No other mutations were identified in the expressed transcript when compared to the B6 cDNA and the published sequence (www.ensembl.org). Thus, repro27 represents a new allele of Golga3, Golga3repro27.
Fig. 1. Genetic mapping and positional cloning of the repro27 mutation.
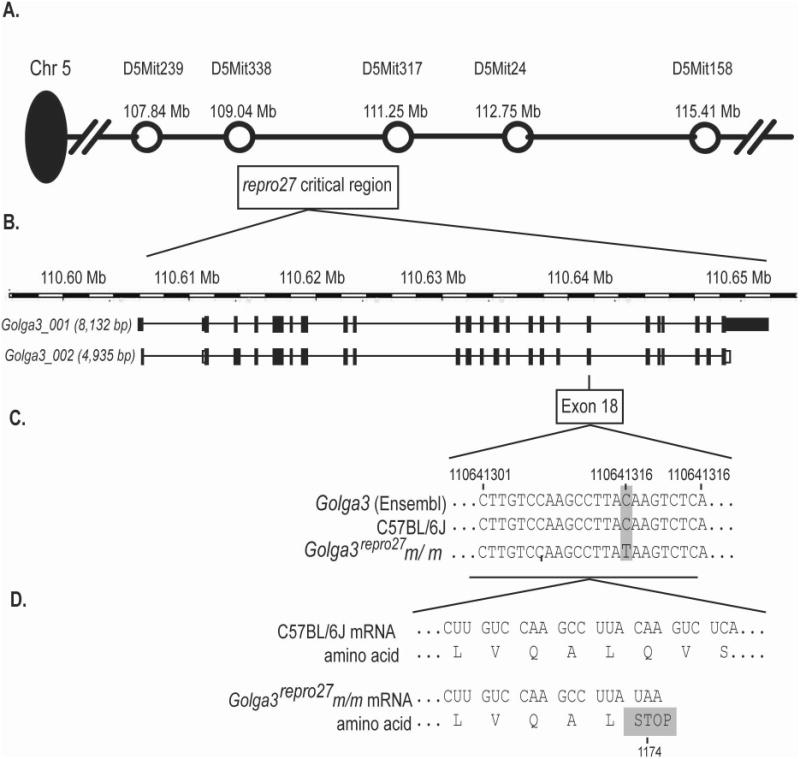
A Map of distal mouse Chr 5 including polymorphic DNA markers and the repro27 critical region (boxed). B. Golga3 genomic structure and the two full-length transcripts, Golga3_001 and Golga3_002 (www.ensembl.org, Oct 2012). C. DNA sequencing of Exon 18 showing the C to T change at position 110,641,316 bp present in Golga3repro27 mice. D. Genetic code and translation of a portion of exon 18 showing the premature termination codon at amino acid position 1174.
Expression of Golga3
Golga3 gene expression was analyzed in congenic C3Fe.B6-Golga3repro27 testes during the first wave of spermatogenesis and in adult testes. Using quantitative PCR, Golga3 mRNA was identified in both C3Fe wildtype controls and Golga3repro27 mutant testes from 4 dpp through adulthood (Fig. 2). Golga3 mRNA levels, expressed as a fold-change over the mean 4 dpp wildtype value, were significantly higher in wildtype testes compared to Golga3repro27 mutants at both 9 and 14 dpp (Fig. 2A). Northern blot analysis identified two transcripts with higher Golga3 expression in C3Fe wildtype testes compared to mutant and heterozygous samples. Golga3 expression was much higher in the adult wildtype testes compared to the 14 dpp sample and the smaller transcript (Golga3-002) was more abundant (Fig. 2B).
Fig. 2. Postnatal testicular expression of Golga3 mRNA.
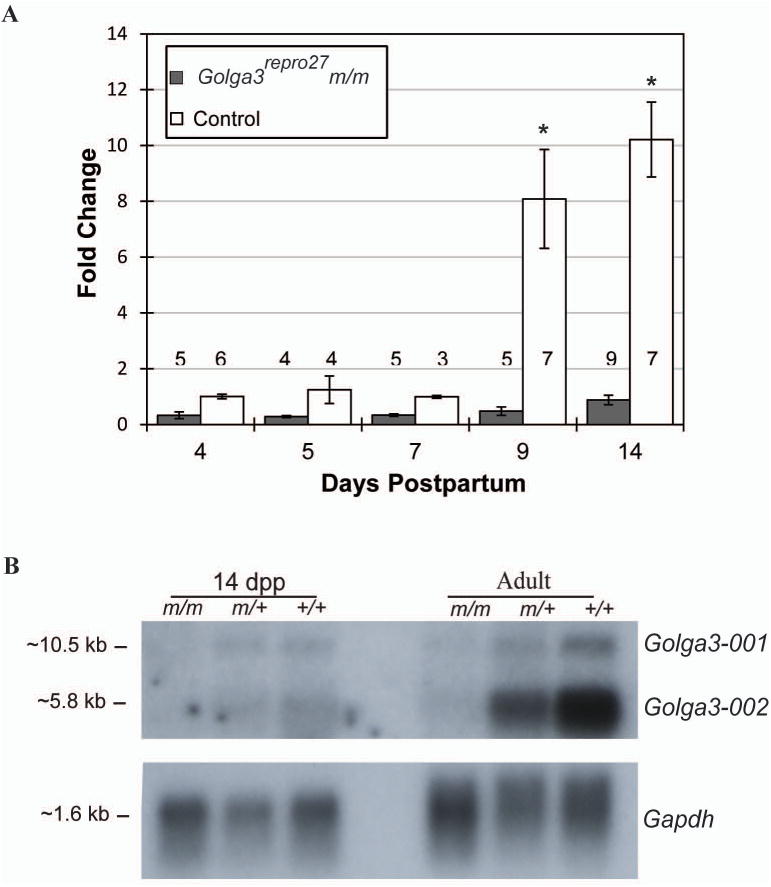
A Quantitative real time PCR. Total RNA from testes collected from congenic C3Fe.B6-Golga3repro27 m/m and C3HeB/FeJ (+/+) mice. ΔCt values were calculated by subtracting β-actin Ct; ΔΔCt values were calculated by subtracting the mean 4 day wildtype ΔCt. Fold change (± SEM) was calculated using the 2-ΔΔCt method. The sample number per group is denoted for each bar. mRNA levels in +/+ were significantly higher than m/m at 9 and 14 dpp (*, p < 0.05). B. Northern blot analysis using 10 μg total RNA isolated from C3Fe.B6-Golga3repro27 (m/m, m/+) and C3HeB/FeJ (+/+) mice.
GOLGA3 protein expression was examined by western blot using antibodies recognizing N- and C-terminal peptides. GOLGA3 protein was not expressed in the testes of congenic Golga3repro27 mutant mice at 15 dpp, whereas it was expressed in testes of heterozygous and wildtype mice (Fig. 3A). GOLGA3 expression was consistently lower in heterozygotes compared to wildtype controls. Fifteen day old mice were chosen for analyses because the testes weights and germ cell numbers among genotypes were not statistically different at this age (supplemental Figs. 2 and 3). The N-terminal antibody is specific to the 167 kD isoform because the exon 3-encoded peptide sequence is missing in the 163 kD isoform. Because the 4 kD difference between the two isoforms is indistinguishable on a 7% gel, it is not possible to distinguish the two isoforms. GOLGA3 is expressed most abundantly in the adult testes with no evidence of protein expression in the brain, heart, or liver of controls or Golga3repro27 mutant mice (Fig 3B-C). In the heart, the C-terminal antibody did occasionally react with a slightly larger unidentified protein product (Fig 3B, panel 3 of right column). Western blots performed with higher protein concentrations (100 μg, compared to 40 μg) and using antibodies supplied by Dr. Matsukuma (1:500 dilution) showed very low levels of expression in the brain and heart (data not shown). There was no evidence of a 129 kDa truncated protein, as predicted from the premature termination codon in exon 18, in testes or other tissues of Golga3repro27 mutant mice.
Fig. 3. GOLGA3 protein expression.
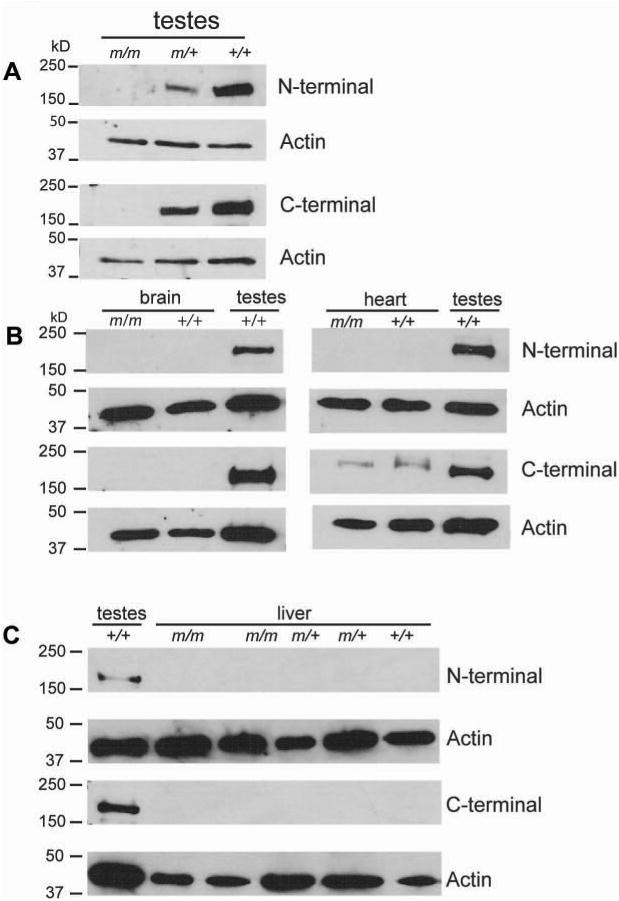
Western blot analysis using N-terminal and C-terminal GOLGA3 antibodies in congenic C3Fe.B6-Golga3repro27 (m/m and m/+), and C3HeB/FeJ (+/+) testes (A, 15 dpp) and adult brain (B), heart (B), and liver (C). Adult wildtype testes samples were used as positive controls and β-actin expression was used as a loading control.
Golga3repro27 mutant mice show defects in male germ cell development
Mutant Golga3repro27 homozygotes exhibited male-specific infertility while females were reproductively viable. The number of litters born and the number of pups per litter in heterozygous and homozygous females did not differ (data not shown). The spermatogenic defects were characterized on both the mixed and congenic strains with no observed differences between strains in testes gross morphological characteristics. Spermatogenesis appeared histologically normal through mid-meiosis during the first wave of spermatogenesis. Defects became visible in late meiosis, by 18 dpp, with significant loss of germ cells, vacuolization, and enlarged lumens in mutants (Fig. 4, “a” arrows). At 21 dpp, Golga3repro27 mutant testes showed the absence of round spermatids (Fig. 4, “c” arrows) while controls contained postmeiotic round spermatids (Fig. 4, “d” arrow).
Fig. 4. Histological analysis of mixed C3Fe;B6-Golga3repro27 testes.
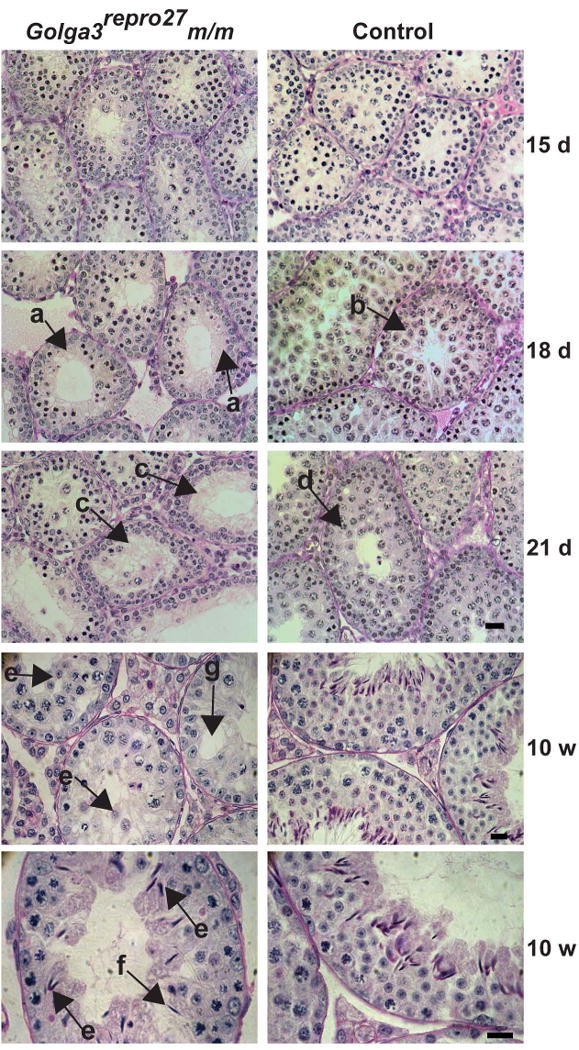
PAS-stained images of seminiferous tubules (15 to 21 days postpartum (d) and at 10 weeks of age (w)). Golga3repro27 m/m showed germ cell depletion at 18 d (denoted by “a” arrows) while control samples contained significantly more late meiotic prophase spermatocytes (“b” arrow). At 21 d, mutants showed depletion of germ cells, vacuolization, enlarged lumens, and the absence of round spermatids (“c” arrows) while controls contain postmeiotic round spermatids (“d” arrow). In 10 w mutant samples, there were very few postmeiotic germ cells (“e” and “f” arrows), decreased tubule size, germ cell deficiency, and vacuolization (“g” arrow). Round (“e” arrows) and elongating spermatids (“f” arrows)contained acrosomes as evidenced by the pink staining. Bar = 20 μm (15-21 dpp) and bar= 25 μm (10 w).
Although the majority of germ cells were lost between 15 and 18 dpp, a few germ cells survived, completed meiosis, and underwent abnormal spermiogenesis. Small numbers of round spermatids were visible in histological sections from 10 week old (10 w) mice. Most of the round and elongating spermatids in mutant mice contained acrosomes as evidenced by PAS staining (Fig. 4, “e” and “f” arrows). Spermatids from mutant mice, analyzed by TEM, showed several defects including paired nuclei during acrosome development (Fig. 5A, “a” arrow), disorganized Golgi complex (Fig. 5A, “b” arrow), abnormal distribution of condensing chromatin, vacuolization in nuclei (Fig. 5B, “d” arrow), mislocalization of mitochondria (Fig. 5B, “e” arrow), and vacuolization around the spermatid head region (Fig. 5C, “f” arrow).
Fig. 5. Transmission EM showing spermiogenesis defects in mixed C3Fe;B6-Golga3repro27 m/m testes.
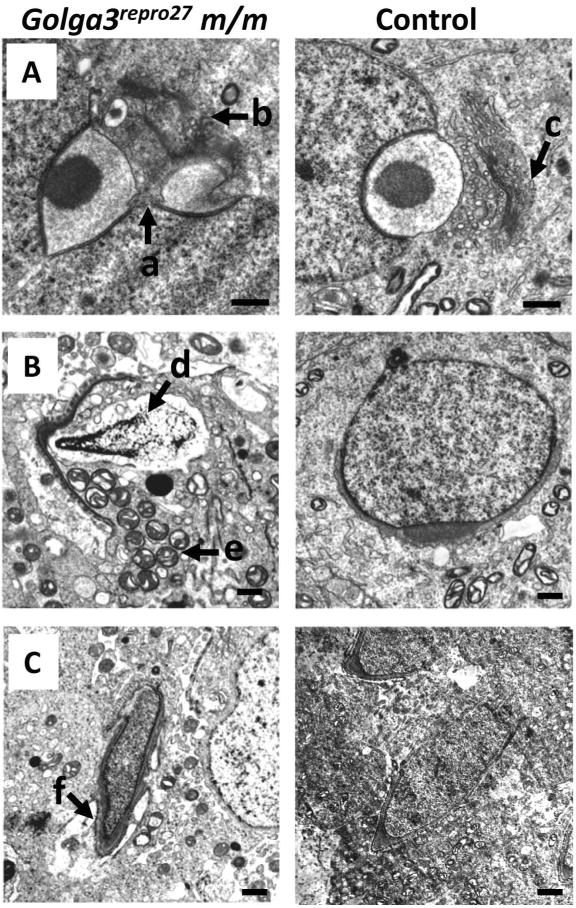
A. Step 3 round spermatids with fused nuclei (“a” arrow), abnormal acrosome development, and a disorganized Golgi complex (“b” arrow) compared to control (“c” arrow); B. Degenerating round spermatid with extensive vacuolization in the nucleus (“d” arrow) and mitochondrial mislocalization (“e” arrow). C. Vacuolization and abnormal Sertoli cells connections (“f” arrow) in elongating spermatid. Bar = 500 nm.
Sperm collected from caudal epididymides of the originally produced mixed C3Fe;B6-Golga3repro27mutants were extremely low in number (roughly 100 sperm cells total found in smears from 3 males) with negligible motility. Morphological defects included abnormally shaped heads, tails without heads, and heads without tails (Fig. 6, top panel). Fertilization potential was determined using a high throughput IVF assay. Control B6 male mouse sperm fertilized 93 of 104 oocytes (89%) and the sperm from each of two C3Fe;B6-Golga3repro27 mutant males fertilized 0 of 65 oocytes and 0 of 122 oocytes, while no sperm were obtained from a third mutant male.
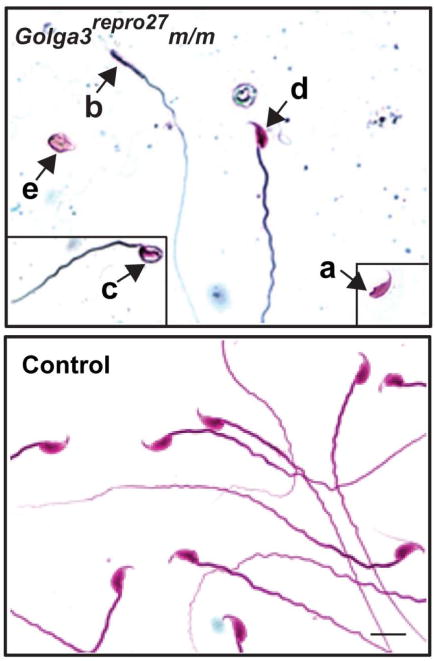
Fig. 6. Morphology of caudal epididymal sperm.
Multiple defects were evident in mixed C3Fe;B6-Golga3repro27 m/m including: normal looking head with no tail (“a” arrow), headless tail (“b” arrow), abnormal head with tail (“c” arrow), normal-appearing sperm (“d” arrow), and abnormal head with no tail (“e” arrow). Bar = 10 μm.
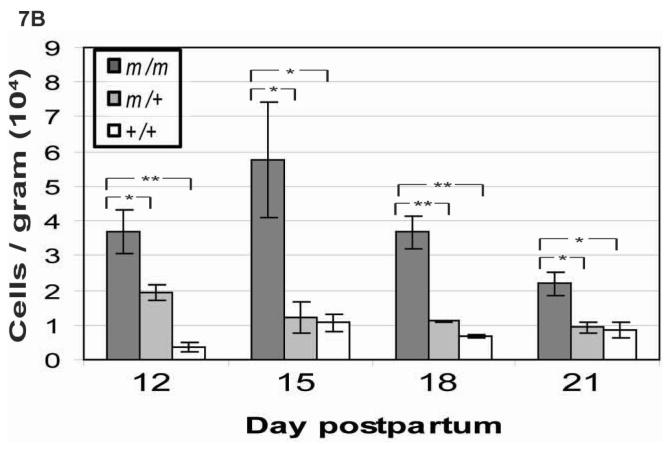
Germ cell loss in Golga3repro27 mutant mice was further supported by morphometric analysis. The testes to body weight ratio (supplemental Fig. 2) and the average total number of germ cells per tubule cross sectional area (supplemental Fig. 3) of 14 and 15 dpp mice was the same in Golga3repro27 mutant and control mice. By 18 dpp, mutant germ cell numbers were significantly lower in mutant than in control mice. Mutant mice continue to have very small testes through adulthood. Quantification of total germ cell numbers per tubule cross section suggested that germ cells were dying in late meiosis during the first wave of spermatogenesis. To evaluate the mechanism of this germ cell loss, DNA fragmentation was assessed using the TUNEL assay. Testes from Golga3repro27 mutant mice between 12 and 21 dpp showed significantly more TUNEL-positive cells compared to heterozygous or wildtype control mice (Fig. 7). Furthermore, at 12 dpp, heterozygous Golga3repro27/+ testes showed significantly more germ cell loss than wildtype mice.
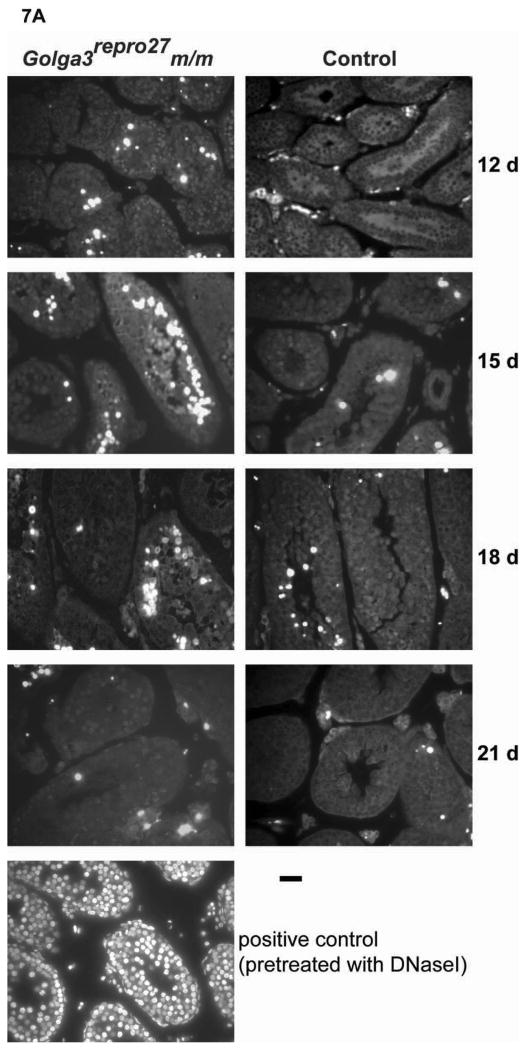
Fig. 7. TUNEL DNA fragmentation analysis during the first wave of spermatogenesis.
A. TUNEL positive cells (white) were more numerous in germ cells in the adluminal compartment of the tubules in mixed C3Fe;B6-Golga3repro27 mutant mice. Wildtype sections pretreated with DNaseI are shown as a positive control. Representative sections photographed by fluorescence microscopy. Bar = 25 μm. B. Quantification of mean total number of TUNEL-stained germ cells/total testis (cells/gram) in Golga3repro27 mutant (m/m), heterozygous (m+), and wild-type (++) mice (n = 3/genotype) at various days postpartum. Values are mean times 104 (± SEM). Significant differences (*, p < 0.05; **, p < 0.01) were observed when mutants (m/m) were compared to controls (m/+, +/+).
Regression analysis of the testes to body weight ratios indicate that mutant mice show a delay in the timing of spermatogenesis (data not shown). This delay is further supported by a delay in the appearance of round and elongating spermatids (supplemental Fig. 4) and an inability to determine the stage of the cycle of the seminiferous tubules because typical germ cell associations among stages of developing spermatogonia, spermatocytes, and spermatids in adult mutant mice were disrupted (Fig. 4, plus additional data analysis not shown).
Defects in Golga3repro27mutant mice appear to be limited to the testes. Testosterone-dependent organs (e.g. seminal vesicles and prostate) appear normal in adult Golga3repro27 mutant mice and analysis of seminal vesicle weights across genotypess from 2 to 20 weeks were similar (supplemental Fig. 5).
Discussion
Here we identify a new genetic aspect of male infertility by showing that the repro27 spermatogenesis defects are associated with a novel mutation in the golgin subfamily A member 3 gene (Golga3). The Golga3repro27 allele, which results in a premature termination codon inserted into exon 18, completely eliminates protein expression and affected male mice exhibit defects in germ cell development that culminate in testicular atrophy, low epididymal sperm concentration, low motility, and unsuccessful in vitro fertilization. Analysis of the first wave of spermatogenesis revealed that spermatogenesis is disrupted primarily in late meiosis, leading to increased cell death and a delay in germ cell maturation. Abnormal spermiogenesis, including head and tail defects, is evident among the surviving germ cells. Together these results call attention to the role in spermatogenesis of the GOLGA3 protein.
The mouse Golga3 gene, located on Chr 5, is ∼50 kb with 24 exons. There are two large transcripts: Golga3-001 (ENSMUST00000112512) is 8,132 bp and encodes a 1,447 amino acid polypeptide (163 kD), and Golga3-002 (ENSMUST00000031477) is 4,805 bp, accounting for a 1,487 residue polypeptide (167 kD). Golga3-001 has a long 3′ untranslated region and is missing exon 3, leading to a smaller protein product. A third transcript, Golga3-005 (ENSMUST00000139611), is 711 bp, contains exons 1 through 3, and encodes a 121 residue polypeptide while transcripts Golga3-003 and Golga3-004, do not encode a protein product (Ensembl, www.ensembl.org, October 2012). Northern analysis identified two transcripts, ∼10.5 and ∼5.8 kb, most likely corresponding to Golga3-001 and Golga3-002, respectively. The larger transcript sizes compared to those reported in Ensembl are most likely the result of estimating the size from the migration of the two ribosomal RNAs. Exon 18, the location of the repro27 mutation, is common to the two full-length protein coding transcripts, Golga3-001 and Golga3-002.
Our study showed that low levels of Golga3 mRNA were present in wildtype testes at 4 dpp and increased significantly by 9 dpp, a developmental expression pattern that corresponds with the onset of meiosis in developing spermatocytes. The smaller Golga3-002 transcript showed the highest expression in adult testes, suggesting expression also in spermatids. Previous expression studies showed a similar pattern except that Lau and coworkers reported a transcript size of ∼1000 bp (Lau et al. 1987; Lau et al. 1989) that may correspond to the Golga3-005 transcript, which would not have been detected with our Golga3 probe. Previous studies using in situ hybridization showed Golga3 mRNA is expressed in postmeiotic spermatids (Kondo & Sutou 1997; Lau et al. 1989). Golga3repro27 mutant mRNA levels were signficantly lower than wildtype and this is in agreement with previous studies with the Golga3Tg(06MGMT)T604Kccri allele (Banu et al. 2002; Matsukuma et al. 1999). The low levels of Golga3 mRNA in Golga3repro27 mutants could be explained by the nonsense-mediated decay survelliance mechanism (NMD) triggered by premature termination codons (Shyu et al. 2008), found also in the testes of the targeted mutation Men1tm1Zqw (Zetoune et al. 2008) and the spontaneous mutation Mdm1arrd2 (Chang et al. 2008).
Observations from using custom peptide antibodies showed that GOLGA3 protein expression was limited to the testes. To control for differences in germ cell composition, we compared protein expression patterns in testes from juvenile mice at fifteen days of age because germ cell populations are not statistically different in composition between mutant and controls, as this time point immediately precedes the onset of significant germ cell loss in mutants. Others have reported low levels of expression in all body tissues (UniProtKB/Swiss-Prot entry P55937) including differential patterns in the brain (Dumin et al. 2006). Very low expression in the brain and heart was found using the Matsukuma antibodies and 100 μg of protein, compared to the 40 μg used with the custom antibodies. Two full-length GOLGA3 isoforms, 163 kD and 167 kD, were indistinguishable in our western blots. We infer that the 167 kD isoform is present in the testes, because our N-terminal antibody, recognizing only the larger isoform, yields a positive signal in control mice. In interesting contrast to this observation, our northern blot results indicate that both transcripts are present, and thus further studies should address the possibility that different cells within the seminiferous tubule express different isoforms.
GOLGA3 is a member of the golgin protein family and located on the cytoplasmic surface of the Golgi complex. The protein's crystal structure has not been determined but there are several functional and compositional domains (Fig. 8). Most of the functional domains have been characterized in the human homolog and appear to be conserved, by amino acid sequence similarity, in the mouse. The Golga3repro27 mutation is located in the C-terminal coiled-coil domain that is proposed to form a rod-like structure that interacts with the Golgi complex. Our preliminary ultrastructural analyses of spermatids revealed defects in acrosome formation, head and tail development, extensive vacuolization around the spermatid head region, and fused heads and tails, consistent with abnormalities of Golgi complex dynamics. Previous studies have shown that golgins are required for normal acrosome and tail formation (Kierszenbaum et al. 2011; Kierszenbaum et al. 2011). Defective acrosome formation was seen in mice lacking the Golgi-associated PDX- and coiled-coil motif-containing protein, GOPC (Yao et al. 2002). In HeLa cells, GOPC protein was shown to directly interact with a specific GOLGA3 isoform (Hicks & Machamer 2005). Vacuolization observed around some spermatids in the Golga3repro27 mutant mice could potentially be due to a lack of adhesion proteins from the trans Golgi network. For example, GOLGA4 (p230/golgin) and GOLGA1 (golgin-97) are required for proper protein targeting of E-cadherin to the basal laminal surface (Lock et al. 2005). Moreover, Golga3repro27 mutant testes exhibited fused heads and tails, perhaps due to incomplete cell division, which also warrants further investigation.
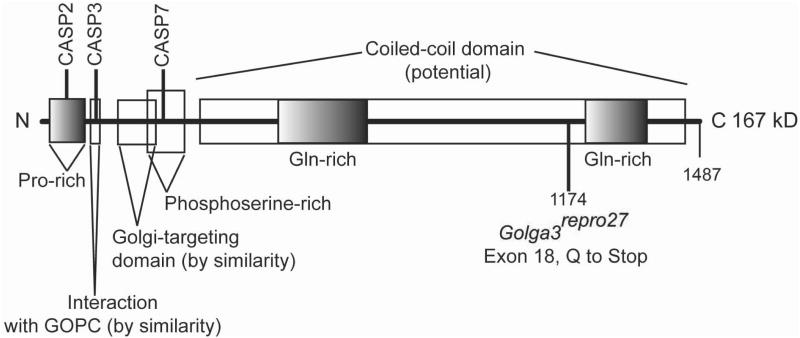
Fig. 8. GOLGA3 protein domains.
The mouse 167 kD GOLGA3 isoform has a number of putative functional and compositional domains. The region interacting with Golgi-associated PDX- and coiled-coil motif-containing protein (GOPC) were identified in human GOLGA3 (Hicks & Machamer 2005) and is inferred in the mouse by amino acid sequence similarity. Figure compiled from data retrieved from UnitProtKB (http://www.uniprot.org/uniprot/Q08378, retrieved October, 2012).
A prominent aspect of the Golga3repro27 mutant phenotype is reduced numbers of postmeiotic germ cells. DNA fragmentation analyses were consistent with an increase in apoptosis and significant germ cell loss by 21 dpp in mutant testes. Increased TUNEL positive cells also were detected in mutant mice carrying the Golga3Tg(06MGMT)T604Kccri allele (Banu et al. 2002). An increase in apoptosis in the absence of functional GOLGA3 appear in contrast with cell culture studies that implicate GOLGA3 in inducing apoptotic pathways through caspase signaling mechanisms. In the brain, GOLGA3 may play a role in ubiquitin-dependent proteosomal degradation pathways (Dumin et al. 2006), and protecting against ischemia-induced apoptosis (Ran et al. 2007). The N-terminal head region contain caspase 2 cleavage sites (Maag et al. 2005; Mancini et al. 2000) and a cryptic nuclear localization signal. Although full-length GOLGA3 is not found in the nucleus, cleaved fragments do accumulate there and suggest GOLGA3 plays a role in the apoptosis signaling pathway (Mancini et al. 2000; Sbodio et al. 2006).
This characterization of the Golga3repro27 allele shows the value of multiple mutant alleles to confirm function and further highlights the importance of GOLGA3 in sperm development. The Golga3Tg(06MGMT)T604Kccri allele led to variable expression of a truncated GOLGA3 protein in different transgenic lines of mice that correlated with defective spermatogenesis. Partial rescue of fertility in the higher expressing lines suggested that the quantity of GOLGA3 in the testes is critical to support spermatogenesis (Banu et al. 2002), and indeed, our analyses of mice homozygous for the Golga3repro27 allele reveal severe defects in mutant testes completely lacking GOLGA3. Although molecular and cellular function of GOLGA3 has been studied primarily in somatic cell lines, our study reveals that the testis is acutely sensitive to the absence of GOLGA3. The Golga3repro27 mutant mice are healthy and appear to have no gross somatic defects. Most interesting, the female mice are fertile, suggesting male germ cell-specific functions of GOLGA3. Here we have developed an important genetic resource, the congenic Golga3repro27 strain where GOLGA3 is in a uniform genetic background, and future experiments with these mutant mice will continue to clarify the role of GOLGA3 in meiosis and spermiogenesis. Golga3repro27 mice will be especially useful in the study of GOLGA3's role in apoptotic signaling and Golgi complex-mediated protein targeting within the seminiferous epithelium.
Supplementary Material
Supplemental Fig. 1. Western blot analysis of 10 week of C3HeB/FeJ wildtype testes (40 μg, 10 weeks of age) using N-terminal and C-terminal GOLGA3 antibodies (1:1500) preabsorbed (left) with peptide 1:1 and unabsorbed (right). Faint bands are evident in the preabsorbed lanes suggesting that the peptide concentration was not high enough to absorb all the antibody or the antibodies recognize a nonspecific protein product at the same molecular weight. Additional western analysis shown in Fig. 3 does not support the latter hypothesis.
Supplemental Fig. 2. Testes/body weight ratios. Average paired testes weights normalized to body weight (TW/BW) in mixed C3Fe;B6-Golga3repro27 mutant mice compared to heterozygous and wildtype controls (± SEM). Mutant TW/BW ratios were significantly lower by 21 days postpartum (*, p < 0.05).
Supplemental Fig. 3. Germ cell quantification. The average total number of germ cells per tubule cross section (± SEM) in mixed C3Fe;B6-Golga3repro27 mutant mice (m/m) compared to heterozygous (m/+) and wildtype (+/+) controls. Twenty tubule cross sections were counted from at least three mice per genotype. The germ cell numbers differ from 12-15 dpp, but were significantly lower in mutant testes compared to controls by 18 days postpartum (*, p < 0.05).
Supplemental Fig. 4. Percentage of post-meiotic germ cells (round and elongating) per total number of germ cells per tubule in mixed C3Fe;B6-Golga3repro27 mutant (m/m) mice compared to heterozygous (m/+) controls (± SEM). Twenty tubule cross sections were counted from at least three mice per genotype.
Supplemental Fig. 5. Seminal vesicle weights in mixed C3Fe;B6-Golga3repro27 mutant mice (m/m) compared to heterozygous (m/+) and wildtype (+/+) controls from 2 to 20 weeks of age. Data represent mean (± SEM) in at least two mice per genotype (most time points represented by at least 4 mice. While the seminal vesicle weight was significantly different in mutant mice at 10 weeks of age (n=6 mice for m/m and m/+, n= 3 for +/+), no other time points were statistically different (*, p < 0.05).
Acknowledgments
We are most grateful to Dr. Shoichi Matsukuma (Kanagawa Cancer Center Research Institute, Nakao, JP) for providing GOLGA3 antibodies and developing the Golga3repro27 allele-specific assay. Drs. Tamara Howard and Helen Hathaway (University of New Mexico Health Sciences Center) for microscopy assistance. Northern blotting was accomplished with assistance from Thomas Jucius, The Jackson Laboratory. Dr. D.N. Rao Veeramachaneni (Animal Reproduction and Biotechnology Laboratory of Colorado State University) provided assistance in evaluating transmission electron microscopy images. Alta Vista Regional Hospital for the use of their film processor and numerous undergraduate students of the Linder lab who maintained the animal colony, performed genotyping, and especially undergraduate Sara Jorgensen Vinceny for histological assistance in quantifying germ cell populations.
This project was supported by grants from the National Center for Research Resources (5p20RR016480-12) and the National Institute of General Medical Sciences (8P20GM103451) from the National Institutes of Health and by Grant Number P520 MD001104 (Research Infrastructure in Minority Institutions) from the NIH National Center for Minority Health and Health Disparities (NCMHD). The Jackson Laboratory Reproductive Genomics Program is supported by NIH P01HD42137. The contents of this manuscript are solely the responsibility of the authors and do not necessarily represent the official views of NIH, NCRR, NIGMS, or NCMHD.
Footnotes
Declaration of Interest: The authors declare that there is no conflict of interest that could be perceived as prejudging the impartiality of the research reported.
References
- Banu Y, Matsuda M, Yoshihara M, Kondo M, Sutou S, Matsukuma S. Golgi matrix protein gene, Golga3/Mea2, rearranged and re-expressed in pachytene spermatocytes restores spermatogenesis in the mouse. Mol Reprod Dev. 2002;61:288–301. doi: 10.1002/mrd.10035. [DOI] [PubMed] [Google Scholar]
- Barr FA. A novel Rab6-interacting domain defines a family of Golgi-targeted coiled-coil proteins. Curr Biol. 1999;9:381–384. doi: 10.1016/s0960-9822(99)80167-5. [DOI] [PubMed] [Google Scholar]
- Bray JD, Chennathukuzhi VM, Hecht NB. Identification and characterization of cDNAs encoding four novel proteins that interact with translin associated factor-X. Genomics. 2002;79:799–808. doi: 10.1006/geno.2002.6779. [DOI] [PubMed] [Google Scholar]
- Chang B, Mandal MN, Chavali VR, Hawes NL, Khan NW, Hurd RE, Smith RS, Davisson ML, Kopplin L, Klein BE, Klein R, Iyengar SK, Heckenlively JR, Ayyagari R. Age-related retinal degeneration (arrd2) in a novel mouse model due to a nonsense mutation in the Mdm1 gene. Hum Mol Genet. 2008;17:3929–3941. doi: 10.1093/hmg/ddn295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark AT, Firozi K, Justice MJ. Mutations in a novel locus on mouse chromosome 11 resulting in male infertility associated with defects in microtubule assembly and sperm tail function. Biol Reprod. 2004;70:1317–1324. doi: 10.1095/biolreprod.103.020628. [DOI] [PubMed] [Google Scholar]
- Dietrich WF, Miller J, Steen R, Merchant MA, Damron-Boles D, Husain Z, Dredge R, Daly MJ, Ingalls KA, O'Connor TJ. A comprehensive genetic map of the mouse genome. Nature. 1996;380:149–152. doi: 10.1038/380149a0. [DOI] [PubMed] [Google Scholar]
- Dumin E, Bendikov I, Foltyn VN, Misumi Y, Ikehara Y, Kartvelishvily E, Wolosker H. Modulation of D-serine levels via ubiquitin-dependent proteasomal degradation of serine racemase. J Biol Chem. 2006;281:20291–20302. doi: 10.1074/jbc.M601971200. [DOI] [PubMed] [Google Scholar]
- Egozcue S, Blanco J, Vendrell JM, Garcia F, Veiga A, Aran B, Barri PN, Vidal F, Egozcue J. Human male infertility: chromosome anomalies, meiotic disorders, abnormal spermatozoa and recurrent abortion. Hum Reprod Update. 2000;6:93–105. doi: 10.1093/humupd/6.1.93. [DOI] [PubMed] [Google Scholar]
- Ferlin A, Raicu F, Gatta V, Zuccarello D, Palka G, Foresta C. Male infertility: role of genetic background. Reprod Biomed Online. 2007;14:734–745. doi: 10.1016/s1472-6483(10)60677-3. [DOI] [PubMed] [Google Scholar]
- Fritzler MJ, Hamel JC, Ochs RL, Chan EK. Molecular characterization of two human autoantigens: unique cDNAs encoding 95- and 160-kD proteins of a putative family in the Golgi complex. J Exp Med. 1993;178:49–62. doi: 10.1084/jem.178.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gifford CA, Racicot K, Clark DS, Austin KJ, Hansen TR, Lucy MC, Davies CJ, Ott TL. Regulation of interferon-stimulated genes in peripheral blood leukocytes in pregnant and bred, nonpregnant dairy cows. J Dairy Sci. 2007;90:274–280. doi: 10.3168/jds.S0022-0302(07)72628-0. [DOI] [PubMed] [Google Scholar]
- Gleeson PA, Anderson TJ, Stow JL, Griffiths G, Toh BH, Matheson F. p230 is associated with vesicles budding from the trans-Golgi network. J Cell Sci. 1996;109(Pt 12):2811–2821. doi: 10.1242/jcs.109.12.2811. [DOI] [PubMed] [Google Scholar]
- Handel MA, Lessard C, Reinholdt L, Schimenti J, Eppig JJ. Mutagenesis as an unbiased approach to identify novel contraceptive targets. Mol Cell Endocrinol. 2006;250:201–205. doi: 10.1016/j.mce.2005.12.046. [DOI] [PubMed] [Google Scholar]
- Hennies HC, Kornak U, Zhang H, Egerer J, Zhang X, Seifert W, Kuhnisch J, Budde B, Natebus M, Brancati F, Wilcox WR, Muller D, Kaplan PB, Rajab A, Zampino G, Fodale V, Dallapiccola B, Newman W, Metcalfe K, Clayton-Smith J, Tassabehji M, Steinmann B, Barr FA, Nurnberg P, Wieacker P, Mundlos S. Gerodermia osteodysplastica is caused by mutations in SCYL1BP1, a Rab-6 interacting golgin. Nat Genet. 2008;40:1410–1412. doi: 10.1038/ng.252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicks SW, Horn TA, McCaffery JM, Zuckerman DM, Machamer CE. Golgin-160 promotes cell surface expression of the beta-1 adrenergic receptor. Traffic. 2006;7:1666–1677. doi: 10.1111/j.1600-0854.2006.00504.x. [DOI] [PubMed] [Google Scholar]
- Hicks SW, Machamer CE. The NH2-terminal domain of Golgin-160 contains both Golgi and nuclear targeting information. J Biol Chem. 2002;277:35833–35839. doi: 10.1074/jbc.M206280200. [DOI] [PubMed] [Google Scholar]
- Hicks SW, Machamer CE. Isoform-specific interaction of golgin-160 with the Golgi-associated protein PIST. J Biol Chem. 2005;280:28944–28951. doi: 10.1074/jbc.M504937200. [DOI] [PubMed] [Google Scholar]
- Kennedy CL, O'Connor AE, Sanchez-Partida LG, Holland MK, Goodnow CC, de Kretser DM, O'Bryan MK. A repository of ENU mutant mouse lines and their potential for male fertility research. Mol Hum Reprod. 2005;11:871–880. doi: 10.1093/molehr/gah251. [DOI] [PubMed] [Google Scholar]
- Kierszenbaum AL, Rivkin E, Tres LL. Cytoskeletal track selection during cargo transport in spermatids is relevant to male fertility. Spermatogenesis. 2011;1:221–230. doi: 10.4161/spmg.1.3.18018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kierszenbaum AL, Rivkin E, Tres LL, Yoder BK, Haycraft CJ, Bornens M, Rios RM. GMAP210 and IFT88 are present in the spermatid golgi apparatus and participate in the development of the acrosome-acroplaxome complex, head-tail coupling apparatus and tail. Dev Dyn. 2011;240:723–736. doi: 10.1002/dvdy.22563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo M, Sutou S. Cloning and molecular characterization of cDNA encoding a mouse male-enhanced antigen-2 (Mea-2): a putative family of the Golgi autoantigen. DNA Seq. 1997;7:71–82. doi: 10.3109/10425179709020154. [DOI] [PubMed] [Google Scholar]
- Lau YF, Chan KM, Kan YW, Goldberg E. Male-enhanced expression and genetic conservation of a gene isolated with an anti-H-Y antibody. Trans Assoc Am Physicians. 1987;100:45–53. [PubMed] [Google Scholar]
- Lau YF, Chan KM, Sparkes R. Male-enhanced antigen gene is phylogenetically conserved and expressed at late stages of spermatogenesis. Proc Natl Acad Sci U S A. 1989;86:8462–8466. doi: 10.1073/pnas.86.21.8462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lessard C, Pendola JK, Hartford SA, Schimenti JC, Handel MA, Eppig JJ. New mouse genetic models for human contraceptive development. Cytogenet Genome Res. 2004;105:222–227. doi: 10.1159/000078192. [DOI] [PubMed] [Google Scholar]
- Linstedt AD, Jesch SA, Mehta A, Lee TH, Garcia-Mata R, Nelson DS, Sztul E. Binding relationships of membrane tethering components. The giantin N terminus and the GM130 N terminus compete for binding to the p115 C terminus. J Biol Chem. 2000;275:10196–10201. doi: 10.1074/jbc.275.14.10196. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Lock JG, Hammond LA, Houghton F, Gleeson PA, Stow JL. E-cadherin transport from the trans-Golgi network in tubulovesicular carriers is selectively regulated by golgin-97. Traffic. 2005;6:1142–1156. doi: 10.1111/j.1600-0854.2005.00349.x. [DOI] [PubMed] [Google Scholar]
- Maag RS, Mancini M, Rosen A, Machamer CE. Caspase-resistant Golgin-160 disrupts apoptosis induced by secretory pathway stress and ligation of death receptors. Mol Biol Cell. 2005;16:3019–3027. doi: 10.1091/mbc.E04-11-0971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancini M, Machamer CE, Roy S, Nicholson DW, Thornberry NA, Casciola-Rosen LA, Rosen A. Caspase-2 is localized at the Golgi complex and cleaves golgin-160 during apoptosis. J Cell Biol. 2000;149:603–612. doi: 10.1083/jcb.149.3.603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda M, Kondo M, Kashiwabara S, Yoshihara M, Sutou S, Matsukuma S. Co-localization of Trax and Mea2 in Golgi complex of pachytene spermatocytes in the mouse. J Histochem Cytochem. 2004;52:1245–1248. doi: 10.1369/jhc.4B6304.2004. [DOI] [PubMed] [Google Scholar]
- Matsukuma S, Kondo M, Yoshihara M, Matsuda M, Utakoji T, Sutou S. Mea2/Golga3 gene is disrupted in a line of transgenic mice with a reciprocal translocation between Chromosomes 5 and 19 and is responsible for a defective spermatogenesis in homozygotes. Mamm Genome. 1999;10:1–5. doi: 10.1007/s003359900932. [DOI] [PubMed] [Google Scholar]
- Matsukuma S, Yoshihara M, Kasai F, Kato A, Yoshida A, Akaike M, Kobayashi O, Nakayama H, Sakuma Y, Yoshida T, Kameda Y, Tsuchiya E, Miyagi Y. Rapid and simple detection of hot spot point mutations of epidermal growth factor receptor, BRAF, and NRAS in cancers using the loop-hybrid mobility shift assay. J Mol Diagn. 2006;8:504–512. doi: 10.2353/jmoldx.2006.060030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matzuk MM, Lamb DJ. Genetic dissection of mammalian fertility pathways. Nat Cell Biol. 2002;4(Suppl):s41–49. doi: 10.1038/ncb-nm-fertilityS41. [DOI] [PubMed] [Google Scholar]
- Mouse Genome Database (MGD) at the Mouse Genome Informatics website. The Jackson Laboratory, Bar Harbor ME. 2012 Oct; ( www.informatics.jax.org)
- Mukherjee S, Shields D. Nuclear import is required for the pro-apoptotic function of the Golgi protein p115. J Biol Chem. 2009;284:1709–1717. doi: 10.1074/jbc.M807263200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munro S. The golgin coiled-coil proteins of the Golgi apparatus. Cold Spring Harb Perspect Biol. 2011;3 doi: 10.1101/cshperspect.a005256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nandi S, Banerjee PP, Zirkin BR. Germ cell apoptosis in the testes of Sprague Dawley rats following testosterone withdrawal by ethane 1,2-dimethanesulfonate administration: relationship to Fas? Biol Reprod. 1999;61:70–75. doi: 10.1095/biolreprod61.1.70. [DOI] [PubMed] [Google Scholar]
- Ran R, Pan R, Lu A, Xu H, Davis RR, Sharp FR. A novel 165-kDa Golgin protein induced by brain ischemia and phosphorylated by Akt protects against apoptosis. Mol Cell Neurosci. 2007;36:392–407. doi: 10.1016/j.mcn.2007.07.014. [DOI] [PubMed] [Google Scholar]
- Russell LD, Ettlin RA, Sinha Hikim AP, Clegge ED. The classification and timing of spermatogenesis. In: Russell LD, editor. Histological and Histophathological Evaluation of the Testis. Cache River Press; St. Louis: 1990. pp. 41–58. [Google Scholar]
- Sbodio JI, Hicks SW, Simon D, Machamer CE. GCP60 preferentially interacts with a caspase-generated golgin-160 fragment. J Biol Chem. 2006;281:27924–27931. doi: 10.1074/jbc.M603276200. [DOI] [PubMed] [Google Scholar]
- Short B, Haas A, Barr FA. Golgins and GTPases, giving identity and structure to the Golgi apparatus. Biochim Biophys Acta. 2005;1744:383–395. doi: 10.1016/j.bbamcr.2005.02.001. [DOI] [PubMed] [Google Scholar]
- Short B, Preisinger C, Korner R, Kopajtich R, Byron O, Barr FA. A GRASP55-rab2 effector complex linking Golgi structure to membrane traffic. J Cell Biol. 2001;155:877–883. doi: 10.1083/jcb.200108079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shyu AB, Wilkinson MF, van Hoof A. Messenger RNA regulation: to translate or to degrade. EMBO J. 2008;27:471–481. doi: 10.1038/sj.emboj.7601977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truett GE, Heeger P, Mynatt RL, Truett AA, Walker JA, Warman ML. Preparation of PCR-quality mouse genomic DNA with hot sodium hydroxide and tris (HotSHOT) Biotechniques. 2000;29(52):54. doi: 10.2144/00291bm09. [DOI] [PubMed] [Google Scholar]
- Williams D, Hicks SW, Machamer CE, Pessin JE. Golgin-160 is required for the Golgi membrane sorting of the insulin-responsive glucose transporter GLUT4 in adipocytes. Mol Biol Cell. 2006;17:5346–5355. doi: 10.1091/mbc.E06-05-0386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav S, Puri S, Linstedt AD. A primary role for Golgi positioning in directed secretion, cell polarity, and wound healing. Mol Biol Cell. 2009;20:1728–1736. doi: 10.1091/mbc.E08-10-1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav S, Puthenveedu MA, Linstedt AD. Golgin160 recruits the dynein motor to position the Golgi apparatus. Dev Cell. 2012;23:153–165. doi: 10.1016/j.devcel.2012.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan W. Male infertility caused by spermiogenic defects: lessons from gene knockouts. Mol Cell Endocrinol. 2009;306:24–32. doi: 10.1016/j.mce.2009.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao R, Ito C, Natsume Y, Sugitani Y, Yamanaka H, Kuretake S, Yanagida K, Sato A, Toshimori K, Noda T. Lack of acrosome formation in mice lacking a Golgi protein, GOPC. Proc Natl Acad Sci U S A. 2002;99:11211–11216. doi: 10.1073/pnas.162027899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zetoune AB, Fontaniere S, Magnin D, Anczukow O, Buisson M, Zhang CX, Mazoyer S. Comparison of nonsense-mediated mRNA decay efficiency in various murine tissues. BMC Genet. 2008;9:83. doi: 10.1186/1471-2156-9-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Fig. 1. Western blot analysis of 10 week of C3HeB/FeJ wildtype testes (40 μg, 10 weeks of age) using N-terminal and C-terminal GOLGA3 antibodies (1:1500) preabsorbed (left) with peptide 1:1 and unabsorbed (right). Faint bands are evident in the preabsorbed lanes suggesting that the peptide concentration was not high enough to absorb all the antibody or the antibodies recognize a nonspecific protein product at the same molecular weight. Additional western analysis shown in Fig. 3 does not support the latter hypothesis.
Supplemental Fig. 2. Testes/body weight ratios. Average paired testes weights normalized to body weight (TW/BW) in mixed C3Fe;B6-Golga3repro27 mutant mice compared to heterozygous and wildtype controls (± SEM). Mutant TW/BW ratios were significantly lower by 21 days postpartum (*, p < 0.05).
Supplemental Fig. 3. Germ cell quantification. The average total number of germ cells per tubule cross section (± SEM) in mixed C3Fe;B6-Golga3repro27 mutant mice (m/m) compared to heterozygous (m/+) and wildtype (+/+) controls. Twenty tubule cross sections were counted from at least three mice per genotype. The germ cell numbers differ from 12-15 dpp, but were significantly lower in mutant testes compared to controls by 18 days postpartum (*, p < 0.05).
Supplemental Fig. 4. Percentage of post-meiotic germ cells (round and elongating) per total number of germ cells per tubule in mixed C3Fe;B6-Golga3repro27 mutant (m/m) mice compared to heterozygous (m/+) controls (± SEM). Twenty tubule cross sections were counted from at least three mice per genotype.
Supplemental Fig. 5. Seminal vesicle weights in mixed C3Fe;B6-Golga3repro27 mutant mice (m/m) compared to heterozygous (m/+) and wildtype (+/+) controls from 2 to 20 weeks of age. Data represent mean (± SEM) in at least two mice per genotype (most time points represented by at least 4 mice. While the seminal vesicle weight was significantly different in mutant mice at 10 weeks of age (n=6 mice for m/m and m/+, n= 3 for +/+), no other time points were statistically different (*, p < 0.05).